Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi
Abstract
1. Introduction
2. Fungal Diversity and Ecology in Antarctic Marine Environments
2.1. Fungal Diversity
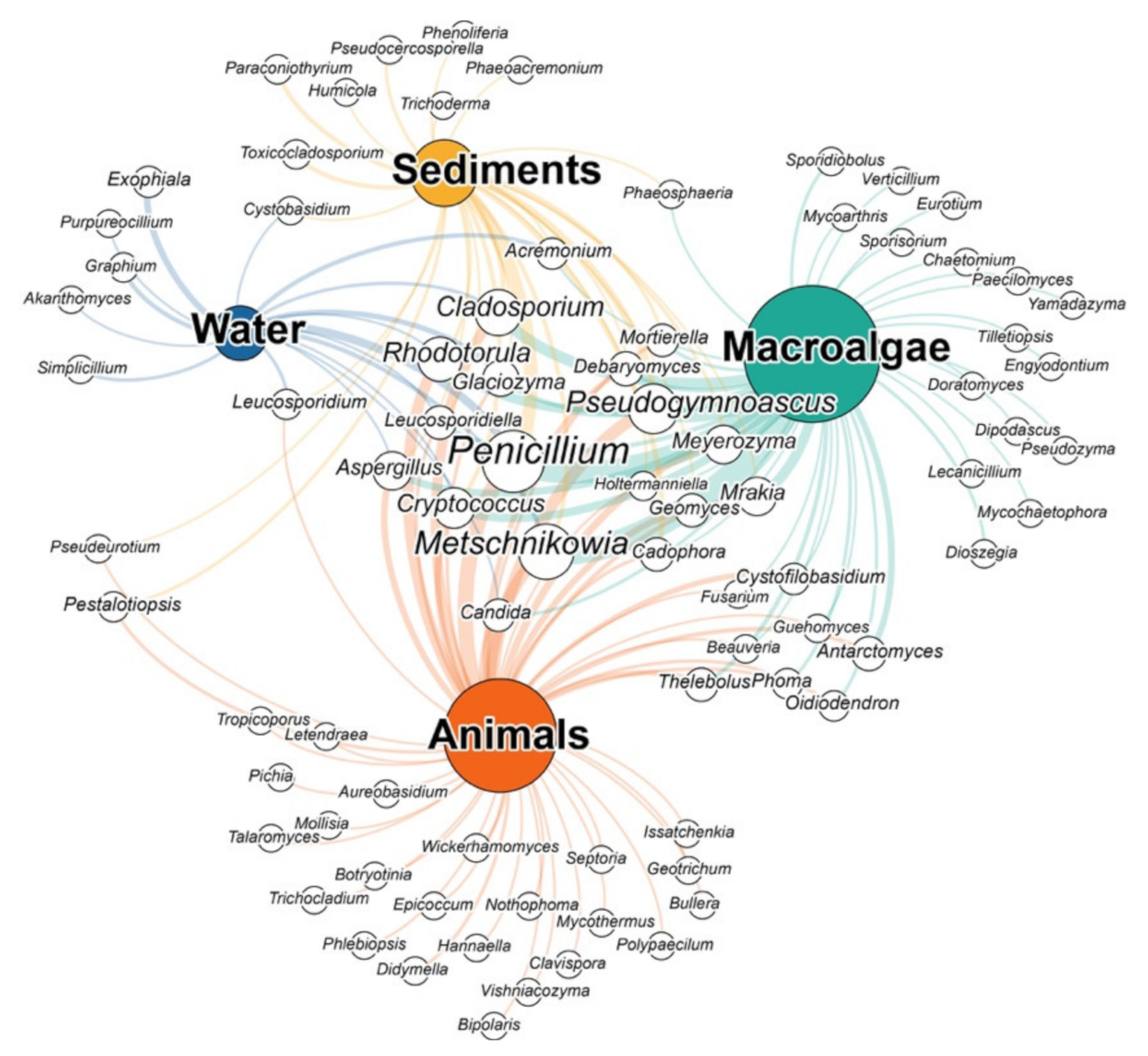
2.1.1. Fungi in Antarctic Marine Environment
2.1.2. Fungi Associated with Antarctic Macroalgae and Animals
2.2. Contribution of Fungi to Ecological Processes in Antarctic Marine Ecosystems
3. Biotechnological Potential of Fungi Inhabiting Marine Antarctic Environments
3.1. Antarctic Marine Fungi: Promising Candidates for Bioprospecting
3.2. Antarctic Fungi as Novel Source of Cold-Active Enzymes
3.3. Emerging Bioprospecting Methods: Pitfalls and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rogers, A.D.; Johnston, N.M.; Murphy, E.J.; Clarke, A. Antarctic Ecosystems; Rogers, A.D., Johnston, N.M., Murphy, E.J., Clarke, A., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2012; ISBN 9781444347241. [Google Scholar]
- Chown, S.L.; Brooks, C.M. The State and Future of Antarctic Environments in a Global Context. Annu. Rev. Environ. Resour. 2019, 44, 1–30. [Google Scholar] [CrossRef]
- Wall, D.H. Biodiversity and ecosystem functioning in terrestrial habitats of Antarctica. Antarct. Sci. 2005, 17, 523–531. [Google Scholar] [CrossRef]
- Peck, L.S. Antarctic marine biodiversity: Adaptations, environments and responses to change. Oceanogr. Mar. Biol. 2018, 56, 105–236. [Google Scholar]
- Clarke, A.; Johnston, N. Antarctic Marine Benthic Diversity. In Oceanography and Marine Biology, An Annual Review; CRC Press: Boca Raton, FL, USA, 2003; Volume 41, pp. 55–57. [Google Scholar]
- De Broyer, C.; Koubbi, P.; Griffiths, H.J.; Raymond, B.; d’Acoz, C.U.; Van de Putte, A.P.; Danis, B.; David, B.; Grant, S.; Gutt, J.; et al. Biogeographic Atlas of the Southern Ocean; Scientific Committee on Antarctic Research: Cambridge, UK, 2014; ISBN 9780948277283. [Google Scholar]
- Griffiths, H.J. Antarctic marine biodiversity—What do we know about the distribution of life in the southern ocean? PLoS ONE 2010, 5, e11683. [Google Scholar] [CrossRef]
- Gutt, J.; Sirenko, B.I.; Smirnov, I.S.; Arntz, W.E. How many macrozoobenthic species might inhabit the Antarctic shelf? Antarct. Sci. 2004, 16, 11–16. [Google Scholar] [CrossRef]
- Gutt, J.; Constable, A.; Cummings, V.; Hosie, G.; McIntyre, T.; Mintenbeck, K.; Murray, A.; Peck, L.; Ropert-Coudert, Y.; Saba, G.K. Vulnerability of Southern Ocean biota to climate change. Antarct. Environ. Portal 2016. [Google Scholar] [CrossRef]
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 5, eaaz0888. [Google Scholar] [CrossRef]
- Clarke, A.; Crame, J.A. Evolutionary dynamics at high latitudes: Speciation and extinction in polar marine faunas. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3655–3666. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Azzaro, M. Diversity and Ecological Roles of Prokaryotes in the Changing Antarctic Marine Environment. In The Ecological Role of Micro-Organisms in the Antarctic Environment; Springer Polar Sciences: Cham, Switzerland; Berlin/Heidelberg, Germany, 2019; pp. 109–131. [Google Scholar]
- Cavicchioli, R. Microbial ecology of Antarctic aquatic systems. Nat. Rev. Microbiol. 2015, 13, 691–706. [Google Scholar] [CrossRef]
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594. [Google Scholar] [CrossRef]
- Wilkins, D.; Yau, S.; Williams, T.J.; Allen, M.A.; Brown, M.V.; Demaere, M.Z.; Lauro, F.M.; Cavicchioli, R. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol. Rev. 2013, 37, 303–335. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Margesin, R.; Feller, G. Biotechnological applications of psychrophiles. Environ. Technol. 2010, 31, 835–844. [Google Scholar] [CrossRef]
- Rizzo, C.; Lo Giudice, A. The variety and inscrutability of polar environments as a resource of biotechnologically relevant molecules. Microorganisms 2020, 8, 1422. [Google Scholar] [CrossRef]
- Correa, T.; Abreu, F. Antarctic microorganisms as sources of biotechnological products. In Physiological and Biotechnological Aspects of Extremophiles; Salwan, R., Sharma, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 269–284. ISBN 978-0-12-818322-9. [Google Scholar]
- Bruno, S.; Coppola, D.; di Prisco, G.; Giordano, D.; Verde, C. Enzymes from Marine Polar Regions and Their Biotechnological Applications. Mar. Drugs 2019, 17, 544. [Google Scholar] [CrossRef]
- Tiquia-Arashiro, S.M.; Grube, M. Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer International Publishing: Cham, Switzerland; Berlin/Heidelberg, Germany, 2019; ISBN 978-3-030-19030-9. [Google Scholar]
- Sayed, A.M.M.; Hassan, M.H.A.H.A.; Alhadrami, H.A.A.; Hassan, H.M.M.; Goodfellow, M.; Rateb, M.E.E. Extreme environments: Microbiology leading to specialized metabolites. J. Appl. Microbiol. 2020, 128, 630–657. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Spiteller, P. Chemical ecology of fungi. Nat. Prod. Rep. 2015, 32, 971–993. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Yu, J.-H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef]
- Rosa, L.H. Fungi of Antarctica: Diversity, Ecology and Biotechnological Applications; Springer International Publishing: Cham, Switzerland; Berlin/Heidelberg, Germany, 2019; ISBN 978-3-030-18367-7. [Google Scholar]
- Yarzábal, L.A. Antarctic Psychrophilic Microorganisms and Biotechnology: History, Current Trends, Applications, and Challenges. In Microbial Models: From Environmental to Industrial Sustainability; Springer: Singapore, 2016; pp. 83–118. [Google Scholar]
- Corinaldesi, C.; Barone, G.; Marcellini, F.; Dell’Anno, A.; Danovaro, R. Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. Mar. Drugs 2017, 15, 118. [Google Scholar] [CrossRef]
- Chávez, R.; Fierro, F.; García-Rico, R.O.; Vaca, I. Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front. Microbiol. 2015, 6, 903. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Amils, R.; Wagner, D.; Mcgenity, T. Life and applications of extremophiles. Environ. Microbiol. 2011, 13, 1903–1907. [Google Scholar] [CrossRef] [PubMed]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10, 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sen, B.; He, Y.; Xie, N.; Wang, G. Spatiotemporal Distribution and Assemblages of Planktonic Fungi in the Coastal Waters of the Bohai Sea. Front. Microbiol. 2018, 9, 584. [Google Scholar] [CrossRef] [PubMed]
- Hassett, B.T.; Vonnahme, T.R.; Peng, X.; Jones, E.B.G.; Heuzé, C. Global diversity and geography of planktonic marine fungi. Bot. Mar. 2020, 63, 121–139. [Google Scholar] [CrossRef]
- López-García, P.; Rodríguez-Valera, F.; Pedrós-Alió, C.; Moreira, D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 2001, 409, 603–607. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.F.; Zhang, Y.Q.; Liu, H.Y.; Yu, L.Y. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Sci. Rep. 2015, 5, 14524. [Google Scholar] [CrossRef]
- Hassett, B.T.; Ducluzeau, A.L.L.; Collins, R.E.; Gradinger, R. Spatial distribution of aquatic marine fungi across the western Arctic and sub-arctic. Environ. Microbiol. 2017, 19, 475–484. [Google Scholar] [CrossRef]
- Turkiewicz, M.; Pazgier, M.; Donachie, S.P.; Kalinowska, H. Invertase and α-glucosidase production by the endemic Antarctic marine yeast Leucosporidium antarcticum. Polish Polar Res. 2005, 26, 125–136. [Google Scholar]
- Gonçalves, V.N.; Vitoreli, G.A.; de Menezes, G.C.A.; Mendes, C.R.B.; Secchi, E.R.; Rosa, C.A.; Rosa, L.H. Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 2017, 21, 1005–1015. [Google Scholar] [CrossRef]
- Ward, N.A.; Robertson, C.L.; Chanda, A.K.; Schneider, R.W. Effects of Simplicillium lanosoniveum on phakopsora pachyrhizi, the soybean rust pathogen, and its use as a biological control agent. Phytopathology 2012, 102, 749–760. [Google Scholar] [CrossRef]
