Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of Our Analysis Idea
2.2. Data Download
2.3. Orthologous Groups and Separation
2.4. Transmembrane Domain (TM) Prediction
2.5. Horizontal Gene Transfer (HGT) Prediction
2.6. Annotation Enrichment
2.7. Phylogram and Heatmap Figure Construction
2.8. Data and Code Availability
3. Results
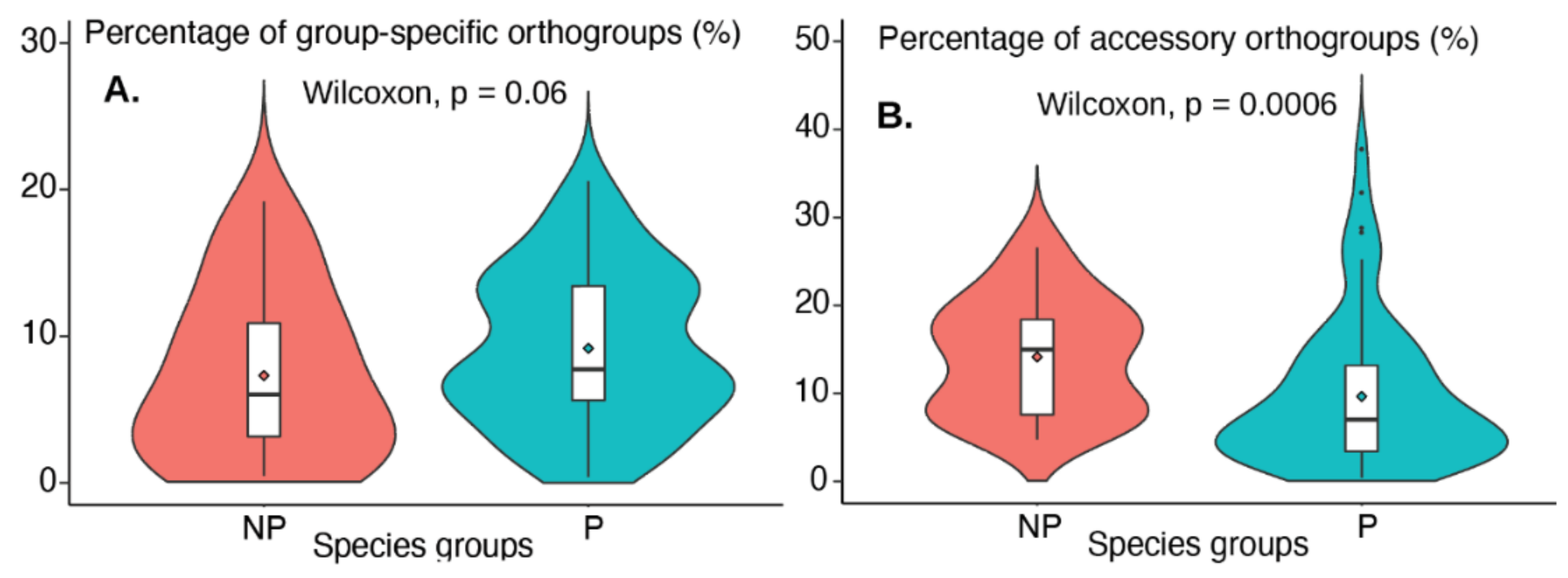
3.1. Orthogroup Categorization Shows That ~20% Orthogroups Are Group-Specific and Accessory
3.2. Species Phylogeny Reveals That Phytopathogenicity Is Not Phylogenetically Determined
3.3. Group-Specific Orthogroups Have More Enriched Functional Terms than Accessory Orthogroups
3.4. Secreted Proteins with Signal Peptides Show Higher Significance in Group-Specific Orthogroups than in Accessory Orthogroups
3.5. HGTs Show Higher Significance and Higher Occurrence in Group-Specific Orthogroups than in Accessory Orthogroups
3.6. Phytopathogenic Fungi Have More Enriched Functional Terms in Group-Specific Orthogroups than Accessory Orthogroups
3.7. More Phytopathogenic Fungi than Non-Phytopathogenic Fungi Are Found to Have a Larger Number of Enriched Functional Terms in Group-Specific Orthogroups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blackwell, M. The Fungi: 1, 2, 3 … 5.1 Million Species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Lutzoni, F.; Kauff, F.; Cox, C.J.; McLaughlin, D.; Celio, G.; Dentinger, C.; Padamsee, M.; Hibbett, D.; James, T.Y.; Baloch, E.; et al. Assembling the Fungal Tree of Life: Progress, Classification, and Evolution of Subcellular Traits. Am. J. Bot. 2004, 91, 1446–1480. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, G.C.; Bisby, G.R.; Kirk, P.M.; Cannon, P.F.; David, J.C.; Stalpers, J.A. Ainsworth and Bisby’s Dictionary of the Fungi, 10th ed.; CABI Bioscience, Ed.; CABI: Wallingford, UK, 2008. [Google Scholar]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.M.; Chandra Nayak, S. Friends or Foes? Emerging Insights from Fungal Interactions with Plants. FEMS Microbiol. Rev. 2016, 40, 182–207. