The Glyoxysomal Protease LON2 Is Involved in Fruiting-Body Development, Ascosporogenesis and Stress Resistance in Sordaria macrospora
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
2.2. Generation of Plasmids
2.3. Construction of the Knockout Strain ΔSmlon2
2.4. Quantitative Analyses of S. macrospora Strains
2.5. Light and Fluorescence Microscopy
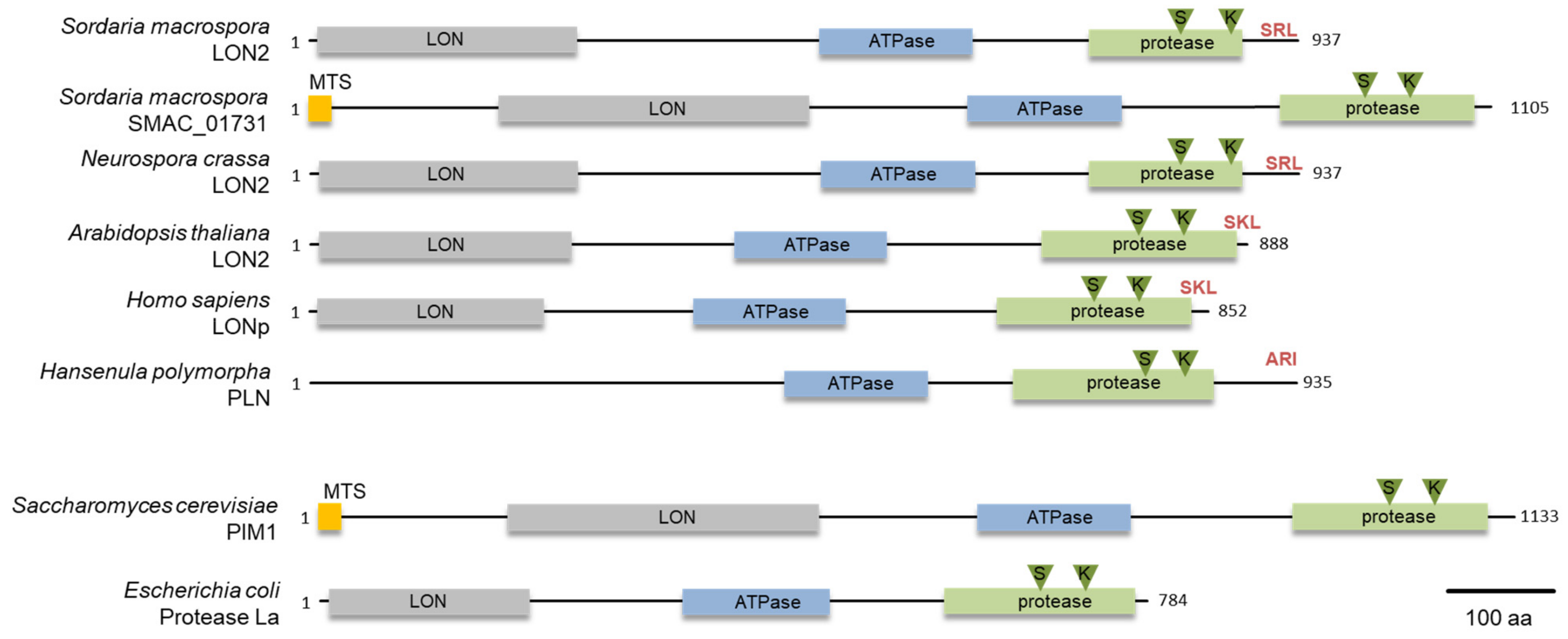
2.6. Protein Domain Analysis and Phylogeny
3. Results
3.1. S. macrospora Encodes Two Lon Proteases
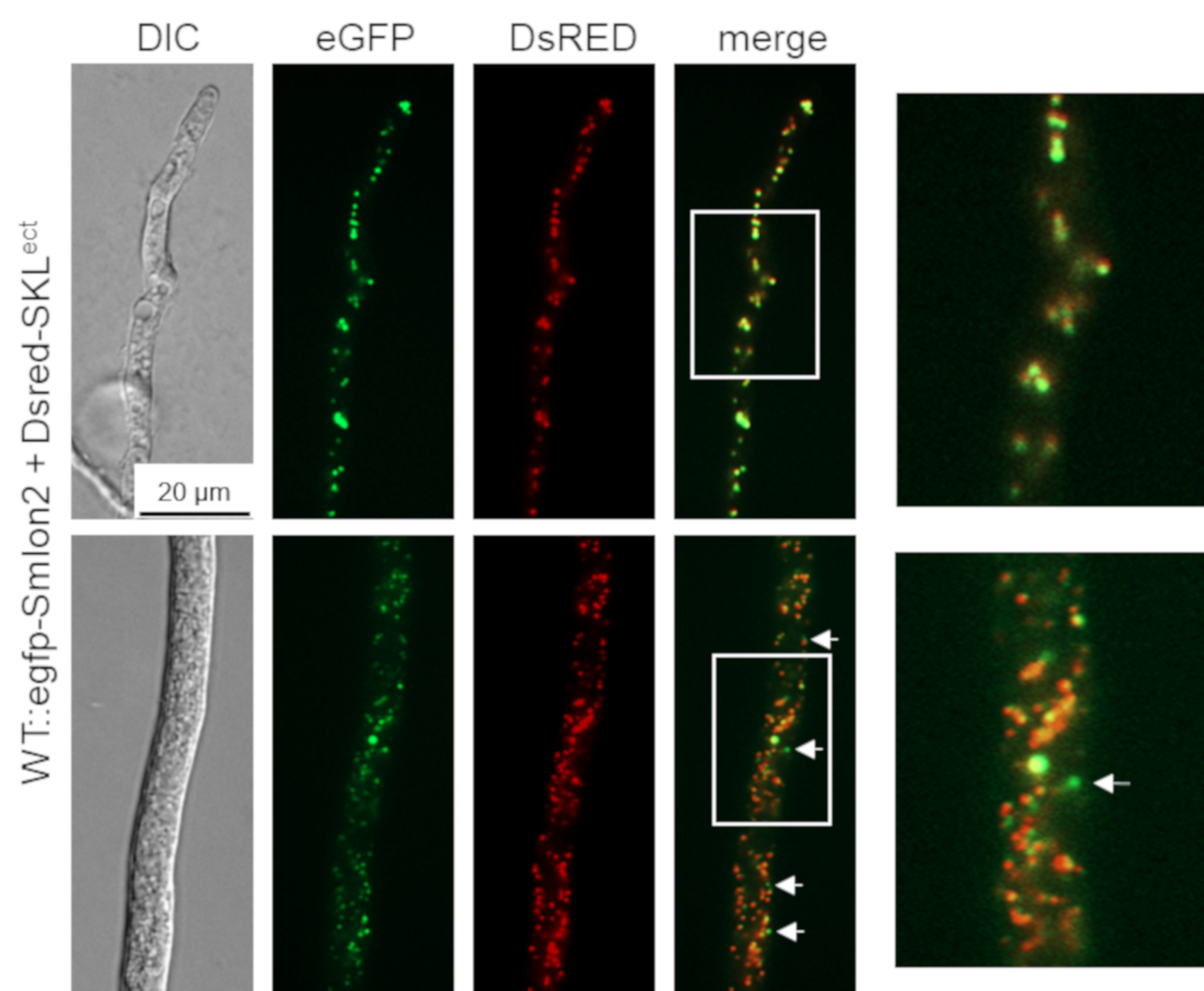
3.2. SmLON2 Localizes Predominantly to Microbodies
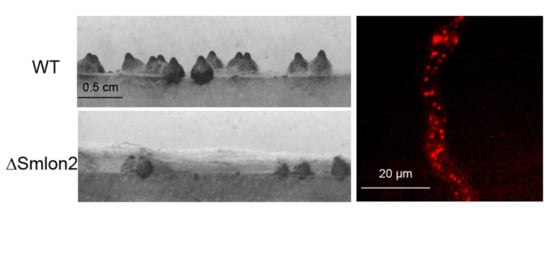
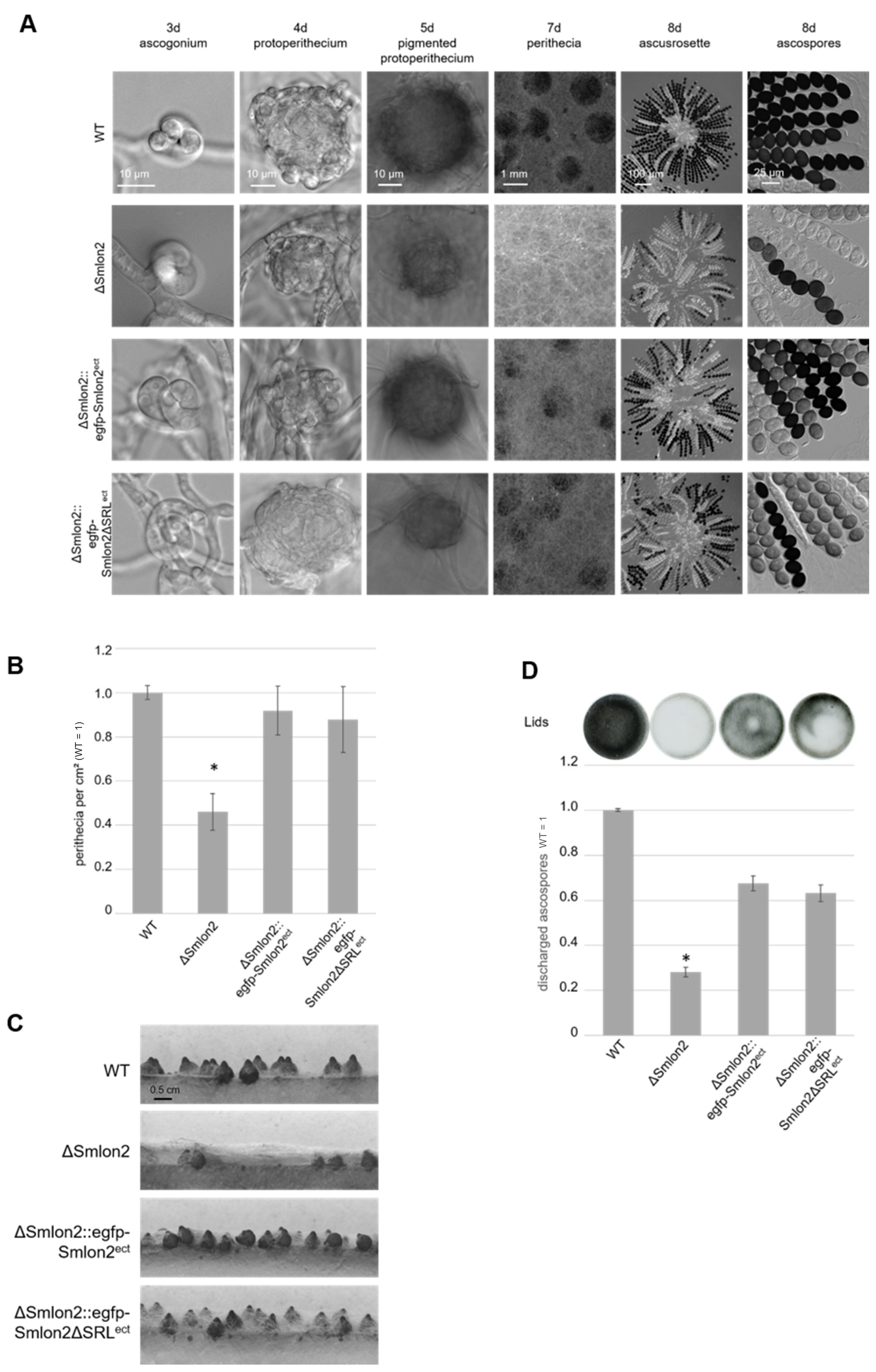
3.3. Deletion of Smlon2 Leads to Defects in Fruiting-Body Development and Ascospore Production
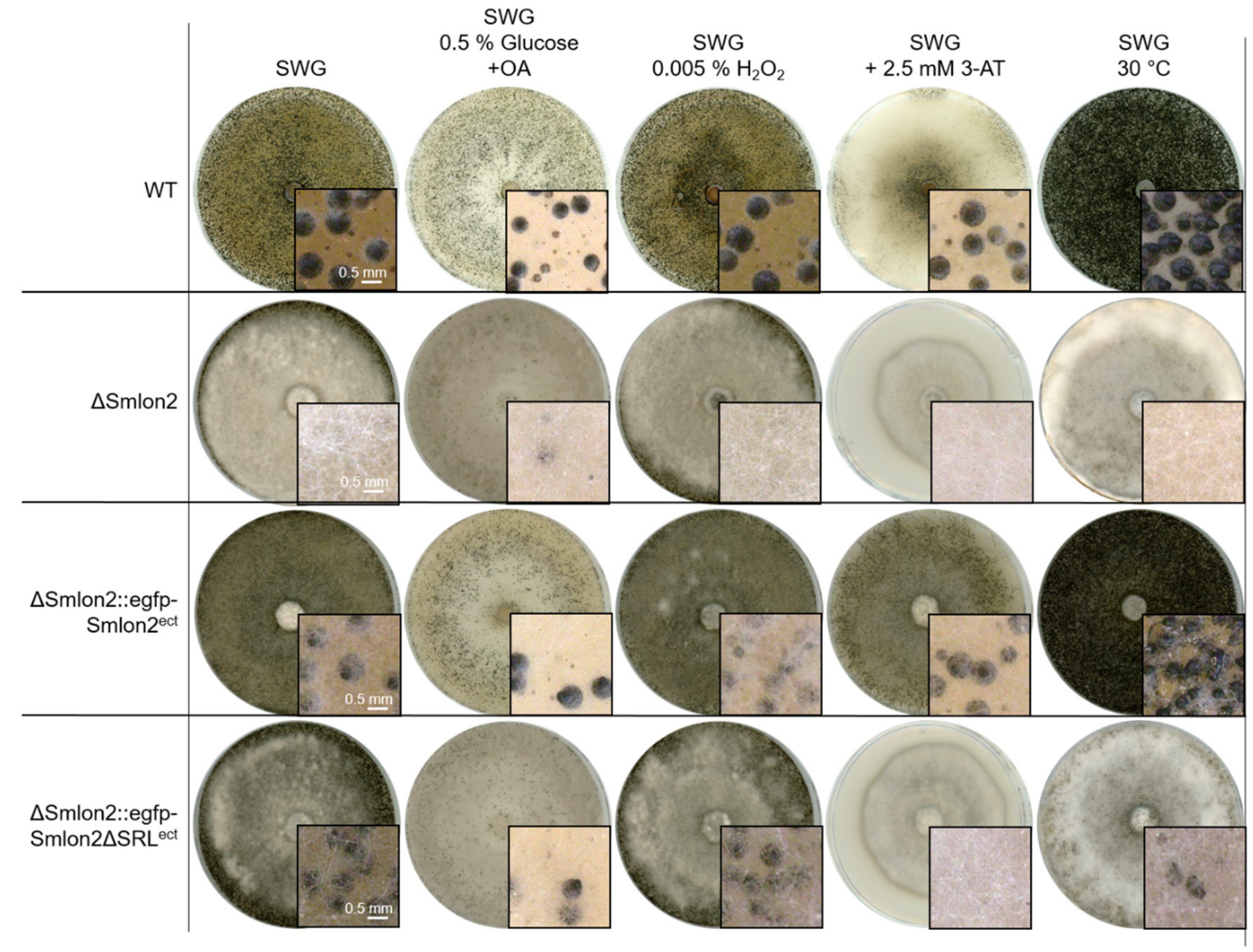
3.4. The Mutant ΔSmlon2 Exhibited an Increased Sensitivity against Oxidative and Nutrient Stresses
3.5. Starvation Stress Resulted in an Enhanced Vacuolar Degradation of Glyoxysomes in the ΔSmlon2 Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peraza-Reyes, L.; Berteaux-Lecellier, V. Peroxisomes and sexual development in fungi. Front. Physiol. 2013, 4, 244. [Google Scholar] [CrossRef]
- Pieuchot, L.; Jedd, G. Peroxisome Assembly and Functional Diversity in Eukaryotic Microorganisms. Annu. Rev. Microbiol. 2012, 66, 237–263. [Google Scholar] [CrossRef] [PubMed]
- Bleichrodt, R.J.; van Veluw, G.J.; Recter, B.; Maruyama, J.; Kitamoto, K.; Wösten, H.A. Hyphal heterogeneity in Aspergillus oryzae is the result of dynamic closure of septa by Woronin bodies. Mol. Microbiol. 2012, 86, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Jedd, G.; Chua, N. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nature 2000, 2, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Girzalsky, W.; Saffian, D.; Erdmann, R. Peroxisomal protein translocation. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1803, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Kempiński, B.; Chełstowska, A.; Poznański, J.; Król, K.; Rymer, Ł.; Frydzińska, Z.; Girzalsky, W.; Skoneczna, A.; Erdmann, R.; Skoneczny, M. The peroxisomal targeting signal 3 (PTS3) of the budding yeast acyl-CoA oxidase is a signal patch. Front. Cell Dev. Biol. 2020, 8, 198. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. The peroxisome: A new cytoplasmic organelle. Proc. R. Soc. Lond. B. Biol. Sci. 1969, 173, 71–83. [Google Scholar]
- Schliebs, W.; Würtz, C.; Kunau, W.-H.; Veenhuis, M.; Rottensteiner, H. A Eukaryote without Catalase-Containing Microbodies: Neurospora crassa Exhibits a Unique Cellular Distribution of Its Four Catalases. Eukaryot. Cell 2006, 5, 1490–1502. [Google Scholar] [CrossRef]
- Bourdais, A.; Bidard, F.; Zickler, D.; Berteaux-Lecellier, V.; Silar, P.; Espagne, E. Wood Utilization Is Dependent on Catalase Activities in the Filamentous Fungus Podospora anserina. PLoS ONE 2012, 7, e29820. [Google Scholar] [CrossRef]
- Pöggeler, S. Evolution of Multicopper Oxidase Genes in Coprophilous and Non-Coprophilous Members of the Order Sordariales. Curr. Genom. 2011, 12, 95–103. [Google Scholar] [CrossRef]
- Teichert, I.; Pöggeler, S.; Nowrousian, M. Sordaria macrospora: 25 years as a model organism for studying the molecular mechanisms of fruiting body development. Appl. Microbiol. Biotechnol. 2020, 104, 3691–3704. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Espagne, E. Sordaria, a model system to uncover links between meiotic pairing and recombination. Semin. Cell Dev. Biol. 2016, 54, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Teichert, I.; Nowrousian, M.; Pöggeler, S.; Kück, U. The Filamentous Fungus Sordaria macrospora as a Genetic Model to Study Fruiting Body Development. Adv. Genet. 2014, 87, 199–244. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Herzog, B.; Voigt, O.; Valerius, O.; Braus, G.H.; Pöggeler, S. NBR1 is involved in selective pexophagy in filamentous ascomycetes and can be functionally replaced by a tagged version of its human homolog. Autophagy 2019, 15, 78–97. [Google Scholar] [CrossRef]