- Guerra, R.S.; do Nascimento, M.M.F.; Miesch, S.; Najafzadeh, M.J.; Ribeiro, R.O.; Ostrensky, A.; de Hoog, G.S.; Vicente, V.A.; Boeger, W.A. Black Yeast Biota in the Mangrove, in Search of the Origin of the Lethargic Crab Disease (LCD). Mycopathologia 2013, 175, 421–430. [Google Scholar] [CrossRef]
- Dayarathne, M.; Jones, E.; Maharachchikumbura, S.; Devadatha, B.; Sarma, V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Hassett, B.T.; Gradinger, R. Chytrids dominate arctic marine fungal communities. Environ. Microbiol. 2016, 18, 2001–2009. [Google Scholar] [CrossRef]
- Hassett, B.T.; Borrego, E.J.; Vonnahme, T.R.; Rämä, T.; Kolomiets, M.V.; Gradinger, R. Arctic marine fungi: Biomass, functional genes, and putative ecological roles. ISME J. 2019, 13, 1484–1496. [Google Scholar] [CrossRef]
- Vaz, A.B.M.; Rosa, L.H.; Vieira, M.L.A.; de Garcia, V.; Brandão, L.R.; Teixeira, L.C.R.S.; Moliné, M.; Libkind, D.; van Broock, M.; Rosa, C.A. The diversity, extracellular enzymatic activities and photoprotective compounds of yeasts isolated in Antarctica. Braz. J. Microbiol. 2011, 42, 937–947. [Google Scholar] [CrossRef]
- Wu, G.; Ma, H.; Zhu, T.; Li, J.; Gu, Q.; Li, D. Penilactones A and B, two novel polyketides from Antarctic deep-sea derived fungus Penicillium crustosum PRB-2. Tetrahedron 2012, 68, 9745–9749. [Google Scholar] [CrossRef]
- Wentzel, L.C.P.; Inforsato, F.J.; Montoya, Q.V.; Rossin, B.G.; Nascimento, N.R.; Rodrigues, A.; Sette, L.D. Fungi from Admiralty Bay (King George Island, Antarctica) Soils and Marine Sediments. Microb. Ecol. 2019, 77, 12–24. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Teixeira, D.R.; Vieira, R.; Lírio, J.M.; Felizardo, J.P.S.; Abuchacra, R.C.; Cardoso, R.P.; Zani, C.L.; Alves, T.M.A.; Junior, P.A.S.; et al. Diversity and bioprospecting of cultivable fungal assemblages in sediments of lakes in the Antarctic Peninsula. Extremophiles 2020, 124, 601–611. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Pinto, O.H.B.; Vieira, R.; Neto, A.A.; Convey, P.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P.E.A.S.; Rosa, L.H. Fungi Present in Antarctic Deep-Sea Sediments Assessed Using DNA Metabarcoding. Microb. Ecol. 2021, 1–8. [Google Scholar] [CrossRef]
- Ha, T.M.; Kim, D.-C.C.; Sohn, J.H.; Yim, J.H.; Oh, H. Anti-Inflammatory and Protein Tyrosine Phosphatase 1B Inhibitory Metabolites from the Antarctic Marine-Derived Fungal Strain Penicillium glabrum SF-7123. Mar. Drugs 2020, 18, 247. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Lin, A.; Gu, Q.; Zhu, T.; Li, D. Four new chloro-eremophilane sesquiterpenes from an antarctic deep-sea derived fungus, Penicillium sp. PR19N-1. Mar. Drugs 2013, 11, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Fan, Z.-W.; Xie, C.-L.; Liu, Q.; Luo, Z.-H.; Liu, G.; Yang, X.-W. Spirograterpene A, a Tetracyclic Spiro-Diterpene with a Fused 5/5/5/5 Ring System from the Deep-Sea-Derived Fungus Penicillium granulatum MCCC 3A00475. J. Nat. Prod. 2017, 80, 2174–2177. [Google Scholar] [CrossRef]
- Laich, F.; Vaca, I.; Chávez, R. Rhodotorula portillonensis sp. nov., a basidiomycetous yeast isolated from Antarctic shallow-water marine sediment. Int. J. Syst. Evol. Microbiol. 2013, 63, 3884–3891. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Dayo-Owoyemi, I.; Nobre, F.S.; Pagnocca, F.C.; Chaud, L.C.S.; Pessoa, A.; Felipe, M.G.A.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef]
- Gonçalves, V.N.; Campos, L.S.; Melo, I.S.; Pellizari, V.H.; Rosa, C.A.; Rosa, L.H. Penicillium solitum: A mesophilic, psychrotolerant fungus present in marine sediments from Antarctica. Polar Biol. 2013, 36, 1823–1831. [Google Scholar] [CrossRef]
- Ren, J.; Yang, Y.; Liu, D.; Chen, W.; Proksch, P.; Shao, B.; Lin, W. Sequential determination of new peptaibols asperelines G-Z12 produced by marine-derived fungus Trichoderma asperellum using ultrahigh pressure liquid chromatography combined with electrospray-ionization tandem mass spectrometry. J. Chromatogr. A 2013, 1309, 90–95. [Google Scholar] [CrossRef]
- Purić, J.; Vieira, G.; Cavalca, L.B.B.; Sette, L.D.D.; Ferreira, H.; Vieira, M.L.C.L.C.; Sass, D.C.C. Activity of Antarctic fungi extracts against phytopathogenic bacteria. Lett. Appl. Microbiol. 2018, 66, 530–536. [Google Scholar] [CrossRef]
- Vieira, G.; Purić, J.; Morão, L.G.G.; dos Santos, J.A.A.; Inforsato, F.J.J.; Sette, L.D.D.; Ferreira, H.; Sass, D.C.C. Terrestrial and marine Antarctic fungi extracts active against Xanthomonas citri subsp. citri. Lett. Appl. Microbiol. 2018, 67, 64–71. [Google Scholar] [CrossRef]
- Li, W.; Luo, D.; Huang, J.; Wang, L.; Zhang, F.; Xi, T.; Liao, J.; Lu, Y. Antibacterial constituents from Antarctic fungus, Aspergillus sydowii SP-1. Nat. Prod. Res. 2018, 32, 662–667. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.H.; Yu, H.B.; Liu, X.Y.; Lu, X.L.; Jiao, B.H. Furanone derivative and sesquiterpene from Antarctic marine-derived fungus Penicillium sp. S-1-18. J. Asian Nat. Prod. Res. 2018, 20, 1108–1115. [Google Scholar] [CrossRef]
- Liu, C.-C.; Zhang, Z.-Z.; Feng, Y.-Y.; Gu, Q.-Q.; Li, D.-H.; Zhu, T.-J. Secondary metabolites from Antarctic marine-derived fungus Penicillium crustosum HDN153086. Nat. Prod. Res. 2019, 33, 414–419. [Google Scholar] [CrossRef]
- Mercantini, R.; Marsella, R.; Moretto, D.; Finotti, E. Keratinophilic fungi in the antarctic environment. Mycopathologia 1993, 122, 169–175. [Google Scholar] [CrossRef]
- Dos Santos, J.A.; Meyer, E.; Sette, L.D. Fungal community in antarctic soil along the retreating collins glacier (Fildes peninsula, King George Island). Microorganisms 2020, 8, 1145. [Google Scholar] [CrossRef]
- Loque, C.P.; Medeiros, A.O.; Pellizzari, F.M.; Oliveira, E.C.; Rosa, C.A.; Rosa, L.H. Fungal community associated with marine macroalgae from Antarctica. Polar Biol. 2010, 33, 641–648. [Google Scholar] [CrossRef]
- Putzke, J.; Pereira, A.B.; Pereira, A.B. Phaeosphaeria deschampsii (Ascomycota): A new parasite species of Deschampsia antarctica (Poaceae) described to Antarctica. An. Acad. Bras. Cienc. 2016, 88, 1967–1969. [Google Scholar] [CrossRef]
- Godinho, V.M.; Furbino, L.E.; Santiago, I.F.; Pellizzari, F.M.; Yokoya, N.S.; Pupo, D.; Alves, T.M.A.; Junior, P.A.S.; Romanha, A.J.; Zani, C.L.; et al. Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J. 2013, 7, 1434–1451. [Google Scholar] [CrossRef]
- Fernandes Duarte, A.W.; Zambrano Passarini, M.R.; Delforno, T.P.; Pellizzari, F.M.; Zecchin Cipro, C.V.; Montone, R.C.; Petry, M.V.; Putzke, J.; Rosa, L.H.; Sette, L.D. Yeasts from macroalgae and lichens that inhabit the South Shetland Islands, Antarctica. Environ. Microbiol. Rep. 2016, 8, 874–885. [Google Scholar] [CrossRef]
- Furbino, L.E.; Godinho, V.M.; Santiago, I.F.; Pellizari, F.M.; Alves, T.M.A.A.; Zani, C.L.; Junior, P.A.S.S.; Romanha, A.J.; Carvalho, A.G.O.O.; Gil, L.H.V.G.V.G.; et al. Diversity Patterns, Ecology and Biological Activities of Fungal Communities Associated with the Endemic Macroalgae Across the Antarctic Peninsula. Microb. Ecol. 2014, 67, 775–787. [Google Scholar] [CrossRef]
- Furbino, L.E.; Pellizzari, F.M.; Neto, P.C.; Rosa, C.A.; Rosa, L.H. Isolation of fungi associated with macroalgae from maritime Antarctica and their production of agarolytic and carrageenolytic activities. Polar Biol. 2018, 41, 527–535. [Google Scholar] [CrossRef]
- Martorell, M.M.; Lannert, M.; Matula, C.V.; Quartino, M.L.; de Figueroa, L.I.C.; Mac Cormack, W.P.; Ruberto, L.A.M. Studies toward the comprehension of fungal-macroalgae interaction in cold marine regions from a biotechnological perspective. Fungal Biol. 2020, 125, 218–230. [Google Scholar] [CrossRef]