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.; Yu, J.H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Birney, E.; Biswas, M.; Bucher, P.; Cerutti, L.; Corpet, F.; Croning, M.D.R.; et al. The InterPro Database, an Integrated Documentation Resource for Protein Families, Domains and Functional Sites. Nucleic Acids Res. 2001, 29, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG Database: An Updated Version Includes Eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Nielsen, H.; Engelbrecht, J.; Brunak, S.; Von Heijne, G. Identification of Prokaryotic and Eukaryotic Signal Peptides and Prediction of Their Cleavage Sites. Protein Eng. 1997, 10. [Google Scholar] [CrossRef]
- Nguyen, M.; Ekstrom, A.; Li, X.; Yin, Y. HGT-Finder: A New Tool for Horizontal Gene Transfer Finding and Application to Aspergillus Genomes. Toxins 2015, 7, 4035–4053. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Almeida Nogueira, M.F.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M.; et al. Fungal Model Systems and the Elucidation of Pathogenicity Determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative Analysis of Fungal Genomes Reveals Different Plant Cell Wall Degrading Capacity in Fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, X.; Pan, Z.; Kale, S.D.; Song, Y.; King, H.; Zhang, Q.; Presley, C.; Deng, X.; Wei, C.I.; et al. Comparative Genome Analyses Reveal Sequence Features Reflecting Distinct Modes of Host-Adaptation between Dicot and Monocot Powdery Mildew 06 Biological Sciences 0604 Genetics. BMC Genom. 2018, 19, 705. [Google Scholar] [CrossRef]
- Le Marquer, M.; San Clemente, H.; Roux, C.; Savelli, B.; Frei Dit Frey, N. Identification of New Signalling Peptides through a Genome-Wide Survey of 250 Fungal Secretomes. BMC Genom. 2019, 20, 64. [Google Scholar]
- Hogenhout, S.A.; Van Der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging Concepts in Effector Biology of Plant-Associated Organisms. Mol. Plant-Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20. [Google Scholar] [CrossRef]
