Integrating Different Lines of Evidence to Establish a Novel Ascomycete Genus and Family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation and Identification
2.2. DNA Extraction, PCR Amplification and DNA Sequencing
2.3. Phylogenetic Analysis
2.4. Fossil Calibration and Divergence Time Estimates
3. Results
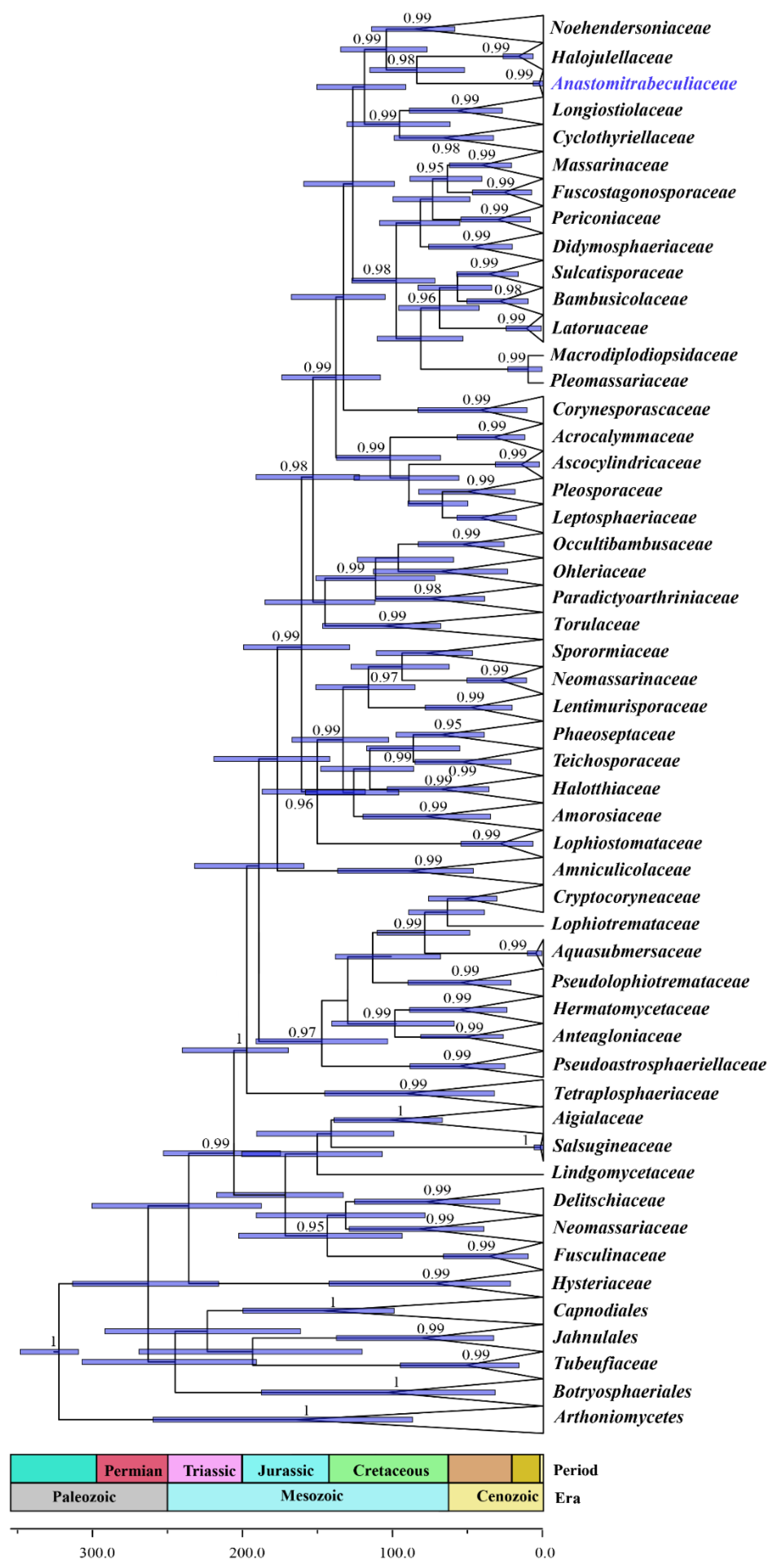
3.1. Phylogenetic Analyses
3.2. Fossil Calibration and Divergence Time Estimates
3.3. Taxonomy
- Anastomitrabeculia Bhunjun, Phukhams. and K.D. Hyde, gen. nov.
- Index Fungorum number: IF556560, Facesoffungi number: FoF 09522.
- Etymology: Referring to the trabeculate pseudoparaphyses anastomosing between the asci and at the apex.
- Anastomitrabeculia didymospora Bhunjun, Phukhams and K.D. Hyde, sp. nov.