- Voigt, O.; Pöggeler, S. Autophagy genesSmatg8andSmatg4are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomyceteSordaria macrospora. Autophagy 2013, 9, 33–49. [Google Scholar] [CrossRef]
- Kumar, S.; Kawalek, A.; Van Der Klei, I.J. Peroxisomal quality control mechanisms. Curr. Opin. Microbiol. 2014, 22, 30–37. [Google Scholar] [CrossRef]
- Howard-Flanders, P.; Simson, E.; Theriot, L. A locus that controls filament formation and sensitivity to radiation in Eschericia coli K12. Genetics 1964, 49, 237–246. [Google Scholar] [CrossRef]
- Chung, C.H.; Goldberg, A.L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc. Natl. Acad. Sci. USA 1981, 78, 4931–4935. [Google Scholar] [CrossRef]
- Lee, I.; Suzuki, C.K. Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2008, 1784, 727–735. [Google Scholar] [CrossRef]
- Rigas, S.; Daras, G.; Tsitsekian, D.; Alatzas, A.; Hatzopoulos, P. Evolution and significance of the Lon gene family in Arabidopsis organelle biogenesis and energy metabolism. Front. Plant Sci. 2014, 5, 145. [Google Scholar] [CrossRef]
- Young, P.G.; Bartel, B. Pexophagy and peroxisomal protein turnover in plants. Biochim. et Biophys. Acta (BBA) Bioenerg. 2016, 1863, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Omi, S.; Nakata, R.; Okamura-Ikeda, K.; Konishi, H.; Taniguchi, H. Contribution of Peroxisome-specific Isoform of Lon Protease in Sorting PTS1 Proteins to Peroxisomes. J. Biochem. 2008, 143, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Aksam, E.B.; Koek, A.; Jourdan, S.; Veenhuis, M.; Van Der Klei, I.J. A Peroxisomal Lon Protease and Peroxisome Degradation by Autophagy Play Key Roles in Vitality ofHansenula polymorphaCells. Autophagy 2007, 3, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewska, M.; Williams, C.; Kikhney, A.; Opaliński, Ł.; Van Roermund, C.W.T.; De Boer, R.; Veenhuis, M.; Van Der Klei, I.J. Peroxisomal Proteostasis Involves a Lon Family Protein That Functions as Protease and Chaperone. J. Biol. Chem. 2012, 287, 27380–27395. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wei, Y.; Xie, X.-L.; Chen, L.-N.; Zhang, S.-H. Mitochondrial and peroxisomal Lon proteases play opposing roles in reproduction and growth but co-function in the normal development, stress resistance and longevity of Thermomyces lanuginosus. Fungal Genet. Biol. 2017, 103, 42–54. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- James, P.; Halladay, J.; Craig, E.A. Genomic Libraries and a Host Strain Designed for Highly Efficient Two-Hybrid Selection in Yeast. Genetics 1996, 144, 1425–1436. [Google Scholar] [CrossRef]
- Walz, M.; Kück, U. Transformation of Sordaria macrospora to hygromycin B resistance: Characterization of transformants by electrophoretic karyotyping and tetrad analysis. Curr. Genet. 1995, 29, 88–95. [Google Scholar] [CrossRef]
- Hoff, B.; Kück, U. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genet. Rep. 2006, 53, 9–11. [Google Scholar]
- Esser, K. Cryptogams—Cyanobacteria, Algae, Fungi, Lichens; Cambridge University Press: London, UK, 1982. [Google Scholar]
- Elleuche, S.; Pöggeler, S. Visualization of peroxisomes via SKL-tagged DsRed protein in Sordaria macrospora. Fungal Genet. Rep. 2008, 55, 8–12. [Google Scholar] [CrossRef]
- Nowrousian, M.; Ringelberg, C.; Dunlap, J.C.; Loros, J.J.; Kück, U. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genom. 2005, 273, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M.; Teichert, I.; Masloff, S.; Kück, U. Whole-Genome Sequencing of Sordaria macrospora Mutants Identifies Developmental Genes. G3 2012, 2, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Bernhards, Y.; Pöggeler, S. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr. Genet. 2011, 57, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Pöggeler, S.; Nowrousian, M.; Ringelberg, C.; Loros, J.J.; Dunlap, J.C.; Kück, U. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genom. 2006, 275, 492–503. [Google Scholar] [CrossRef]