- Entwistle, S.; Li, X.; Yin, Y. Orphan Genes Shared by Pathogenic Genomes Are More Associated with Bacterial Pathogenicity. mSystems 2019, 4. [Google Scholar] [CrossRef]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Pan-Genome Analyses of Model Fungal Species. Microb. Genom. 2019, 5, e000243. [Google Scholar] [CrossRef]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Pangloss: A Tool for Pan-Genome Analysis of Microbial Eukaryotes. Genes 2019, 10, 521. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous Identification of Fungi: Where Do We Stand and How Accurate and Precise Is Fungal DNA Barcoding? IMA Fungus 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm Portal: Gearing up for 1000 Fungal Genomes. Nucleic Acids Res. 2014, 42, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Yin, Y.; Fischer, D. On the Origin of Microbial ORFans: Quantifying the Strength of the Evidence for Viral Lateral Transfer. BMC Evol. Biol. 2006, 6, 63. [Google Scholar] [CrossRef][Green Version]
- Ekman, D.; Elofsson, A. Identifying and Quantifying Orphan Protein Sequences in Fungi. J. Mol. Biol. 2010, 396, 396–405. [Google Scholar] [CrossRef]
- Carvunis, A.R.; Rolland, T.; Wapinski, I.; Calderwood, M.A.; Yildirim, M.A.; Simonis, N.; Charloteaux, B.; Hidalgo, C.A.; Barbette, J.; Santhanam, B.; et al. Proto-Genes and de Novo Gene Birth. Nature 2012, 487, 370–374. [Google Scholar] [CrossRef]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a Fully Resolved Fungal Tree of Life. Annu. Rev. Microbiol. 2020, 74, 291–313. [Google Scholar] [CrossRef]
- Haridas, S.; Albert, R.; Binder, M.; Bloem, J.; LaButti, K.; Salamov, A.; Andreopoulos, B.; Baker, S.E.; Barry, K.; Bills, G.; et al. 101 Dothideomycetes Genomes: A Test Case for Predicting Lifestyles and Emergence of Pathogens. Stud. Mycol. 2020, 96, 141–153. [Google Scholar] [CrossRef]
- Gardiner, D.M.; McDonald, M.C.; Covarelli, L.; Solomon, P.S.; Rusu, A.G.; Marshall, M.; Kazan, K.; Chakraborty, S.; McDonald, B.A.; Manners, J.M. Comparative Pathogenomics Reveals Horizontally Acquired Novel Virulence Genes in Fungi Infecting Cereal Hosts. PLoS Pathog. 2012, 8, e1002952. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; Christiansen, F.B.; Hansen, T.T.; Dutheil, J.Y.; Schierup, M.H. Fusion of Two Divergent Fungal Individuals Led to the Recent Emergence of a Unique Widespread Pathogen Species. Proc. Natl. Acad. Sci. USA 2012, 109, 10954–10959. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; Bataillon, T.; Dutheil, J.Y.; Hansen, T.T.; Li, R.; Zala, M.; McDonald, B.A.; Wang, J.; Schierup, M.H. The Making of a New Pathogen: Insights from Comparative Population Genomics of the Domesticated Wheat Pathogen Mycosphaerella Graminicola and Its Wild Sister Species. Genome Res. 2011, 21, 2157–2166. [Google Scholar] [CrossRef]
- Moolhuijzen, P.M.; See, P.T.; Oliver, R.P.; Moffat, C.S. Genomic Distribution of a Novel Pyrenophora Tritici-Repentis ToxA Insertion Element. PLoS ONE 2018, 13, e0206586. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q.; Li, C.; Bi, Y.; Fu, X.; Wang, R. Novel Haplotypes and Networks of AVR-Pik Alleles in Magnaporthe Oryzae. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef]
- Menardo, F.; Praz, C.R.; Wicker, T.; Keller, B. Rapid Turnover of Effectors in Grass Powdery Mildew (Blumeria Graminis). BMC Evol. Biol. 2017, 17, 223. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Tiwari, P.; Bae, H. Horizontal Gene Transfer and Endophytes: An Implication for the Acquisition of Novel Traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef]
- Coelho, M.A.; Gonçalves, C.; Sampaio, J.P.; Gonçalves, P. Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Qiu, H.; Cai, G.; Luo, J.; Bhattacharya, D.; Zhang, N. Extensive Horizontal Gene Transfers between Plant Pathogenic Fungi. BMC Biol. 2016, 14, 41. [Google Scholar] [CrossRef]
- Whittaker, C.A.; Hynes, R.O. Distribution and Evolution of von Willebrand/Integrin A Domains: Widely Dispersed Domains with Roles in Cell Adhesion and Elsewhere. Mol. Biol. Cell 2002, 13, 3369–3387. [Google Scholar] [CrossRef]
- Yang, F.; Li, W.; Jørgensen, H.J.L. Transcriptional Reprogramming of Wheat and the Hemibiotrophic Pathogen Septoria Tritici during Two Phases of the Compatible Interaction. PLoS ONE 2013, 8, e81606. [Google Scholar] [CrossRef]
- Yang, F.; Li, W.; Derbyshire, M.; Larsen, M.R.; Rudd, J.J.; Palmisano, G. Unraveling Incompatibility between Wheat and the Fungal Pathogen Zymoseptoria Tritici through Apoplastic Proteomics. BMC Genom. 2015, 16, 362. [Google Scholar] [CrossRef]