- Index Fungorum number: IF556559; Facesoffungi number: FoF 09523 Figure 3.
- Etymology: Referring to the didymosporous ascospores.
- Holotype–MFLU 20-0694.
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimot, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Hongsanan, S.; Ryberg, M.; Ariyawansa, H.A.; Chomnunti, P.; Bahkali, A.H.; Hyde, K.D. The evolution of Massarineae with Longipedicellataceae fam. nov. Mycosphere 2016, 7, 1713–1731. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Checa, J.; Blanco, M.N.; Olariaga, I.; Tello, S.; Voglmayr, H. A preliminary account of the Cucurbitariaceae. Stud. Mycol. 2018, 90, 71–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.G.; Lin, C.G.; Liu, J.K.; Samarakoon, M.C.; Hongsanan, S.; Bhat, D.J.; Hyde, K.D.; McKenzie, E.H.; Jumpathong, J. Lentimurisporaceae, a new Pleosporalean family with divergence times estimates. Cryptogam. Mycol. 2018, 39, 259–283. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; Mckenzie, E.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Pem, D.; Bhat, D.J.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Ramesh, C. Loculoascomycetes from India. Front. Fungal Divers. India 2003, 457–479. [Google Scholar]
- Kruys, Å.; Eriksson, O.E.; Wedin, M. Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol. Res. 2006, 110, 527–536. [Google Scholar] [CrossRef]
- Zhang, Y.; Schoch, C.L.; Fournier, J.; Crous, P.W.; De Gruyter, J.; Woudenberg, J.H.C.; Hirayama, K.; Tanaka, K.; Pointing, S.B.; Spatafora, J.W.; et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009, 64, 85–102. [Google Scholar] [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jeewon, R.; Phillips, A.J.; Maharachchikumbura, S.S.; Ryberg, M.; Liu, Z.Y.; Zhao, Q. Ranking higher taxa using divergence times: A case study in Dothideomycetes. Fungal Divers. 2017, 84, 75–99. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Wanasinghe, D.N.; Papizadeh, M.; Goonasekara, I.D.; Camporesi, E.; Bhat, D.J.; Mckenzie, E.H.C.; Phillips, A.J.L.; Diederich, P.; et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 2016, 77, 1–316. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Rajeshkumar, K.C.; Hawksworth, D.L.; Madrid, H.; Kirk, P.M.; Braun, U.; Singh, R.V.; Crous, P.W.; Kukwa, M.; et al. Notes for genera: Ascomycota. Fungal Divers. 2017, 86, 1–594. [Google Scholar] [CrossRef]
- Gratani, L.; Crescente, M.F.; Varone, L.; Fabrini, G.; Digiulio, E. Growth pattern and photosynthetic activity of different bamboo species growing in the Botanical Garden of Rome. Flora-Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 77–84. [Google Scholar] [CrossRef]
- Kelchner, S.A.; Group, B.P. Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 2013, 67, 404–413. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal Divers. 2016, 82, 1–105. [Google Scholar] [CrossRef]
- Kirschner, R.; Yang, Z.L.; Zhao, Q.; Feng, B. Ovipoculum album, a new anamorph with gelatinous cupulate bulbilliferous conidiomata from China and with affinities to the Auriculariales (Basidiomycota). Fungal Divers. 2009, 43, 55–65. [Google Scholar] [CrossRef]
- Hyde, K.D.; Zhou, D.; McKenzie, E.; Ho, W.; Dalisay, T. Vertical distribution of saprobic fungi on bamboo culms. Fungal Divers. 2002, 11, 109–118. [Google Scholar]
- Hyde, K.D.; Zhou, D.; Dalisay, T. Bambusicolous fungi: A review. Fungal Divers. 2002, 9, 1–14. [Google Scholar]
- Prieto, M.; Wedin, M. Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS ONE 2013, 8, e65576. [Google Scholar] [CrossRef] [PubMed]
- Hongsanan, S.; Sánchez-Ramírez, S.; Crous, P.W.; Ariyawansa, H.A.; Zhao, R.L.; Hyde, K.D. The evolution of fungal epiphytes. Mycosphere 2016, 7, 1690–1712. [Google Scholar] [CrossRef]