- Alker, A.P.; Smith, G.W.; Kim, K. Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. Hydrobiologia 2001, 460, 105–111. [Google Scholar] [CrossRef]
- Van Dover, C.L.; Ward, M.E.; Scott, J.L.; Underdown, J.; Anderson, B.; Gustafson, C.; Whalen, M.; Carnegie, R.B. A fungal epizootic in mussels at a deep-sea hydrothermal vent. Mar. Ecol. 2007, 28, 54–62. [Google Scholar] [CrossRef]
- Cawthorn, R.J. Diseases of American lobsters (Homarus americanus): A review. J. Invertebr. Pathol. 2011, 106, 71–78. [Google Scholar] [CrossRef]
- Amend, A. From Dandruff to Deep-Sea Vents: Malassezia-like Fungi Are Ecologically Hyper-diverse. PLoS Pathog. 2014, 10, e1004277. [Google Scholar] [CrossRef]
- Nagahama, T.; Hamamoto, M.; Nakase, T.; Horikoshi, K. Rhodotorula lamellibrachii sp. nov., a new yeast species from a tubeworm collected at the deep-sea floor in Sagami Bay and its phylogenetic analysis. Antonie Leeuwenhoek 2001, 80, 317–323. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, B.; Sun, W.; Zhang, F.; Li, Z. Phylogenetically diverse endozoic fungi in the South China Sea sponges and their potential in synthesizing bioactive natural products suggested by PKS gene and cytotoxic activity analysis. Fungal Divers. 2013, 58, 127–141. [Google Scholar] [CrossRef]
- Nguyen, M.T.H.D.; Thomas, T. Diversity, host-specificity and stability of sponge-associated fungal communities of co-occurring sponges. PeerJ 2018, 2018, e4965. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, S. Animals in Coastal Benthic Ecosystem and Aquaculture Systems. In Fungi in Coastal and Oceanic Marine Ecosystems; Raghukumar, S., Ed.; Springer International Publishing: Cham, Switzerland; Berlin/Heidelberg, Germany, 2017; pp. 163–183. ISBN 978-3-319-54304-8. [Google Scholar]
- Laich, F.; Chávez, R.; Vaca, I. Leucosporidium escuderoi f.a., sp. nov., a basidiomycetous yeast associated with an Antarctic marine sponge. Antonie Leeuwenhoek 2014, 105, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Vaca, I.; Faúndez, C.; Maza, F.; Paillavil, B.; Hernández, V.; Acosta, F.; Levicán, G.; Martínez, C.; Chávez, R. Cultivable psychrotolerant yeasts associated with Antarctic marine sponges. World J. Microbiol. Biotechnol. 2013, 29, 183–189. [Google Scholar] [CrossRef]
- Kim, D.C.; Lee, H.S.; Ko, W.; Lee, D.S.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; Oh, H. Anti-inflammatory effect of methylpenicinoline from a marine isolate of Penicillium sp. (SF-5995): Inhibition of NF-κB and MAPK pathways in lipopolysaccharide-induced RAW264.7 macrophages and BV2 microglia. Molecules 2014, 19, 18073–18089. [Google Scholar] [CrossRef]
- Henríquez, M.; Vergara, K.; Norambuena, J.; Beiza, A.; Maza, F.; Ubilla, P.; Araya, I.; Chávez, R.; San-Martín, A.; Darias, J.; et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 2014, 30, 65–76. [Google Scholar] [CrossRef]
- Cui, X.; Zhu, G.; Liu, H.; Jiang, G.; Wang, Y.; Zhu, W. Diversity and function of the Antarctic krill microorganisms from Euphausia superba. Sci. Rep. 2016, 6, 36496. [Google Scholar] [CrossRef]
- Wang, F.; Sheng, J.; Chen, Y.; Xu, J. Microbial diversity and dominant bacteria causing spoilage during storage and processing of the Antarctic krill, Euphausia superba. Polar Biol. 2021, 44, 163–171. [Google Scholar] [CrossRef]
- Moreno-Pino, M.; Cristi, A.; Gillooly, J.F.; Trefault, N. Characterizing the microbiomes of Antarctic sponges: A functional metagenomic approach. Sci. Rep. 2020, 10, 645. [Google Scholar] [CrossRef]
- Coda, R.; Cassone, A.; Rizzello, C.G.; Nionelli, L.; Cardinali, G.; Gobbetti, M. Antifungal activity of Wickerhamomyces anomalus and Lactobacillus plantarum during sourdough fermentation: Identification of novel compounds and long-term effect during storage of wheat bread. Appl. Environ. Microbiol. 2011, 77, 3484–3492. [Google Scholar] [CrossRef]
- Walker, A.K.; Robicheau, B.M. Fungal diversity and community structure from coastal and barrier island beaches in the United States Gulf of Mexico. Sci. Rep. 2021, 11, 3889. [Google Scholar] [CrossRef]
- Nagano, Y.; Nagahama, T.; Hatada, Y.; Nunoura, T.; Takami, H.; Miyazaki, J.; Takai, K.; Horikoshi, K. Fungal diversity in deep-sea sediments—The presence of novel fungal groups. Fungal Ecol. 2010, 3, 316–325. [Google Scholar] [CrossRef]
- Rédou, V.; Ciobanu, M.C.; Pachiadaki, M.G.; Edgcomb, V.; Alain, K.; Barbier, G.; Burgaud, G. In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol. Ecol. 2014, 90, 908–921. [Google Scholar] [CrossRef]
- Singh, P.; Raghukumar, C.; Verma, P.; Shouche, Y. Fungal Community Analysis in the Deep-Sea Sediments of the Central Indian Basin by Culture-Independent Approach. Microb. Ecol. 2011, 61, 507–517. [Google Scholar] [CrossRef]
- Gao, Y.; Du, X.; Xu, W.; Fan, R.; Zhang, X.; Yang, S.; Chen, X.; Lv, J.; Luo, Z. Fungal Diversity in Deep Sea Sediments from East Yap Trench and Their Denitrification Potential. Geomicrobiol. J. 2020, 37, 848–858. [Google Scholar] [CrossRef]
- Vargas-Gastélum, L.; Riquelme, M. The Mycobiota of the Deep Sea: What Omics Can Offer. Life 2020, 10, 292. [Google Scholar] [CrossRef]
- Barone, G.; Rastelli, E.; Corinaldesi, C.; Tangherlini, M.; Danovaro, R.; Dell’Anno, A. Benthic deep-sea fungi in submarine canyons of the Mediterranean Sea. Prog. Oceanogr. 2018, 168, 57–64. [Google Scholar] [CrossRef]
- Yang, S.; Xu, W.; Gao, Y.; Chen, X.; Luo, Z.H. Fungal diversity in deep-sea sediments from Magellan seamounts environment of the western Pacific revealed by high-throughput Illumina sequencing. J. Microbiol. 2020, 58, 841–852. [Google Scholar] [CrossRef]
- Alexander, E.; Stock, A.; Breiner, H.W.; Behnke, A.; Bunge, J.; Yakimov, M.M.; Stoeck, T. Microbial eukaryotes in the hypersaline anoxic L’Atalante deep-sea basin. Environ. Microbiol. 2009, 11, 360–381. [Google Scholar] [CrossRef]
- Bernhard, J.M.; Kormas, K.; Pachiadaki, M.G.; Rocke, E.; Beaudoin, D.J.; Morrison, C.; Visscher, P.T.; Cobban, A.; Starczak, V.R.; Edgcomb, V.P. Benthic protists and fungi of Mediterranean deep hypsersaline anoxic basin redoxcline sediments. Front. Microbiol. 2014, 5, 605. [Google Scholar] [CrossRef]
- Stock, A.; Breiner, H.W.; Pachiadaki, M.; Edgcomb, V.; Filker, S.; La Cono, V.; Yakimov, M.M.; Stoeck, T. Microbial eukaryote life in the new hypersaline deep-sea basin Thetis. Extremophiles 2012, 16, 21–34. [Google Scholar] [CrossRef]
- Nagahama, T.; Takahashi, E.; Nagano, Y.; Abdel-Wahab, M.A.; Miyazaki, M. Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ. Microbiol. 2011, 13, 2359–2370. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.P.; Cao, H.L.; Shek, C.S.; Tian, R.M.; Wong, Y.H.; Batang, Z.; Al-Suwailem, A.; Qian, P.-Y.Y. Diversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red Sea. Front. Microbiol. 2014, 5, 37. [Google Scholar] [CrossRef]
- Nagano, Y.; Miura, T.; Nishi, S.; Lima, A.O.; Nakayama, C.; Pellizari, V.H.; Fujikura, K. Fungal diversity in deep-sea sediments associated with asphalt seeps at the Sao Paulo Plateau. Deep Sea Res. Part II Top. Stud. Oceanogr. 2017, 146, 59–67. [Google Scholar] [CrossRef]
- Gadanho, M.; Sampaio, J.P. Occurrence and diversity of yeasts in the mid-atlantic ridge hydrothermal fields near the Azores Archipelago. Microb. Ecol. 2005, 50, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Burgaud, G.; Arzur, D.; Durand, L.; Cambon-Bonavita, M.-A.; Barbier, G. Marine culturable yeasts in deep-sea hydrothermal vents: Species richness and association with fauna. FEMS Microbiol. Ecol. 2010, 73, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Velez, P.; Gasca-Pineda, J.; Riquelme, M. Cultivable fungi from deep-sea oil reserves in the Gulf of Mexico: Genetic signatures in response to hydrocarbons. Mar. Environ. Res. 2020, 153, 104816. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.N.; Carvalho, C.R.; Johann, S.; Mendes, G.; Alves, T.M.; Zani, C.L.; Junior, P.A.S.S.; Murta, S.M.F.F.; Romanha, A.J.; Cantrell, C.L.; et al. Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol. 2015, 38, 1143–1152. [Google Scholar] [CrossRef]