- Laohavisit, A.; Mortimer, J.C.; Demidchik, V.; Coxon, K.M.; Stancombe, M.A.; Macpherson, N.; Brownlee, C.; Hofmann, A.; Webb, A.A.R.; Miedema, H.; et al. Zea Mays Annexins Modulate Cytosolic Free Ca2+ and Generate a Ca2+-Permeable Conductance. Plant Cell 2009, 21, 479–493. [Google Scholar] [CrossRef]
- Skowyra, D.; Craig, K.L.; Tyers, M.; Elledge, S.J.; Harper, J.W. F-Box Proteins Are Receptors That Recruit Phosphorylated Substrates to the SCF Ubiquitin-Ligase Complex. Cell 1997, 91, 209–219. [Google Scholar] [CrossRef]
- Moriya, S.; Terakami, S.; Okada, K.; Shimizu, T.; Adachi, Y.; Katayose, Y.; Fujisawa, H.; Wu, J.; Kanamori, H.; Yamamoto, T.; et al. Identification of Candidate Genes Responsible for the Susceptibility of Apple (Malus × Domestica Borkh.) to Alternaria Blotch. BMC Plant Biol. 2019, 19, 132. [Google Scholar] [CrossRef]
- Price, C.T.D.; Abu Kwaik, Y. Exploitation of Host Polyubiquitination Machinery through Molecular Mimicry by Eukaryotic-like Bacterial F-Box Effectors. Front. Microbiol. 2010, 1, 122. [Google Scholar] [CrossRef]
- Pedersen, C.; van Themaat, E.V.L.; McGuffin, L.J.; Abbott, J.C.; Burgis, T.A.; Barton, G.; Bindschedler, L.V.; Lu, X.; Maekawa, T.; Weßling, R.; et al. Structure and Evolution of Barley Powdery Mildew Effector Candidates. BMC Genom. 2012, 13, 694. [Google Scholar] [CrossRef]
- Olombrada, M.; Lázaro-Gorines, R.; López-Rodríguez, J.C.; Martínez-Del-Pozo, Á.; Oñaderra, M.; Maestro-López, M.; Lacadena, J.; Gavilanes, J.G.; García-Ortega, L. Fungal Ribotoxins: A Review of Potential Biotechnological Applications. Toxins 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Jin, Q.; Xu, J.R.; Liu, H. Identification of a Fungi-Specific Lineage of Protein Kinases Closely Related to Tyrosine Kinases. PLoS ONE 2014, 9, e89813. [Google Scholar] [CrossRef] [PubMed]
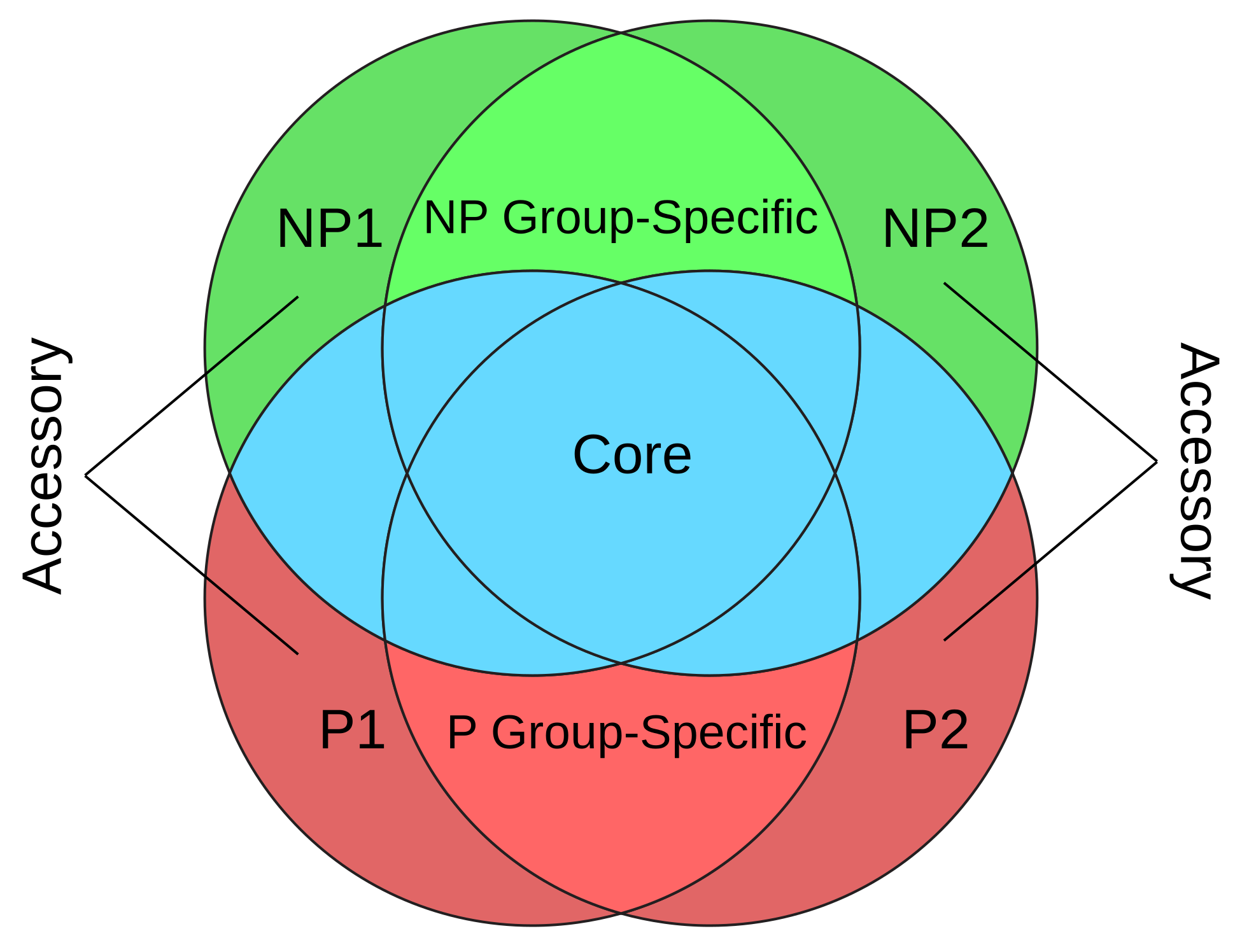
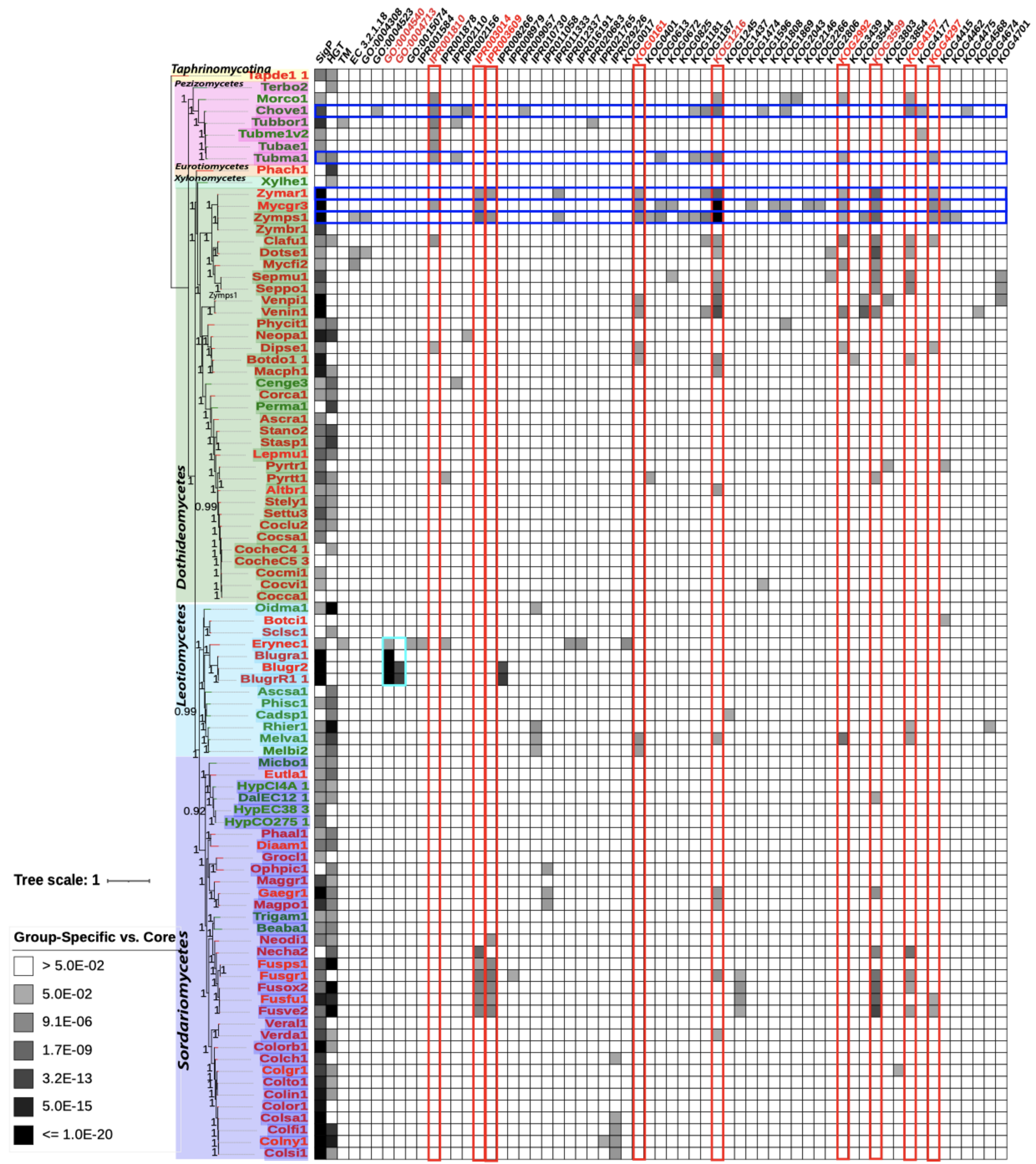

| Enriched Terms | Description | # of Species (% of Total 92) | # of P Species (% of Total 68) | # of NP Species (% of Total 24) |
|---|---|---|---|---|
| Signal Pep | Signal peptide | 79 (85.9%) | 60 (88.2%) * | 19 (79.2%) |
| HGT | horizontal gene transfer | 55 (59.8%) | 38 (55.9%) | 17 (70.8%) |