- Kawabata, T.; Inoue, H. Detection of physical interactions by immunoprecipitation of FLAG- and HA-tagged proteins expressed at the his-3 locus in Neurospora crassa. Fungal Genet. Newsl. 2007, 54, 5–8. [Google Scholar] [CrossRef][Green Version]
- Lichius, A.; Read, N.D. A versatile set of Lifeact-RFP expression plasmids for live-cell imaging of F-actin in filamentous fungi. Fungal Genet. Rep. 2010, 57, 8–14. [Google Scholar] [CrossRef][Green Version]
- Pöggeler, S.; Masloff, S.; Hoff, B.; Mayrhofer, S.; Kück, U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 2003, 43, 54–61. [Google Scholar] [CrossRef]
- Klix, V.; Nowrousian, M.; Ringelberg, C.; Loros, J.J.; Dunlap, J.C.; Poggeler, S. Functional Characterization of MAT1-1-Specific Mating-Type Genes in the Homothallic Ascomycete Sordaria macrospora Provides New Insights into Essential and Nonessential Sexual Regulators. Eukaryot. Cell 2010, 9, 894–905. [Google Scholar] [CrossRef]
- Bloemendal, S.; Bernhards, Y.; Bartho, K.; Dettmann, A.; Teichert, I.; Pöggeler, S.; Voigt, O.; Seiler, S.; Wolters, D.A.; Kück, U. A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 2012, 84, 310–323. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved Prediction of Mitochondrial Targeting Sequences and Their Cleavage Sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Han, M.V.; Zmasek, C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinform. 2009, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M.; Stajich, J.E.; Chu, M.; Engh, I.; Espagne, E.; Halliday, K.J.; Kamerewerd, J.; Kempken, F.; Knab, B.; Kuo, H.-C.; et al. De novo Assembly of a 40 Mb Eukaryotic Genome from Short Sequence Reads: Sordaria macrospora, a Model Organism for Fungal Morphogenesis. PLoS Genet. 2010, 6, e1000891. [Google Scholar] [CrossRef] [PubMed]