- Hongsanan, S.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Samarakoon, M.C.; Jeewon, R.; Zhao, Q.; Al-Sadi, A.M.; Bahkali, A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017, 84, 25–41. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Hyde, K.D.; Promputtha, I.; Hongsanan, S.; Ariyawansa, H.A.; Maharachchikumbura, S.S.N.; Daranagama, D.A.; Stadler, M.; Mapook, A. Evolution of Xylariomycetidae (Ascomycota: Sordariomycetes). Mycosphere 2016, 7, 1746–1761. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Hyde, K.D.; Hongsanan, S.; McKenzie, E.H.; Ariyawansa, H.A.; Promputtha, I.; Zeng, X.Y.; Tian, Q.; Liu, J.K. Divergence time calibrations for ancient lineages of Ascomycota classification based on a modern review of estimations. Fungal Divers. 2019, 96, 285–346. [Google Scholar] [CrossRef]
- Hyde, K.D.; Maharachchikumbura, S.S.; Hongsanan, S.; Samarakoon, M.C.; Lücking, R.; Pem, D.; Harishchandra, D.; Jeewon, R.; Zhao, R.L.; Xu, J.C.; et al. The ranking of fungi: A tribute to David L. Hawksworth on his 70th birthday. Fungal Divers. 2017, 84, 1–23. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The faces of fungi database: Fungal names linked with morphology, molecular and human attributes. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences, evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Pinnoi, A.; Jeewon, R.; Sakayaroj, J.; Hyde, K.D.; Jones, E.B.G. Berkleasmium crunisia sp. nov. and its phylogenetic affinities to the Pleosporales based on 18S and 28S rDNA sequence analyses. Mycologia 2007, 99, 378–384. [Google Scholar] [CrossRef]
- Beimforde, C.; Feldberg, K.; Nylinder, S.; Rikkinen, J.; Tuovila, H.; Dörfelt, H.; Gube, M.; Jackson, D.J.; Reitner, J.; Seyfullah, L.J.; et al. Estimating the Phanerozoic history of the Ascomycota lineages: Combining fossil and molecular data. Mol. Phylogenet. Evol. 2014, 78, 386–398. [Google Scholar] [CrossRef]
- Pratibha, J.; Prabhugaonkar, A.; Hyde, K.D.; Bhat, D.J. Phylogenetic placement of Bahusandhika, Cancellidium and Pseudoepicoccum (asexual Ascomycota). Phytotaxa 2014, 176, 68–80. [Google Scholar] [CrossRef][Green Version]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C.; Boonmee, S.; Camporesi, E.; Hashimoto, A.; et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony, Version 4.0 b10; Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 August 2020).
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Gernhard, T.; Hartmann, K.; Steel, M. Stochastic properties of generalised Yule models, with biodiversity applications. J. Math. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Gueidan, C.; Ruibal, C.; De Hoog, G.S.; Schneider, H. Rock-inhabiting fungi originated during periods of dry climate in the late Devonian and middle Triassic. Fungal Biol. 2011, 115, 987–996. [Google Scholar] [CrossRef]
- Pérez-Ortega, S.; Garrido-Benavent, I.; Grube, M.; Olmo, R.; de los Ríos, A. Hidden diversity of marine borderline lichens and a new order of fungi: Collemopsidiales (Dothideomyceta). Fungal Divers. 2016, 80, 285–300. [Google Scholar] [CrossRef]
- Mindell, R.A.; Stockey, R.A.; Beard, G.; Currah, R.S. Margaretbarromyces dictyosporus gen. sp. nov.: A permineralized corticolous ascomycete from the Eocene of Vancouver Island, British Columbia. Mycol. Res. 2007, 111, 680–684. [Google Scholar] [CrossRef]
- Berbee, M.; Taylor, J.W. Dating the molecular clock in fungi-how close are we? Fungal Biol. Rev. 2010, 24, 1–16. [Google Scholar] [CrossRef]
- Taylor, T.N.; Krings, M.; Taylor, E.L. Ascomycota. In Fossil fungi; Academic Press: San Diego, CA, USA, 2015. [Google Scholar]