- Bochdansky, A.B.; Clouse, M.A.; Herndl, G.J. Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 2017, 11, 362–373. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Coelho, L.C.; Vieira, R.; Neto, A.A.; Zani, C.L.; Alves, T.M.A.A.; Junior, P.A.S.S.; Murta, S.M.F.F.; Barbosa, E.C.; Oliveira, J.G.; et al. Cultivable fungi present in deep-sea sediments of Antarctica: Taxonomy, diversity, and bioprospecting of bioactive compounds. Extremophiles 2020, 24, 227–238. [Google Scholar] [CrossRef]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Barone, G.; Varrella, S.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Danovaro, R.; Corinaldesi, C. Marine Fungi: Biotechnological Perspectives from Deep-Hypersaline Anoxic Basins. Diversity 2019, 11, 113. [Google Scholar] [CrossRef]
- Grossart, H.P.; Rojas-Jimenez, K. Aquatic fungi: Targeting the forgotten in microbial ecology. Curr. Opin. Microbiol. 2016, 31, 140–145. [Google Scholar] [CrossRef]
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128. [Google Scholar] [CrossRef]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef]
- Comeau, A.M.; Vincent, W.F.; Bernier, L.; Lovejoy, C. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 2016, 6, 30120. [Google Scholar] [CrossRef]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 15, 1261605. [Google Scholar] [CrossRef]
- Frenken, T.; Alacid, E.; Berger, S.A.; Bourne, E.C.; Gerphagnon, M.; Grossart, H.P.; Gsell, A.S.; Ibelings, B.W.; Kagami, M.; Küpper, F.C.; et al. Integrating chytrid fungal parasites into plankton ecology: Research gaps and needs. Environ. Microbiol. 2017, 19, 3802–3822. [Google Scholar] [CrossRef]
- Lepelletier, F.; Karpov, S.A.; Alacid, E.; Le Panse, S.; Bigeard, E.; Garcés, E.; Jeanthon, C.; Guillou, L. Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a Chytrid Infecting Marine Dinoflagellates. Protist 2014, 165, 230–244. [Google Scholar] [CrossRef]
- Sime-Ngando, T. Phytoplankton Chytridiomycosis: Fungal Parasites of Phytoplankton and Their Imprints on the Food Web Dynamics. Front. Microbiol. 2012, 3, 361. [Google Scholar] [CrossRef]
- Scholz, B.; Küpper, F.C.; Vyverman, W.; Karsten, U. Eukaryotic pathogens (Chytridiomycota and Oomycota) infecting marine microphytobenthic diatoms—A methodological comparison. J. Phycol. 2014, 50, 1009–1019. [Google Scholar] [CrossRef]
- Gleason, F.H.; Kagami, M.; Lefevre, E.; Sime-Ngando, T. The ecology of chytrids in aquatic ecosystems: Roles in food web dynamics. Fungal Biol. Rev. 2008, 22, 17–25. [Google Scholar] [CrossRef]
- Gachon, C.M.M.; Sime-Ngando, T.; Strittmatter, M.; Chambouvet, A.; Kim, G.H. Algal diseases: Spotlight on a black box. Trends Plant Sci. 2010, 15, 633–640. [Google Scholar] [CrossRef]
- Pusceddu, A.; Dell’Anno, A.; Fabiano, M.; Danovaro, R. Quantity and bioavailability of sediment organic matter as signatures of benthic trophic status. Mar. Ecol. Prog. Ser. 2009, 375, 41–52. [Google Scholar] [CrossRef]
- Jebaraj, C.S.; Raghukumar, C.; Behnke, A.; Stoeck, T. Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol. Ecol. 2010, 71, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Dethier, M.N.; Brown, A.S.; Burgess, S.; Eisenlord, M.E.; Galloway, A.W.E.; Kimber, J.; Lowe, A.T.; O’Neil, C.M.; Raymond, W.W.; Sosik, E.A.; et al. Degrading detritus: Changes in food quality of aging kelp tissue varies with species. J. Exp. Mar. Bio. Ecol. 2014, 460, 72–79. [Google Scholar] [CrossRef]
- Crowther, T.W.; Grossart, H.P. The role of bottom-up and top-down interactions in determining microbial and fungal diversity and function. In Trophic Ecology: Bottom-Up and Top-Down Interactions Across Aquatic and Terrestrial Systems; Hanley, T.C., La Pierre, K.J., Eds.; Cambridge University Press: Cambridge, UK, 2015; pp. 260–287. ISBN 9781139924856. [Google Scholar]
- Attermeyer, K.; Premke, K.; Hornick, T.; Hilt, S.; Grossart, H.P. Ecosystem-level studies of terrestrial carbon reveal contrasting bacterial metabolism in different aquatic habitats. Ecology 2013, 94, 2754–2766. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.C.O.; Safar, S.V.B.; Marques, A.R.; Medeiros, A.O.; Santos, A.R.O.; Carvalho, C.; Lachance, M.-A.; Sampaio, J.P.; Rosa, C.A. The diversity and extracellular enzymatic activities of yeasts isolated from water tanks of Vriesea minarum, an endangered bromeliad species in Brazil, and the description of Occultifur brasiliensis f.a., sp. nov. Antonie Leeuwenhoek 2015, 107, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Krauss, G.J.; Solé, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Bärlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef]
- Bhadury, P.; Bik, H.; Lambshead, J.D.; Austen, M.C.; Smerdon, G.R.; Rogers, A.D. Molecular diversity of fungal phylotypes Co-Amplified alongside nematodes from coastal and Deep-Sea marine environments. PLoS ONE 2011, 6, e26445. [Google Scholar] [CrossRef]
- Sapir, A.; Dillman, A.R.; Connon, S.A.; Grupe, B.M.; Ingels, J.; Mundo-Ocampo, M.; Levin, L.A.; Baldwin, J.G.; Orphan, V.J.; Sternberg, P.W. Microsporidia-nematode associations in methane seeps reveal basal fungal parasitism in the deep sea. Front. Microbiol. 2014, 5, 43. [Google Scholar] [CrossRef]
- Dayton, P.K. Ecology of Kelp Communities. Annu. Rev. Ecol. Syst. 1985, 16, 215–245. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Pang, K.-L. Marine Fungi: And Fungal-Like Organisms; Walter de Gruyter: Berlin, Germany, 2012; ISBN 3110264064. [Google Scholar]
- Richards, T.A.; Jones, M.D.M.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef]
- Zuccaro, A.; Mitchell, J.I. Fungal communities of seaweeds. In The Fungal Community; CRC Press: Boca Raton, USA, 2005; pp. 533–579. [Google Scholar]
- Egan, S.; Harder, T.; Burke, C.; Steinberg, P.; Kjelleberg, S.; Thomas, T. The seaweed holobiont: Understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 2013, 37, 462–476. [Google Scholar] [CrossRef]
- Singh, R.P.; Kumari, P.; Reddy, C.R.K. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol. 2015, 99, 1571–1586. [Google Scholar] [CrossRef]
- Wichard, T.; Beemelmanns, C. Role of Chemical Mediators in Aquatic Interactions across the Prokaryote–Eukaryote Boundary. J. Chem. Ecol. 2018, 44, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Rämä, T.; Hassett, B.T.; Bubnova, E. Arctic marine fungi: From filaments and flagella to operational taxonomic units and beyond. Bot. Mar. 2017, 60, 433–452. [Google Scholar] [CrossRef]
- Castro-Sowinski, S. The Ecological Role of Micro-Organisms in the Antarctic Environment; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 3030027864. [Google Scholar]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Omar, S.M.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Corinaldesi, C. New perspectives in benthic deep-sea microbial ecology. Front. Mar. Sci. 2015, 2, 17. [Google Scholar] [CrossRef]