- Rotanova, T.V.; Andrianova, A.G.; Kudzhaev, A.M.; Li, M.; Botos, I.; Wlodawer, A.; Gustchina, E. New insights into structural and functional relationships between LonA proteases and ClpB chaperones. FEBS Open Bio 2019, 9, 1536–1551. [Google Scholar] [CrossRef] [PubMed]
- Lametschwandtner, G.; Brocard, C.; Fransen, M.; Van Veldhoven, P.; Berger, J.; Hartig, A. The Difference in Recognition of Terminal Tripeptides as Peroxisomal Targeting Signal 1 between Yeast and Human Is Due to Different Affinities of Their Receptor Pex5p to the Cognate Signal and to Residues Adjacent to It. J. Biol. Chem. 1998, 273, 33635–33643. [Google Scholar] [CrossRef]
- Farmer, L.M.; Rinaldi, M.A.; Young, P.G.; Danan, C.H.; Burkhart, S.E.; Bartel, B. Disrupting Autophagy Restores Peroxisome Function to an Arabidopsis lon2 Mutant and Reveals a Role for the LON2 Protease in Peroxisomal Matrix Protein Degradation. Plant Cell 2013, 25, 4085–4100. [Google Scholar] [CrossRef]
- Goto-Yamada, S.; Mano, S.; Nakamori, C.; Kondo, M.; Yamawaki, R.; Kato, A.; Nishimura, M. Chaperone and Protease Functions of LON Protease 2 Modulate the Peroxisomal Transition and Degradation with Autophagy. Plant Cell Physiol. 2014, 55, 482–496. [Google Scholar] [CrossRef]
- Luce, K.; Osiewacz, H.D. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nature 2009, 11, 852–858. [Google Scholar] [CrossRef]
- Adam, C.; Picard, M.; Déquard-Chablat, M.; Sellem, C.H.; Denmat, S.H.-L.; Contamine, V. Biological Roles of the Podospora anserina Mitochondrial Lon Protease and the Importance of Its N-Domain. PLoS ONE 2012, 7, e38138. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Espíndola, R.; Suaste-Olmos, F.; Peraza-Reyes, L. Dynamic Regulation of Peroxisomes and Mitochondria during Fungal Development. J. Fungi 2020, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Takano-Rojas, H.; Zickler, D.; Peraza-Reyes, L. Peroxisome dynamics during development of the fungus Podospora anserina. Mycologia 2016, 108, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Espíndola, R.; Takano-Rojas, H.; Suaste-Olmos, F.; Peraza-Reyes, L. Distinct contributions of the peroxisome-mitochondria fssion machinery during sexual development of the fungus Podospora anserina. Front. Microbiol. 