- Cohen, K.M.; Finney, S.C.; Gibbard, P.L.; Fan, J.X. The ICS International Chronostratigraphic Chart. Episodes 2013, 36, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Willis, K.J. State of the World’s Fungi 2018; Royal Botanic Gardens, Kew: London, UK, 2018. [Google Scholar]
- Hyde, K.D.; Jeewon, R.; Chen, Y.J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.B.; Lin, C.G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020, 103, 219–271. [Google Scholar] [CrossRef]
- Giraldo, A.; Crous, P.W.; Schumacher, R.K.; Cheewangkoon, R.; Ghobad-Nejhad, M.; Langer, E. The Genera of Fungi—G3: Aleurocystis, Blastacervulus, Clypeophysalospora, Licrostroma, Neohendersonia and Spumatoria. Mycol. Prog. 2017, 16, 325–348. [Google Scholar] [CrossRef]
- Liew, E.C.Y.; Aptroot, A.; Hyde, K.D. Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol. Phylogenet. Evol. 2000, 16, 392–402. [Google Scholar] [CrossRef]
- Phookamsak, R.; Norphanphoun, C.; Tanaka, K.; Dai, D.Q.; Luo, Z.L.; Liu, J.K.; Su, H.Y.; Bhat, D.J.; Bahkali, A.H.; Mortimer, P.E.; et al. Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 2015, 74, 143–197. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016, 85, 35–64. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Chethana, K.W.T.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Zhang, S.N.; Hyde, K.D.; Jones, E.B.G.; Jeewon, R.; Cheewangkoon, R.; Liu, J.K. Striatiguttulaceae, a new pleosporalean family to accommodate Longicorpus and Striatiguttula gen. nov. from palms. MycoKeys 2019, 49, 99–129. [Google Scholar] [CrossRef]
- Chaw, S.M.; Chang, C.C.; Chen, H.L.; Li, W.H. Dating the Monocot–Dicot divergence and the origin of core Eudicots using whole chloroplast genomes. J. Mol. Evol. 2004, 58, 424–441. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Hyde, K.D.; Alves, A.; Liu, J.K. Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019, 94, 1–22. [Google Scholar] [CrossRef]
- De Vienne, D.M.; Refrégier, G.; López-Villavicencio, M.; Tellier, A.; Hood, M.E.; Giraud, T. Cospeciation vs host-shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 2013, 198, 347–385. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; McKenzie, E.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Marc, S.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Zeng, X.Y.; Jeewon, R.; Hongsanan, S.; Hyde, K.D.; Wen, T.C. Unravelling evolutionary relationships between epifoliar Meliolaceae and angiosperms. J. Syst. Evol. 2020, in press. [Google Scholar] [CrossRef]



| Taxon | Strain Number | GenBank Accession Numbers | ||
|---|---|---|---|---|
| LSU | SSU | TEF1α | ||
| Acrocalymma aquatica | MFLUCC 11-0208 | JX276952 | JX276953 | - |
| Acrocalymma fici | CBS 317.76 | KP170712 | - | - |
| Acrocalymma medicaginis | CPC 24340 | KP170713 | - | - |
| Acrocalymma medicaginis | CPC 24341 | KP170714 | - | - |
| Acrocalymma medicaginis | CPC 24345 | KP170718 | - | - |
| Acrocalymma pterocarpi | MFLUCC 17-0926 | MK347949 | MK347840 | - |
| Aigialus grandis | BCC 20000 | GU479775 | GU479739 | GU479839 |
| Aigialus mangrovis | BCC 33563 | GU479776 | GU479741 | GU479840 |
| Aigialus parvus | BCC 18403 | GU479778 | GU479743 | GU479842 |
| Aigialus rhizophorae | BCC 33572 | GU479780 | GU479745 | GU479844 |
| Aliquandostipite khaoyaiensis | CBS 118232 | GU301796 | - | GU349048 |
| Amniculicola immersa | CBS 123083 | FJ795498 | GU456295 | GU456273 |
| Amniculicola lignicola | CBS 123094 | EF493861 | EF493863 | - |
| Amniculicola parva | CBS 123092 | GU301797 | GU296134 | GU349065 |
| Amorosia littoralis | NN 6654 | AM292055 | AM292056 | - |
| Anastomitrabeculia didymospora | MFLUCC 16-0412 | MW412978 | MW412977 | MW411338 |