- Chambergo, F.S.; Valencia, E.Y. Fungal biodiversity to biotechnology. Appl. Microbiol. Biotechnol. 2016, 100, 2567–2577. [Google Scholar] [CrossRef] [PubMed]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Ul Arifeen, M.Z.; Ma, Y.N.; Xue, Y.R.; Liu, C.H. Deep-sea fungi could be the new arsenal for bioactive molecules. Mar. Drugs 2020, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Margesin, R. Cold-Adapted Yeasts: A Lesson from the Cold and a Challenge for the XXI Century. In Cold-Adapted Yeasts: Biodiversity, Adaptation Strategies and Biotechnological Significance; Buzzini, P., Margesin, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–22. ISBN 978-3-642-39681-6. [Google Scholar]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Bernan, V.S.; Greenstein, M.; Maiese, W.M. Marine microorganisms as a source of new natural products. Adv. Appl. Microbiol. 1997, 43, 57–90. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Carroll, A.R.; Munro, M.M.H.G.; Prinsep, M.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 35, 8–53. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Bonugli-Santos, R.C.; dos Santos Vasconcelos, M.R.; Passarini, M.R.Z.; Vieira, G.A.L.; Lopes, V.C.P.; Mainardi, P.H.; dos Santos, J.A.; de Azevedo Duarte, L.; Otero, I.V.R.; da Silva Yoshida, A.M.; et al. Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6, 269. [Google Scholar] [CrossRef]
- Imhoff, J.F. Natural products from marine fungi—Still an underrepresented resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef]
- Gomes, N.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can Some Marine-Derived Fungal Metabolites Become Actual Anticancer Agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef]
- Varrella, S.; Tangherlini, M.; Corinaldesi, C. Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest. Mar. Drugs 2020, 18, 91. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine Fungi: A Source of Potential Anticancer Compounds: A Source of Potential Anticancer Compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef]
- Núñez-Pons, L.; Shilling, A.; Verde, C.; Baker, B.J.; Giordano, D. Marine Terpenoids from Polar Latitudes and Their Potential Applications in Biotechnology. Mar. Drugs 2020, 18, 401. [Google Scholar] [CrossRef]
- Frisvad, J.C. Cold-Adapted Fungi as a Source for Valuable Metabolites. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 381–387. ISBN 978-3-540-74335-4. [Google Scholar]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular Enzymes and Bioactive Compounds from Antarctic Terrestrial Fungi for Bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef]
- Lin, A.; Wu, G.; Gu, Q.; Zhu, T.; Li, D. New eremophilane-type sesquiterpenes from an Antarctic deep-sea derived fungus, Penicillium sp. PR19 N-1. Arch. Pharm. Res. 2014, 37, 839–844. [Google Scholar] [CrossRef]
- ChemSpider Search and Share Chemistry. Available online: https://www.chemspider.com/ (accessed on 7 May 2021).
- Huang, J.-N.; Zou, Q.; Chen, J.; Xu, S.-H.; Luo, D.; Zhang, F.-G.; Lu, Y.-Y. Phenols and diketopiperazines isolated from Antarctic-derived fungi, Penicillium citreonigrum SP-6. Phytochem. Lett. 2018, 27, 114–118. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Li, Y.Y.; Huang, Y.J.; Su, W.J.; Shen, Y.M. Three new sesquiterpenoids from Xylaria sp. NCY2. Helv. Chim. Acta 2008, 91, 46–52. [Google Scholar] [CrossRef]
- Figueroa, L.; Jiménez, C.; Rodríguez, J.; Areche, C.; Chávez, R.; Henríquez, M.; de la Cruz, M.; Díaz, C.; Segade, Y.; Vaca, I. 3-Nitroasterric Acid Derivatives from an Antarctic Sponge-Derived Pseudogymnoascus sp. Fungus. J. Nat. Prod. 2015, 78, 919–923. [Google Scholar] [CrossRef]
- Li, Y.; Sun, B.; Liu, S.; Jiang, L.; Liu, X.; Zhang, H.; Che, Y. Bioactive Asterric Acid Derivatives from the Antarctic Ascomycete Fungus Geomyces sp. J. Nat. Prod. 2008, 71, 1643–1646. [Google Scholar] [CrossRef]
- Ren, J.; Xue, C.; Tian, L.; Xu, M.; Chen, J.; Deng, Z.; Proksch, P.; Lin, W. Asperelines A−F, Peptaibols from the Marine-Derived Fungus Trichoderma asperellum. J. Nat. Prod. 2009, 72, 1036–1044. [Google Scholar] [CrossRef]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D.; et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Nandanwar, S.K.; Borkar, S.B.; Lee, J.H.; Kim, H.J. Taking Advantage of Promiscuity of Cold-Active Enzymes. Appl. Sci. 2020, 10, 8128. [Google Scholar] [CrossRef]
- Sočan, J.; Purg, M.; Åqvist, J. Computer simulations explain the anomalous temperature optimum in a cold-adapted enzyme. Nat. Commun. 2020, 11, 2644. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kokare, C. Chapter 11—Microbial Enzymes of Use in Industry. In Biotechnology of Microbial Enzymes Production, Biocatalysis and Industrial Applications; Brahmachari, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 267–298. ISBN 978-0-12-803725-6. [Google Scholar]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6, 174. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef]
- Buzzini, P.; Branda, E.; Goretti, M.; Turchetti, B. Psychrophilic yeasts from worldwide glacial habitats: Diversity, adaptation strategies and biotechnological potential. FEMS Microbiol. Ecol. 2012, 82, 217–241. [Google Scholar] [CrossRef]
- Flam, F. The chemistry of life at the margins. Science 1994, 265, 471–472. [Google Scholar] [CrossRef]
- Dumorné, K.; Córdova, D.C.; Astorga-Eló, M.; Renganathan, P. Extremozymes: A potential source for industrial applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar] [CrossRef]
- Bjelic, S.; Brandsdal, B.O.; Åqvist, J. Cold adaptation of enzyme reaction rates. Biochemistry 2008, 47, 10049–10057. [Google Scholar] [CrossRef]
- Feller, G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys. Condens. Matter 2010, 22, 323101. [Google Scholar] [CrossRef]
- Feller, G. Psychrophilic Enzymes: From Folding to Function and Biotechnology. Scientifica 2013, 2013, 512840. [Google Scholar] [CrossRef]
- Gerday, C. Psychrophily and Catalysis. Biology 2013, 2, 719–741. [Google Scholar] [CrossRef]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef]
- Martínez-Martínez, M.; Bargiela, R.; Ferrer, M. Metagenomics and the Search for Industrial Enzymes. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications; Brahmachari, G., Demain, A.L., Adrio, J.L., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 167–184. [Google Scholar]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and Applications of Extremozymes from Deep-Sea Extremophilic Microorganisms: A Mini Review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef]
- Castilla, I.A.; Woods, D.F.; Reen, F.J.; O’Gara, F. Harnessing marine biocatalytic reservoirs for green chemistry applications through metagenomic technologies. Mar. Drugs 2018, 16, 227. [Google Scholar] [CrossRef]
- Martorell, M.M.; Ruberto, L.A.M.; de Figueroa, L.I.C.; Mac Cormack, W.P. Antarctic Yeasts as a Source of Enzymes for Biotechnological Applications. In Fungi of Antarctica; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 285–304. [Google Scholar]
- Arora, N.K.; Panosyan, H. Extremophiles: Applications and roles in environmental sustainability. Environ. Sustain. 2019, 2, 217–218. [Google Scholar] [CrossRef]
- Burhan, H.; Ravinder, S.R.; Deepak, C.; Poonam, S.; Fayaz, A.M.; Sanjay, S.; Ishfaq, A. Psychrophilic yeasts and their biotechnological applications—A review. Afr. J. Biotechnol. 2014, 13, 2188–2197. [Google Scholar] [CrossRef][Green Version]
- Feller, G. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. 2003, 60, 648–662. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Gomes, J.; Steiner, W. The biocatalytic potential of extremophiles and extremozymes. Food Technol. Biotechnol. 2004, 42, 223–235. [Google Scholar]
- Dalmaso, G.Z.L.; Ferreira, D.; Vermelho, A.B.; Zamith, G.; Dalmaso, L.; Ferreira, D.; Vermelho, A.B.; Dalmaso, G.Z.L.; Ferreira, D.; Vermelho, A.B. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef]
- Kuddus, M. Cold-active enzymes in food biotechnology: An updated mini review. J. Appl. Biol. Biotechnol. 2018, 6, 58–63. [Google Scholar]