| KOG1216 | Von Willebrand factor and related coagulation proteins | 20 (21.7%) | 16 (23.5%) * | 4 (16.7%) |
| KOG3599 | Ca2+-modulated nonselective cation channel polycystin | 17 (18.5%) | 16 (23.5%) * | 1 (4.2%) |
| KOG4157 | beta-1,6-N-acetylglucosaminyltransferase, contains WSC domain | 13 (14.1%) | 9 (13.2%) | 4 (16.7%) |
| KOG2992 | Nucleolar GTPase/ATPase p130 | 10 (10.9%) | 7 (10.3%) | 3 (12.5%) |
| IPR001810 | Cyclin-like F-box | 9 (9.8%) | 3 (4.4%) | 6 (25.0%) |
| IPR003014 | N/apple PAN | 9 (9.8%) | 9 (13.2%) * | 0 |
| KOG0161 | Myosin class II heavy chain | 9 (9.8%) | 7 (10.3%) * | 2 (8.3%) |
| IPR003609 | Apple-like | 8 (8.7%) | 8 (11.8%) * | 0 |
| KOG4297 | C-type lectin | 8 (8.7%) | 7 (10.3%) * | 1 (4.2%) |
| JGI ID * | Species | Species Type | Taxonomy Class | # of Enriched Terms |
|---|---|---|---|---|
| Zymps1 | Zymoseptoria pseudotritici | P | Dothideomycetes | 21 |
| Mycgr3 | Mycosphaerella graminicola | P | Dothideomycetes | 16 |
| Chove1 | Choiromyces venosus | NP | Pezizomycetes | 14 |
| Tubma1 | Tuber magnatum | NP | Pezizomycetes | 10 |
| Zymar1 | Zymoseptoria ardabiliae | P | Dothideomycetes | 10 |
| Clafu1 | Cladosporium fulvum | P | Dothideomycetes | 9 |
| Erynec1 | Erysiphe necator | P | Leotiomycetes | 9 |
| Fusgr1 | Fusarium graminearum | P | Sordariomycetes | 8 |
| Fusve2 | Fusarium verticillioides | P | Sordariomycetes | 8 |
| Venin1 | Venturia inaequalis | P | Dothideomycetes | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterson, D.; Li, T.; Calvo, A.M.; Yin, Y. Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity. J. Fungi 2021, 7, 337. https://doi.org/10.3390/jof7050337
Peterson D, Li T, Calvo AM, Yin Y. Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity. Journal of Fungi. 2021; 7(5):337. https://doi.org/10.3390/jof7050337
Chicago/Turabian StylePeterson, Daniel, Tang Li, Ana M. Calvo, and Yanbin Yin. 2021. "Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity" Journal of Fungi 7, no. 5: 337. https://doi.org/10.3390/jof7050337
APA StylePeterson, D., Li, T., Calvo, A. M., & Yin, Y. (2021). Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Phytopathogenicity. Journal of Fungi, 7(5), 337. https://doi.org/10.3390/jof7050337