| Anastomitrabeculia didymospora | MFLUCC 16-0417 | MW413899 | MW413898 | MW411339 |
| Angustimassarina populi | MFLUCC 13-0034 | KP888642 | KP899128 | KR075164 |
| Angustimassarina quercicola | MFLUCC 14-0506 | KP888638 | KP899124 | KR075169 |
| Anteaglonium abbreviatum | ANM 925a | GQ221877 | - | - |
| Anteaglonium globosum | SMH 5283 | GQ221911 | - | GQ221919 |
| Anteaglonium parvulum | MFLUCC 14-0821 | KU922915 | KU922916 | - |
| Antealophiotrema brunneosporum | CBS 123095 | LC194340 | LC194298 | LC194382 |
| Aquasubmersa japonica | HHUF 30468 | LC061586 | LC061581 | - |
| Aquasubmersa japonica | HHUF 30469 | LC061587 | LC061582 | - |
| Aquasubmersa mircensis | MFLUCC 11-0401 | JX276955 | JX276956 | - |
| Arthonia dispersa | UPSC 2583 | AY571381 | AY571379 | - |
| Ascocratera manglicola | BCC 09270 | GU479782 | GU479747 | GU479846 |
| Ascocylindrica marina | MD6011 | KT252905 | KT252907 | - |
| Ascocylindrica marina | MD6012 | KT252906 | - | - |
| Ascocylindrica marina | MF416 | MK007123 | MK007124 | - |
| Bahusandhika indica | GUFCC 18001 | KF460274 | - | - |
| Bambusicola massarinia | MFLUCC 11-0389 | JX442037 | JX442041 | - |
| Berkleasmium micronesicum | BCC 8141 | DQ280272 | DQ280268 | - |
| Berkleasmium nigroapicale | BCC 8220 | DQ280273 | DQ280269 | - |
| Bimuria novae-zelandiae | CBS 107.79 | AY016356 | AY016338 | DQ471087 |
| Botryosphaeria dothidea | CBS 115476 | AY928047 | EU673173 | AY236898 |
| Brevicollum hyalosporum | MAFF 243400 | LC271239 | LC271236 | LC271245 |
| Brevicollum hyalosporum | MFLUCC 17-0071 | MG602200 | MG602202 | MG739516 |
| Brevicollum hyalosporum | PUFNI 17628 | MH918671 | - | - |
| Brevicollum versicolor | HHUF 30591 | LC271240 | LC271237 | LC271246 |
| Capnodium salicinum | CBS 131.34 | DQ678050 | DQ677997 | - |
| Cladosporium cladosporioides | CBS 170.54 | DQ678057 | DQ678004 | - |
| Clematidis italica | MFLUCC 15-0084 | KU842381 | KU842382 | - |
| Corynespora cassiicola | CBS 100822 | GU301808 | GU296144 | GU349052 |
| Corynespora smithii | CABI 5649b | GU323201 | - | GU349018 |
| Crassiparies quadrisporus | HHUF 30590 | LC271241 | LC271238 | LC271248 |
| Crassiparies quadrisporus | HHUF 30409 | LC100025 | LC100017 | - |
| Crassiperidium octosporum | KT 2144 | LC373108 | LC373084 | LC373120 |
| Crassiperidium octosporum | KT 2894 | LC373109 | LC373085 | LC373121 |
| Crassiperidium octosporum | KT 3008 | LC373110 | LC373086 | LC373122 |
| Crassiperidium octosporum | KT 3029 | LC373111 | LC373087 | LC373123 |
| Crassiperidium octosporum | KT 3046 | LC373112 | LC373088 | LC373124 |
| Crassiperidium octosporum | KT 3188 | LC373113 | LC373089 | LC373125 |
| Crassiperidium octosporum | KT 3468 | LC373114 | LC373090 | LC373126 |
| Crassiperidium octosporum | KT 3604 | LC373115 | LC373091 | LC373127 |
| Crassiperidium octosporum | KT 3605 | LC373116 | LC373092 | LC373128 |
| Crassiperidium octosporum | MM 9 | LC373117 | LC373093 | LC373129 |
| Crassiperidium quadrisporum | KT 27981 | LC373118 | LC373094 | LC373130 |
| Crassiperidium quadrisporum | KT 27982 | LC373119 | LC373095 | LC373131 |
| Cryptoclypeus oxysporus | HHUF 30507 | LC194345 | LC194303 | LC194390 |
| Cryptocoryneum akitaense | MAFF 245365 | LC194348 | LC194306 | LC096136 |
| Cryptocoryneum japonicum | MAFF 245370 | LC194356 | LC194314 | LC096144 |
| Cryptocoryneum longicondensatum | MAFF 245374 | LC194360 | LC194318 | LC096148 |
| Cyclothyriella rubronotata | CBS 141486 | KX650544 | KX650507 | KX650519 |
| Cyclothyriella rubronotata | CBS 121892 | KX650541 | - | KX650516 |
| Cyclothyriella rubronotata | CBS 385.39 | MH867543 | - | - |
| Cyclothyriella rubronotata | CBS 419 85 | GU301875 | - | GU349002 |
| Delitschia didyma | UME 31411 | DQ384090 | AF242264 | - |