- Blamey, J.M.; Fischer, F.; Meyer, H.P.; Sarmiento, F.; Zinn, M. Enzymatic Biocatalysis in Chemical Transformations: A Promising and Emerging Field in Green Chemistry Practice. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications; Brahmachari, G., Demain, A.L., Adrio, J.L., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 347–403. ISBN 9780128037461. [Google Scholar]
- Fenice, M. The Psychrotolerant Antarctic Fungus Lecanicillium muscarium CCFEE 5003: A Powerful Producer of Cold-Tolerant Chitinolytic Enzymes. Molecules 2016, 21, 447. [Google Scholar] [CrossRef]
- Barghini, P.; Moscatelli, D.; Garzillo, A.M.V.; Crognale, S.; Fenice, M. High production of cold-tolerant chitinases on shrimp wastes in bench-top bioreactor by the Antarctic fungus Lecanicillium muscarium CCFEE 5003: Bioprocess optimization and characterization of two main enzymes. Enzym. Microb. Technol. 2013, 53, 331–338. [Google Scholar] [CrossRef]
- Ramli, A.N.M.; Mahadi, N.M.; Shamsir, M.S.; Rabu, A.; Joyce-Tan, K.H.; Murad, A.M.A.; Md. Illias, R. Structural prediction of a novel chitinase from the psychrophilic Glaciozyma antarctica PI12 and an analysis of its structural properties and function. J. Comput. Aided Mol. Des. 2012, 26, 947–961. [Google Scholar] [CrossRef]
- Teoh, C.P.; Koh, S.P.; Ling, C.M.W.V. Characterisation of an Antarctic Yeast, Glaciozyma antarctica PI12. Borneo Int. J. Biotechnol. 2020, 1, 89. [Google Scholar] [CrossRef]
- Bharudin, I.; Abu Bakar, M.F.; Hashim, N.H.F.; Mat Isa, M.N.; Alias, H.; Firdaus-Raih, M.; Md Illias, R.; Najimudin, N.; Mahadi, N.M.; Abu Bakar, F.D.; et al. Unravelling the adaptation strategies employed by Glaciozyma antarctica PI12 on Antarctic sea ice. Mar. Environ. Res. 2018, 137, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.; Hashim, N.H.F.; Mahadi, M.N.; Murad, A.M. The Cold-Active Endo-β-1,3(4)-Glucanase from a Marine Psychrophilic Yeast, Glaciozyma antarctica PI12: Heterologous Expression, Biochemical Characterisation, and Molecular Modeling. Int. J. Appl. Biol. Pharm. Technol. 2021, 12, 279–300. [Google Scholar] [CrossRef]
- Mohammadi, S.; Parvizpour, S.; Razmara, J.; Abu Bakar, F.D.; Illias, R.M.; Mahadi, N.M.; Murad, A.M.A. Structure Prediction of a Novel Exo-β-1,3-Glucanase: Insights into the Cold Adaptation of Psychrophilic Yeast Glaciozyma antarctica PI12. Interdiscip. Sci. Comput. Life Sci. 2018, 10, 157–168. [Google Scholar] [CrossRef]
- Hashim, N.H.F.; Mahadi, N.M.; Illias, R.M.; Feroz, S.R.; Abu Bakar, F.D.; Murad, A.M.A. Biochemical and structural characterization of a novel cold-active esterase-like protein from the psychrophilic yeast Glaciozyma antarctica. Extremophiles 2018, 22, 607–616. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Barato, M.B.; Nobre, F.S.; Polezel, D.A.; de Oliveira, T.B.; dos Santos, J.A.; Rodrigues, A.; Sette, L.D. Production of cold-adapted enzymes by filamentous fungi from King George Island, Antarctica. Polar Biol. 2018, 41, 2511–2521. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Lopes, A.M.; Molino, J.V.D.; Pessoa, A.; Sette, L.D. Liquid-liquid extraction of lipase produced by psychrotrophic yeast Leucosporidium scottii L117 using aqueous two-phase systems. Sep. Purif. Technol. 2015, 156, 215–225. [Google Scholar] [CrossRef]
- Carrasco, M.; Rozas, J.M.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012, 12, 251. [Google Scholar] [CrossRef]
- Tsuji, M.; Yokota, Y.; Shimohara, K.; Kudoh, S.; Hoshino, T. An Application of Wastewater Treatment in a Cold Environment and Stable Lipase Production of Antarctic Basidiomycetous Yeast Mrakia blollopis. PLoS ONE 2013, 8, e59376. [Google Scholar] [CrossRef]
- Poveda, G.; Gil-Durán, C.; Vaca, I.; Levicán, G.; Chávez, R. Cold-active pectinolytic activity produced by filamentous fungi associated with Antarctic marine sponges. Biol. Res. 2018, 51, 28. [Google Scholar] [CrossRef]
- Yu, P.; Wang, X.-T.; Liu, J.-W. Purification and characterization of a novel cold-adapted phytase from Rhodotorula mucilaginosa strain JMUY14 isolated from Antarctic. J. Basic Microbiol. 2015, 55, 1029–1039. [Google Scholar] [CrossRef]
- Lario, L.D.; Chaud, L.; das Graças Almeida, M.; Converti, A.; Durães Sette, L.; Pessoa, A.; Duraes Sette, L.; Pessoa, A.; Durães Sette, L.; Pessoa, A. Production, purification, and characterization of an extracellular acid protease from the marine Antarctic yeast Rhodotorula mucilaginosa L7. Fungal Biol. 2015, 119, 1129–1136. [Google Scholar] [CrossRef]
- Chaud, L.C.S.; Lario, L.D.; Bonugli-Santos, R.C.; Sette, L.D.; Pessoa Junior, A.; Felipe, M.d.G.d.A. Improvement in extracellular protease production by the marine antarctic yeast Rhodotorula mucilaginosa L7. New Biotechnol. 2016, 33, 807–814. [Google Scholar] [CrossRef]
- Turkiewicz, M.; Pazgier, M.; Kalinowska, H.; Bielecki, S. A cold-adapted extracellular serine proteinase of the yeast Leucosporidium antarcticum. Extremophiles 2003, 7, 435–442. [Google Scholar] [CrossRef]
- Alias, N.; Ahmad Mazian, M.; Salleh, A.B.; Basri, M.; Rahman, R.N.Z.R.A. Molecular Cloning and Optimization for High Level Expression of Cold-Adapted Serine Protease from Antarctic Yeast Glaciozyma antarctica PI12. Enzym. Res. 2014, 2014, 197938. [Google Scholar] [CrossRef]
- Glodowsky, A.P.; Ruberto, L.A.; Martorell, M.M.; Mac Cormack, W.P.; Levin, G.J. Cold active transglutaminase from antarctic Penicillium chrysogenum: Partial purification, characterization and potential application in food technology. Biocatal. Agric. Biotechnol. 2020, 29, 101807. [Google Scholar] [CrossRef]
- Gil-Durán, C.; Ravanal, M.-C.; Ubilla, P.; Vaca, I.; Chávez, R. Heterologous expression, purification and characterization of a highly thermolabile endoxylanase from the Antarctic fungus Cladosporium sp. Fungal Biol. 2018, 122, 875–882. [Google Scholar] [CrossRef]
- Del-Cid, A.; Ubilla, P.; Ravanal, M.-C.M.-C.C.; Medina, E.; Vaca, I.; Levican, G.; Eyzaguirre, J.; Chavez, R.; Levicán, G.; Eyzaguirre, J.; et al. Cold-Active Xylanase Produced by Fungi Associated with Antarctic Marine Sponges. Appl. Biochem. Biotechnol. 2014, 172, 524–532. [Google Scholar] [CrossRef]
- Azhar, M. Cloning, Expression and Analysis of α-Amylase Gene from Psychrophilic Yeast Leucosporidium antarcticum PI12. Ph.D. Thesis, Universiti Teknologi Malaysia, Skudai, Malaysia, 2012. [Google Scholar]
- Song, C.; Liu, G.L.; Xu, J.L.; Chi, Z.M. Purification and characterization of extracellular β-galactosidase from the psychrotolerant yeast Guehomyces pullulans 17-1 isolated from sea sediment in Antarctica. Process Biochem. 2010, 45, 954–960. [Google Scholar] [CrossRef]
- Erich, S.; Kuschel, B.; Schwarz, T.; Ewert, J.; Böhmer, N.; Niehaus, F.; Eck, J.; Lutz-Wahl, S.; Stressler, T.; Fischer, L. Novel high-performance metagenome β-galactosidases for lactose hydrolysis in the dairy industry. J. Biotechnol. 2015, 210, 27–37. [Google Scholar] [CrossRef]
- Ugidos-Rodríguez, S.; Matallana-González, M.C.; Sánchez-Mata, M.C. Lactose malabsorption and intolerance: A review. Food Funct. 2018, 9, 4056–4068. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Gerday, C.; Feller, G.; Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar]
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Guerrand, D. Lipases industrial applications: Focus on food and agroindustries. OCL 2017, 24, D403. [Google Scholar] [CrossRef]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. Biomed. Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef]
- Kavitha, M. Cold active lipases—An update. Front. Life Sci. 2016, 9, 226–238. [Google Scholar] [CrossRef]
- Vakhlu, J.; Kour, A. Yeast lipases: Enzyme purification, biochemical properties and gene cloning. Electron. J. Biotechnol. 2006, 9, 717–3458. [Google Scholar] [CrossRef]