2020, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Ast, J.; Stiebler, A.C.; Freitag, J.; Bölker, M. Dual targeting of peroxisomal proteins. Front. Physiol. 2013, 4, 297. [Google Scholar] [CrossRef] [PubMed]
- Teichert, I.; Dahlmann, T.A.; Kück, U.; Nowrousian, M. RNA Editing During Sexual Development Occurs in Distantly Related Filamentous Ascomycetes. Genome Biol. Evol. 2017, 9, 855–868. [Google Scholar] [CrossRef]
- Blank-Landeshammer, B.; Teichert, I.; Märker, R.; Nowrousian, M.; Kück, U.; Sickmann, A. Combination of Proteogenomics with PeptideDe NovoSequencing Identifies New Genes and Hidden Posttranscriptional Modifications. mBio 2019, 10, e02367-19. [Google Scholar] [CrossRef]
- Teichert, I. Adenosine to inosine mRNA editing in fungi and how it may relate to fungal pathogenesis. PLoS Pathog. 2018, 14, e1007231. [Google Scholar] [CrossRef]


| Strain | Genotype | Source |
|---|---|---|
| Escherichia coli | ||
| MACH1 | ΔrecA1398, endA1, tonA, Φ80ΔlacM15, ΔlacX74, hsdR, (rK-mK+) | Invitrogen |
| Saccharomyces cerevisiae | ||
| PJ69-4A | MATa, trp1-901, leu2-3,112, ura3-52, his3-200, ga14Δ, ga18OΔ LYS2::GALl-HIS3, GAL2-ADE2, met2::GAL7-lacZ | [28] |
| Sordaria macrospora | ||
| DSM997 | Wild type (WT) | DSMZ |
| S23442 | mutation in fus1-1 gene, brownish ascospores | [34] |
| Δku70 | Δku70::natR | [36] |
| WT::1783-1ect | ectopic integration of p1783-1 into DSM997; hygR, ssi, fertile, Pgpd::egfp::TtrpC | [15] |
| WT::RHN1ect | ectopic integration of pRHN1 into DSM997; natR, ssi, fertile, Pgpd::Dsred::TtrpC | [14] |
| WT::TagRFP-Tect | ectopic integration of ptRFP_nat into DSM997; natR, ssi, fertile, Pccg1::Tagrfp-t::TtrpC | This study |
| WT::Dsred-SKLect | ectopic integration of pDsred-SKL into S48977; natR, ssi, fertile, Pgpd::Dsred-SKL::TtrpC | [32] |
| ΔSmlon2 | ΔSmlon2::hygR, ssi | This study |