| Delitschia winteri | CBS 225.62 | DQ678077 | DQ678026 | DQ677922 |
| Dendrographa decolorans | Ertz 5003 | AY548815 | AY548809 | - |
| Dendrographa leucophaea f. minor | AF279382 | AF279381 | - | |
| Dendryphion europaeum | CPC 22943 | KJ869203 | - | - |
| Dendryphion europaeum | CPC 23231 | NG_059120 | - | - |
| Dendryphion nanum | MFLUCC 16-0975 | MG208132 | - | MG207983 |
| Didymosphaeria rubi-ulmifolii | MFLUCC 14-0023 | KJ436586 | KJ436588 | - |
| Dissoconium aciculare | CBS 204.89 | GU214419 | GU214523 | - |
| Ernakulamia cochinensis | PRC 3992 | LT964670 | - | - |
| Flavomyces fulophazii | CBS 135761 | KP184040 | KP184082 | - |
| Fuscostagonospora cytisi | MFLUCC 16-0622 | KY770978 | KY770977 | KY770979 |
| Fuscostagonospora sasae | CBS 139687 | AB807548 | AB797258 | - |
| Fusculina eucalyptorum | CBS 145083 | MK047499 | - | - |
| Gordonomyces mucovaginatus | CBS 127273 | JN712552 | ||
| Halojulella avicenniae | JK 5326A | GU479790 | GU479756 | - |
| Halojulella avicenniae | BCC 20173 | GU371822 | GU371830 | GU371815 |
| Halojulella avicenniae | PUFD542 | MK026757 | MK026754 | - |
| Halojulella avicenniae | BCC 18422 | GU371823 | GU371831 | GU371816 |
| Halojulella avicenniae | BCC28357 | KC555567 | KC555565 | - |
| Halojulella avicenniae | GR00584 | KC555568 | KC555566 | - |
| Halotthia posidoniae | BBH 22481 | GU479786 | GU479752 | - |
| Helminthosporium aquaticum | MFLUCC 15-0357 | KU697306 | KU697310 | - |
| Helminthosporium velutinum | MAFF 243854 | AB807530 | AB797240 | - |
| Helminthosporium velutinum | MFLUCC 13-0243 | KU697305 | - | - |
| Helminthosporium velutinum | MFLUCC 15-0423 | KU697304 | - | - |
| Hermatomyces iriomotensis | HHUF 30518 | LC194367 | LC194325 | LC194394 |
| Hermatomyces tectonae | MFLUCC 14-1140 | KU764695 | KU712465 | KU872757 |
| Hermatomyces thailandica | MFLUCC 14-1143 | KU764692 | KU712468 | KU872754 |
| Hobus wogradensis | TI | KX650546 | KX650508 | KX650521 |
| Hysterium angustatum | CBS 236.34 | FJ161180 | GU397359 | FJ161096 |
| Hysterium angustatum | MFLUCC 16-0623 | MH535893 | MH535885 | MH535878 |
| Jahnula seychellensis | SS2113 | EF175665 | EF175643 | - |
| Latorua caligans | CBS 576.65 | KR873266 | - | - |
| Latorua grootfonteinensis | CBS 369.72 | KR873267 | - | - |
| Lentimurispora urniformis | MFLUCC 18-0497 | MH179144 | MH179160 | MH188055 |
| Leptosphaeria doliolum | CBS 505.75 | GQ387576 | GQ387515 | GU349069 |
| Leptoxyphium cacuminum | MFLUCC 10-0049 | JN832602 | JN832587 | - |
| Leucaenicola phraeana | MFLUCC 18-0472 | MK348003 | MK347892 | - |
| Lignosphaeria fusispora | MFLUCC 11-0377 | KP888646 | - | - |
| Lignosphaeria thailandica | MFLUCC 11-0376 | KP888645 | - | - |
| Lindgomyces ingoldianus | ATCC 200398 | AB521736 | AB521719 | - |
| Longiostiolum tectonae | MFLUCC 12 0562 | KU764700 | KU712459 | - |
| Lophiotrema eburnoides | HHUF 30079 | LC001707 | LC001706 | - |
| Lophiotrema nucula | CBS 627.86 | GU301837 | GU296167 | GU349073 |
| Macrodiplodiopsis desmazieri | CBS 140062 | KR873272 | - | - |
| Magnicamarosporium diospyricola | MFLUCC 16-0419 | KY554212 | KY554211 | KY554209 |
| Massarina eburnea | CBS 473.64 | GU301840 | GU296170 | - |
| Massariosphaeria phaeospora | CBS 611.86 | GU301843 | GU296173 | - |
| Mauritiana rhizophorae | BCC 28866 | GU371824 | GU371832 | GU371817 |
| Medicopsis romeroi | CBS 122784 | EU754208 | EU754109 | KF015679 |
| Medicopsis romeroi | CBS 252.60 | EU754207 | EU754108 | KF015678 |
| Medicopsis romeroi | CBS 132878 | KF015622 | KF015648 | KF015682 |
| Murispora rubicunda | IFRD 2017 | FJ795507 | GU456308 | GU456289 |
| Neoastrosphaeriella krabiensis | MFLUCC 11-0025 | JN846729 | JN846739 | - |