- Domínguez De María, P.; Carboni-Oerlemans, C.; Tuin, B.; Bargeman, G.; Van Der Meer, A.; Van Gemert, R. Biotechnological applications of Candida antarctica lipase A: State-of-the-art. J. Mol. Catal. B Enzym. 2005, 37, 36–46. [Google Scholar] [CrossRef]
- Szczesna-Antczak, M.; Kamińska, J.; Florczak, T.; Turkiewicz, M. Cold-active yeast lipases: Recent issues and future prospects. In Cold-Adapted Yeasts: Biodiversity, Adaptation Strategies and Biotechnological Significance; Springer: Berlin/Heidelberg, Germany, 2013; pp. 353–375. ISBN 9783642396816. [Google Scholar]
- Joseph, B.; Ramteke, P.W.; Thomas, G. Cold active microbial lipases: Some hot issues and recent developments. Biotechnol. Adv. 2008, 26, 457–470. [Google Scholar] [CrossRef]
- Martorell, M.M.; Ruberto, L.A.M.; Fernández, P.M.; Castellanos de Figueroa, L.I.; Mac Cormack, W.P. Bioprospection of cold-adapted yeasts with biotechnological potential from Antarctica. J. Basic Microbiol. 2017, 57, 504–516. [Google Scholar] [CrossRef]
- Cabrera, M.Á.; Blamey, J.M. Biotechnological applications of archaeal enzymes from extreme environments. Biol. Res. 2018, 51, 37. [Google Scholar] [CrossRef]
- Furhan, J. Adaptation, production, and biotechnological potential of cold-adapted proteases from psychrophiles and psychrotrophs: Recent overview. J. Genet. Eng. Biotechnol. 2020, 18, 36. [Google Scholar] [CrossRef]
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef]
- Sharma, K.M.; Kumar, R.; Panwar, S.; Kumar, A. Microbial alkaline proteases: Optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017, 15, 115–126. [Google Scholar] [CrossRef]
- Grossart, H.P.; Wurzbacher, C.; James, T.Y.; Kagami, M. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol. 2016, 19, 28–38. [Google Scholar] [CrossRef]
- Overy, D.P.; Rämä, T.; Oosterhuis, R.; Walker, A.K.; Pang, K.L. The neglected marine fungi, sensu stricto, and their isolation for natural products’ discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef]
- Vester, J.K.; Glaring, M.A.; Stougaard, P. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles 2015, 19, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.-H. Phylogenetic Identification and In Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.; Lloyd, K.; Wrighton, K. The bright side of microbial dark matter: Lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Ambrosino, L.; Tangherlini, M.; Colantuono, C.; Esposito, A.; Sangiovanni, M.; Miralto, M.; Sansone, C.; Chiusano, M.L. Bioinformatics for Marine Products: An Overview of Resources, Bottlenecks, and Perspectives. Mar. Drugs 2019, 17, 576. [Google Scholar] [CrossRef] [PubMed]
- Barone, R.; De Santi, C.; Palma Esposito, F.; Tedesco, P.; Galati, F.; Visone, M.; Di Scala, A.; De Pascale, D. Marine metagenomics, a valuable tool for enzymes and bioactive compounds discovery. Front. Mar. Sci. 2014, 1, 38. [Google Scholar] [CrossRef]
- Madhavan, A.; Sindhu, R.; Parameswaran, B.; Sukumaran, R.K.; Pandey, A. Metagenome Analysis: A Powerful Tool for Enzyme Bioprospecting. Appl. Biochem. Biotechnol. 2017, 183, 636–651. [Google Scholar] [CrossRef]
- Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and methods to access novel antibiotics from actinomycetes. Antibiotics 2018, 7, 44. [Google Scholar] [CrossRef]
- Zhang, M.M.; Qiao, Y.; Ang, E.L.; Zhao, H. Using natural products for drug discovery: The impact of the genomics era. Expert Opin. Drug Discov. 2017, 12, 475–487. [Google Scholar] [CrossRef]
- Skellam, E. Strategies for Engineering Natural Product Biosynthesis in Fungi. Trends Biotechnol. 2019, 37, 416–427. [Google Scholar] [CrossRef]
- Goyal, D.; Swaroop, S.; Pandey, J. Harnessing the Genetic Diversity and Metabolic Potential of Extremophilic Microorganisms through the Integration of Metagenomics and Single-Cell Genomics. In Extremophilic Microbes and Metabolites—Diversity, Bioprespecting and Biotechnological Applications; IntechOpen: London, UK, 2021. [Google Scholar]
- Milshteyn, A.; Schneider, J.S.; Brady, S.F. Mining the metabiome: Identifying novel natural products from microbial communities. Chem. Biol. 2014, 21, 1211–1223. [Google Scholar] [CrossRef]

| Fungal Taxa | Product | Bioactivity | Source | Ref. |
|---|---|---|---|---|
| Penicillium citrinum OUCMDZ4136 | 2,4-Dihydroxy-3,5,6-trimethylbenzoic acid; Citreorosein; Pinselin; Citrinin; Dihydrocitrinone; Pennicitrinone A; Quinolactacin A1 | Cytotoxic activities against MCF-7, A549, K562 cell lines | Antarctic krill Euphasia superba | [83] |
| Penicillium citreonigrum SP-6 | Diketopiperazine, phenols | Inhibitory activity against HCT116 cancer cell line | Marine sediment, Great Wall Station | [165] |
| Penicillium crustosum HDN153086 | Diketopiperazine | Cytotoxic activities against K562 cell line | Marine sediment, Pridz Bay | [61] |
| Penicillium crustosum PRB-2 | Penilactone A | NF-KB inhibitory activities of HCT-8, Bel-7402, BGC-823, A549 and A2780 tumor cell lines | Deep-sea sediment, Prydz Bay | [46] |
| Penicillium glabrum SF-7123 | Citromycetin derivative, neuchromenin; myxotrichin C, deoxyfunicone; | Anti-inflammatory; tyrosine phosphatase 1B inhibition | Marine sediment, Ross Sea | [50] |
| Penicillium granulatum MCCC 3A00475 | Spirograterpene A | Antiallergic effect on immunoglobulin E (IgE)-mediated rat mast RBL-2H3 cells | Deep-sea sediment, Prydz Bay | [52] |
| Penicillium sp. PR19N-1 | Chlorinated eremophilane sesquiterpenes, eremofortine C, eremophilane-type sesquiterpenes, eremophilane-type lactam | Cytotoxic activity against HL-60 and A549 cancer cell lines | Deep-sea sediment, Prydz Bay | [51,163] |
| Penicillium sp. S-1–18 | Butanolide A, guignarderemophilane F, xylarenone A | Butanolide: inhibitory activity against tyrosine phosphatase 1B; xylarenone A: antitumor activity against HeLa and HepG2 cells and growth-inhibitory effects against pathogenic microbes | Sea-bed sediment | [60,166] |
| Penicillium sp. UFMGCB 6034 and UFMGCB 6120 | Aromatic compounds | Antifungal and trypanocidal activities | Macroalgae: Palmaria decipiens and Monostroma hariotii | [66] |
| Pseudogymnoascus sp. | Pseudogymnoascin A, B, C, 3-nitroasterric acid; Geomycins B, C | Antibacterial and antifungal activities | Sponge genus Hymeniacidon | [167,168] |
| Trichoderma asperellum | Asperelines A-F, peptaibols | Not assayed | Marine sediment, Penguin Island | [56,169] |
| Enzyme | Reaction | Fungi | Source of (Isolate) Sample | Applications/Potential Uses | Ref. |
|---|---|---|---|---|---|
| Carragenase (EC 3.2.1.83) | Hydrolysis of 1,4-β-linkages between galactose 4-sulfate and 3,6-anhydro-galactose to produce kappa-carrageenans | Pseudogymnoascus sp. UFMGCB 10054 | Macroalga: Iridaea cordata, | Biomedical field, textile industry, bioethanol production, and detergent additive | [69] |
| Cellulase (EC 3.2.1.4) | Cellulose hydrolysis into glucose | Cystofilobasidium infirmominiatum 071209-E8-C1-liblev; Metschnikowia australis, Rhodotorula glacialis; Candida spencermartinsiae, Leucosporidiella creatinivora, Leucosporidium scottii | Marine sponge: Tedania; marine sediments; seawater | Food industry, animal feed, beer and wine, textile and laundry, pulp and paper industry, agriculture, biofuel, pharmaceutical industries, and waste management | [45,80] |
| Chitinase (EC 3.2.1.14) | Cleavage of glycosidic linkages in chitin and chitodextrins generating chitooligosaccharides | Lecanicillium muscarium CCFEE-5003; Glaciozyma antarctica PI12 | Shrimp wastes; seawater | Cosmetic, pharmaceutic fields, fermentation research, and biomedicine | [45,198,199,200,201,202] |