| WT::egfp-Smlon2ect+Dsred-SKLect | ectopic integration of pDsred-SKL and pegfp-Smlon2 into DSM997; natR, hygR ssi, fertile, Pgpd::Dsred-SKL::TtrpC; PSmlon2:: ORF egfp+Smlon2::TSmlon2 | This study |
| ΔSmlon2::egfp-Smlon2ect | ectopic integration of pegfp-Smlon2 into ΔSmlon2; natR, hygR, ssi, fertile, PSmlon2:: ORF egfp+Smlon2::TSmlon2 | This study |
| ΔSmlon2::Tagrfp-t-Smlon2ect | ectopic integration of ptrfp-Smlon2 into ΔSmlon2; natR, hygR, ssi, fertile, PSmlon2:: ORF trfp+Smlon2::TSmlon2 | This study |
| ΔSmlon2::egfp-Smlon2ΔSRLtect | ectopic integration of pegfp-Smlon2ΔSRL into ΔSmlon2; natR, hygR, ssi, fertile, PSmlon2:: ORF egfp+Smlon2 deletion of aa 935-937 (SRL)::TSmlon2 | This study |
| ΔSmlon2::Dsred-SKLect | ectopic integration of pDsred-SKL into ΔSmlon2; natR, hygR, ssi, fertile, Pgpd::Dsred-SKL::TtrpC | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Werner, A.; Otte, K.; Stahlhut, G.; Hanke, L.M.; Pöggeler, S. The Glyoxysomal Protease LON2 Is Involved in Fruiting-Body Development, Ascosporogenesis and Stress Resistance in Sordaria macrospora. J. Fungi 2021, 7, 82. https://doi.org/10.3390/jof7020082
Werner A, Otte K, Stahlhut G, Hanke LM, Pöggeler S. The Glyoxysomal Protease LON2 Is Involved in Fruiting-Body Development, Ascosporogenesis and Stress Resistance in Sordaria macrospora. Journal of Fungi. 2021; 7(2):82. https://doi.org/10.3390/jof7020082
Chicago/Turabian StyleWerner, Antonia, Kolja Otte, Gertrud Stahlhut, Leon M. Hanke, and Stefanie Pöggeler. 2021. "The Glyoxysomal Protease LON2 Is Involved in Fruiting-Body Development, Ascosporogenesis and Stress Resistance in Sordaria macrospora" Journal of Fungi 7, no. 2: 82. https://doi.org/10.3390/jof7020082
APA StyleWerner, A., Otte, K., Stahlhut, G., Hanke, L. M., & Pöggeler, S. (2021). The Glyoxysomal Protease LON2 Is Involved in Fruiting-Body Development, Ascosporogenesis and Stress Resistance in Sordaria macrospora. Journal of Fungi, 7(2), 82. https://doi.org/10.3390/jof7020082