| Neohendersonia kickxii | CBS 112403 | KX820266 | - | - |
| Neohendersonia kickxii | CBS 122938 | KX820268 | - | - |
| Neohendersonia kickxii | CBS 114276 | KX820267 | - | - |
| Neohendersonia kickxii | CPC 24865 | KX820270 | - | - |
| Neohendersonia kickxii | CBS 122941 | KX820269 | - | - |
| Neomassaria fabacearum | MFLUCC 16-1875 | KX524145 | KX524147 | KX524149 |
| Neomassaria formosana | NTUCC 17-007 | MH714756 | MH714759 | MH714762 |
| Neomassarina chromolaenae | MFLUCC 17-1480 | MT214466 | MT214419 | MT235785 |
| Neomassarina pandanicola | MFLUCC 16-0270 | MG298945 | - | MG298947 |
| Neomassarina thailandica | MFLUCC 10-0552 | KX672157 | KX672160 | KX672163 |
| Neomassarina thailandica | MFLUCC 17-1432 | MT214467 | MT214420 | MT235786 |
| Neotorula aquatica | MFLUCC 150342 | KU500576 | KU500583 | - |
| Neotorula submersa | KUMCC 15-0280 | KX789217 | - | - |
| Occultibambusa bambusae | MFLUCC 13-0855 | KU863112 | KU872116 | - |
| Occultibambusa pustula | MFLUCC 11-0502 | KU863115 | KU872118 | - |
| Ohleria modesta | MGC | KX650562 | - | KX650533 |
| Ohleria modesta | CBS 141480 | KX650563 | KX650513 | KX650534 |
| Paradictyoarthrinium diffractum | MFLUCC 13-0466 | KP744498 | KP753960 | - |
| Paradictyoarthrinium diffractum | MFLUCC 12-0557 | KP744497 | - | - |
| Paradictyoarthrinium hydei | MFLUCC 13-0465 | MG747497 | - | - |
| Paradictyoarthrinium tectonicola | MFLUCC 13-0465 | KP744500 | KP753961 | - |
| Paradictyoarthrinium tectonicola | MFLUCC 12-0556 | KP744499 | - | - |
| Periconia thailandica | MFLUCC 17-0065 | KY753888 | KY753889 | - |
| Phaeoseptum aquaticum | CBS 123113 | JN644072 | - | - |
| Phaeoseptum terricola | MFLUCC 10-0102 | MH105779 | MH105780 | MH105781 |
| Phyllosticta capitalensis | CBS 226.77 | KF206289 | KF766300 | - |
| Piedraia hortae | CBS 480.64 | GU214466 | - | - |
| Polyplosphaeria fusca | CBS 125425 | AB524607 | AB524466 | AB524822 |
| Preussia lignicola | CBS 363.69 | DQ384098 | DQ384087 | - |
| Preussia lignicola | CBS 264.69 | GU301872 | GU296197 | GU349027 |
| Pseudoastrosphaeriella bambusae | MFLUCC 11-0205 | KT955475 | KT955455 | KT955437 |
| Pseudoastrosphaeriella longicolla | MFLUCC 11-0171 | KT955476 | KT955456 | KT955438 |
| Pseudoastrosphaeriella thailandensis | MFLUCC 10-0553 | KT955477 | KT955456 | KT955439 |
| Pseudolophiotrema elymicola | HHUF 28984 | LC194381 | LC194339 | LC194418 |
| Pseudomassariosphaeria bromicola | MFLUCC 15-0031 | KT305994 | KT305996 | KT305999 |
| Pseudotetraploa curviappendiculata | CBS 125426 | AB524610 | AB524469 | AB524825 |
| Quadricrura septentrionalis | CBS 125428 | AB524617 | AB524476 | AB524832 |
| Racodium rupestre | L346 | EU048583 | EU048575 | - |
| Racodium rupestre | L424 | EU048582 | EU048577 | - |
| Ramusculicola thailandica | MFLUCC 13-0284 | KP888647 | KP899131 | KR075167 |
| Rimora mangrovei | JK 5246A | GU301868 | GU296193 | |
| Roccella fuciformis | Tehler 8171 | FJ638979 | - | - |
| Rostriconidium aquaticum | KUMCC 15-0297 | MG208144 | - | MG207995 |
| Rostriconidium aquaticum | MFLUCC 16-1113 | MG208143 | - | MG207994 |
| Salsuginea ramicola | KT 2597.1 | GU479800 | GU479767 | GU479861 |
| Salsuginea ramicola | CBS 125781 | MH877872 | - | - |
| Scorias spongiosa | CBS 325.33 | MH866910 | GU214696 | - |
| Seriascoma didymospora | MFLUCC 11-0179 | KU863116 | KU872119 | - |
| Sigarispora arundinis | JCM 13550 | AB618998 | AB618679 | LC001737 |
| Sigarispora ravennica | MFLUCC 14-0005 | KP698414 | KP698415 | - |
| Splanchnonema platani | CBS 222.37 | KR909316 | KR909318 | KR909319 |
| Sporidesmioides thailandica | KUMCC 16-0012 | KX437758 | KX437760 | KX437767 |
| Sporidesmioides thailandica | MFLUCC 13-0840 | NG_059703 | NG_061242 | KX437766 |