| Endo-β-1,3(4)-glucanase (EC 3.2.1.6) | Endohydrolysis of (1→3)- or (1→4)-linkages in β-D-glucans | Glaciozyma antarctica PI12 | Seawater | Brewing and animal, feed-stuff industry, biofuel production, and pharmaceuticals | [202,203,204] |
| Esterase (EC 3.1.1.1) | Hydrolyis of short acyl-chain soluble esters | Cryptococcus victoriae, Metschnikowia australis, Rhodotorula glacialis, Leucosporidium scottii, Leucosporidiella creatinivora; Glaciozyma antarctica | Marine sediments; seawater, sea ice | Paper bleaching, bioremediation, degradation, and removal of xenobiotics and toxic compounds | [45,205] |
| Invertase (EC 3.2.1.26) | Hydrolysis of the terminal non-reducing β-fructofuranoside residue in sucrose, raffinose and related β-D-fructofuranosides | Glaciozyma antarctica 17 (formerly Leucosporidium antarcticum) | Seawater | Beverage, confectionary, bakery, invert sugar, high fructose syrup, artificial honey, calf feed, food for honeybees | [38] |
| Laccase (EC 1.10.3.2) | Oxidation of phenolic compound like lignin | Cadophora malorum A2B, Cadophora malorum AS2A, Cadophora luteo-olivacea P1 | Marine sediments | Biosensors, microfuel and bioelectrocatalysis, food, pharmaceutic, cosmetic, pulp and paper, textile industries, and bioremediation | [206] |
| Lignin peroxidase (EC 1.11.1.14) | Oxidative breakdown of lignin | Cadophora malorum M7, Cadophora sp. OB-4B | Marine sediments | Pulp and paper, cosmetics (treatment of hyperpigmentation, and skin-lightening through melanin oxidation), textile, bioremediation (degradation of azo, heterocyclic, reactive, and polymeric dyes, xenobiotic, and pesticides), and bioethanol production | [206] |
| Lipase (EC 3.1.1.3) | Hydrolysis of long-chain triacylglycerol substances with the formation of an alcohol and a carboxylic acid | Leucosporidium scottii L117, Metschnikowia sp. CRM1589; Mrakia blollopis SK-4; Cystofilobasidium infirmominiatum 071209-E8-C1-IIa-lev and isolate 131209-E2A-C1-II-lev; Metschnikowia australis 131209-E3-C1-(GPY)-lev and isolate 131209-E2A-C4-II-lev; Rhodotorula pinicola 071209-E4-C9-lev; Candida zeylanoides, Cryptococcus victoriae, Leucosporidiella creatinivora, Leucosporidium scottii, Candida sake, Candida spencermartinsiae | Marine sediments; Algal mat in sediment; marine sponges: Tedania, Hymeniacidon, Dendrilla; Seawater | Food, beverage, detergent, biofuel production, animal feed, textiles, leather, paper processing, and cosmetic industry | [45,47,48,207,208,209] |
| L-asparaginase (EC 3.5.1.1) | Degradation of asparagine into ammonia and aspartate | Cosmospora sp 0B4B, Cosmospora sp 0B1B, Cosmospora sp 0B2, Geomyces sp. S2B | Marine sediments | Food industry and medical applications as anti-cancer, antimicrobial, infectious diseases, autoimmune diseases | [206] |
| Pectinase (EC 3.2.1.15) | Hydrolisis of polysaccharides to produce pectate and other galacturonans | Geomyces sp. strain F09-T3-2, Pseudogymnoascus sp., Cladosporium sp. F09-T12-1, Cryptococcus victoriae, Leucosporidiella muscorum, Metschnikowia australis, Rhodotorula glacialis; Leucosporidiella creatinivora, Leucosporidium scottii | Marine sponges; marine sediments; Seawater | Food and textile industry, coffee and tea fermentation, wine processing, oil extraction, vegetable and fruit processing industry for juice clarification, color, and yield enhancer. Applications in paper and pulp making, recycling of wastepaper, pretreatment of pectic wastewaters, and retting of plant fibers | [45,82,210] |
| Phytase (EC 3.1.3.26) | Hydrolysis of phytate to produce phosphorylated myo-inositol derivatives | Rhodotorula mucilaginosa JMUY14 | Deep-sea sediments | Food and feed industry, pharmaceutical use as neuro protective agents, anti-inflammatory, antioxidant and anti-cancer agents | [211] |
| Protease (EC 3.4) | Cleavage of peptide bonds | Rhodotorula mucilaginosa L7; Pseudogymnoascus sp. CRM1533, Leucosporidiella muscorum; Leucosporidiella sp. 131209-E2A-C3-II-lev, Leucosporidiella creatinivora 071209-E8-C4-II-lev; Rhodotorula glacialis; Leucosporidiella creatinivora, Leucosporidium scottii | Marine macroalgae; marine sediments; marine sponges: Tedania, Hymeniacidon; Seawater | Food, feed, pharmacology (anticancer and antihemolytic activity) cosmetic (keratin-based preparation) industries, cleaning processes (e. g. detergent additive), waste management | [45,47,48,212,213] |
| Protease (Subtilase) (EC 3.4.21) | Cleavage of peptide bonds | Glaciozyma antarctica 17 (formerly Leucosporidium antarcticum) | Sub-glacial waters (depth of 200 m) | Food and beverage industries | [214,215] |
| Transglutami-nase (EC 2.3.2.13) | Acyl transfer reaction between gamma-carboxyamide groups of glutamine residues in proteins and various primary amines | Penicillium chrysogenum | Marine macroalga Gigartinas kosttbergii | Food, pharmaceutical, leather, textile, biotechnology industry, biomedical research | [216] |
| Xylanase (EC 3.2.1.8) | Hydrolysis of the main chain of xylan to oligosaccharides, which in turn are degraded to xylose | Cladosporium sp.; Penicillium sp. E2B Penicillium sp. N5, Penicillium sp. E2-1 | Marine sponge; marine sediments | Food (bread making), feed, paper and pulp industries, and also used to increase the sugar recovery from agricultural residues for biofuel production | [206,217,218] |
| α-amylase (EC 3.2.1.1) | Cleavage of α-1,4-glycosidic linkages within starch molecules, which generate smaller polymers of glucose units | Glaciozyma antarctica PII12 (formerly Leucosporidium antarcticum); Cystofilobasidium infirmominiatum 071209-E8-C1-IIa-lev, 131209-E2A-C1-II-lev, 131209-E2A-C5-II-lev and isolate 071209-E8-C1-IIb-lev; Metschnikowia australis 071209-E8-C3-II-lev and isolate 071209-E8-C1-II-lev; Leucosporidiella sp. 131209-E2A-C3-II-lev | Seawater; marine sponges: Tedania, Hymeniacidon | Pharmaceutical and chemical industry; employed as additives in processed food, in detergents for cold washing, in waste-water treatment, in bioremediation in cold climates, and in molecular biology protocols | [80,202,219] |
| β-agarase (EC 3.2.1.81) | Hydrolysis of beta-(1–>4) linkages of agarose to produce oligosaccharides | Penicillium sp., Cladosporium sp. 2, Penicillium sp., Pseudogymnoascus sp. UFMGCB 10054, Doratomyces sp. | Macroalgae: Ascoseira mirabilis, Georgiella confluens, Iridaea cordata, Palmaria decipiens | Food, cosmetic, medical industries, and as a tool enzyme for biological, physiological, and cytological studies | [69] |
| β-galactosidase (EC 3.2.1.23) | Hydrolysis of lactose into its constituent monosaccharides | Tausonia pullulans 17-1 (formerly Guehomyces pullulans) | Marine sediments | Food, biofuel, and agricultural industries; surfactant production | [220] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varrella, S.; Barone, G.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Corinaldesi, C. Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi. J. Fungi 2021, 7, 391. https://doi.org/10.3390/jof7050391
Varrella S, Barone G, Tangherlini M, Rastelli E, Dell’Anno A, Corinaldesi C. Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi. Journal of Fungi. 2021; 7(5):391. https://doi.org/10.3390/jof7050391
Chicago/Turabian StyleVarrella, Stefano, Giulio Barone, Michael Tangherlini, Eugenio Rastelli, Antonio Dell’Anno, and Cinzia Corinaldesi. 2021. "Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi" Journal of Fungi 7, no. 5: 391. https://doi.org/10.3390/jof7050391
APA StyleVarrella, S., Barone, G., Tangherlini, M., Rastelli, E., Dell’Anno, A., & Corinaldesi, C. (2021). Diversity, Ecological Role and Biotechnological Potential of Antarctic Marine Fungi. Journal of Fungi, 7(5), 391. https://doi.org/10.3390/jof7050391