| Sporormia fimetaria | UPS:Dissing Gr.81.194 | GQ203729 | - | - |
| Sporormiella minima | CBS 52450 | DQ468046 | - | DQ468003 |
| Stagonospora pseudocaricis | CBS 135132 | KF251762 | KF251259 | KF252741 |
| Stemphylium vesicarium | CBS 191.86 | DQ247804 | DQ247812 | DQ471090 |
| Stemphylium vesicarium | CBS 714.68 | DQ678049 | DQ767648 | DQ677888 |
| Sulcatispora acerina | KT2982 | LC014610 | LC014605 | LC014615 |
| Sulcosporium thailandicum | MFLUCC 12-0004 | KT426563 | KT426564 | - |
| Teichospora quercus | CBS 143396 | MH107966 | - | MH108030 |
| Tetraplosphaeria sasicola | KT 563 | AB524631 | AB524490 | AB524838 |
| Torula gaodangensis | MFLUCC 17-0234 | NG_059827 | NG_063641 | - |
| Torula herbarum | CBS 111855 | KF443386 | KF443391 | KF443403 |
| Triplosphaeria maxima | MAFF 239682 | AB524637 | AB524496 | - |
| Tubeufia chiangmaiensis | MFLUCC 11-0514 | KF301538 | KF301543 | KF301557 |
| Tubeufia javanica | MFLUCC 12-0545 | KJ880036 | KJ880035 | KJ880037 |
| Vargamyces aquaticus | CBS 639.63 | KY853539 | - | - |
| Vargamyces aquaticus | HKUCC 10830 | DQ408575 | - | - |
| Versicolorisporium triseptatum | HHUF 28815 | AB330081 | AB524501 | - |
| Westerdykella dispersa | CBS 297.56 | MH869191 | - | - |
| Westerdykella ornata | CBS 379.55 | GU301880 | GU296208 | GU349021 |
| Xenomassariosphaeria rosae | MFLUCC 15-0179 | MG829092 | MG829192 | - |
| Nodes | Node Age | Geological Time Period |
|---|---|---|
| Arthoniomycetes–Dothideomycetes | 323 (310–349) | Carboniferous |
| Dothideomycetes crown group | 263 (216–313) | Permian |
| Hysteriales–Pleosporales | 236 (188–300) | Triassic |
| Pleosporales crown group | 206 (171–254) | Triassic |
| Capnodiales crown group | 147 (99–200) | Jurassic |
| Anastomitrabeculiaceae stem group | 84 (52–116) | Cretaceous |
| Aigialaceae–Aigialus sp. | 37 (18–56) | Eocene |
| Anastomitrabeculiaceae crown group | 2.6 (0.19–6.61) | Neogene |
| Families | Crown Age | Stem Age |
|---|---|---|
| Aigialaceae | 102 | 141 |
| Amniculicolaceae | 90 | 177 |
| Anastomitrabeculiaceae | 2.6 | 84 |
| Anteagloniaceae | 52 | 98 |
| Bambusicolaceae | 29 | 57 |
| Cyclothyriellaceae | 66 | 95 |
| Delitschiaceae | 78 | 131 |
| Didymosphaeriaceae | 47 | 81 |
| Fuscostagonosporaceae | 26 | 63 |
| Lindgomycetaceae | 31 | 92 |
| Neomassariaceae | 82 | 131 |
| Pseudoastrosphaeriellaceae | 56 | 147 |
| Tetraplosphaeriaceae | 91 | 189 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhunjun, C.S.; Phukhamsakda, C.; Jeewon, R.; Promputtha, I.; Hyde, K.D. Integrating Different Lines of Evidence to Establish a Novel Ascomycete Genus and Family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales. J. Fungi 2021, 7, 94. https://doi.org/10.3390/jof7020094
Bhunjun CS, Phukhamsakda C, Jeewon R, Promputtha I, Hyde KD. Integrating Different Lines of Evidence to Establish a Novel Ascomycete Genus and Family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales. Journal of Fungi. 2021; 7(2):94. https://doi.org/10.3390/jof7020094
Chicago/Turabian StyleBhunjun, Chitrabhanu S., Chayanard Phukhamsakda, Rajesh Jeewon, Itthayakorn Promputtha, and Kevin D. Hyde. 2021. "Integrating Different Lines of Evidence to Establish a Novel Ascomycete Genus and Family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales" Journal of Fungi 7, no. 2: 94. https://doi.org/10.3390/jof7020094
APA StyleBhunjun, C. S., Phukhamsakda, C., Jeewon, R., Promputtha, I., & Hyde, K. D. (2021). Integrating Different Lines of Evidence to Establish a Novel Ascomycete Genus and Family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales. Journal of Fungi, 7(2), 94. https://doi.org/10.3390/jof7020094








