New Xerophilic Species of Penicillium from Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Fungal Isolation
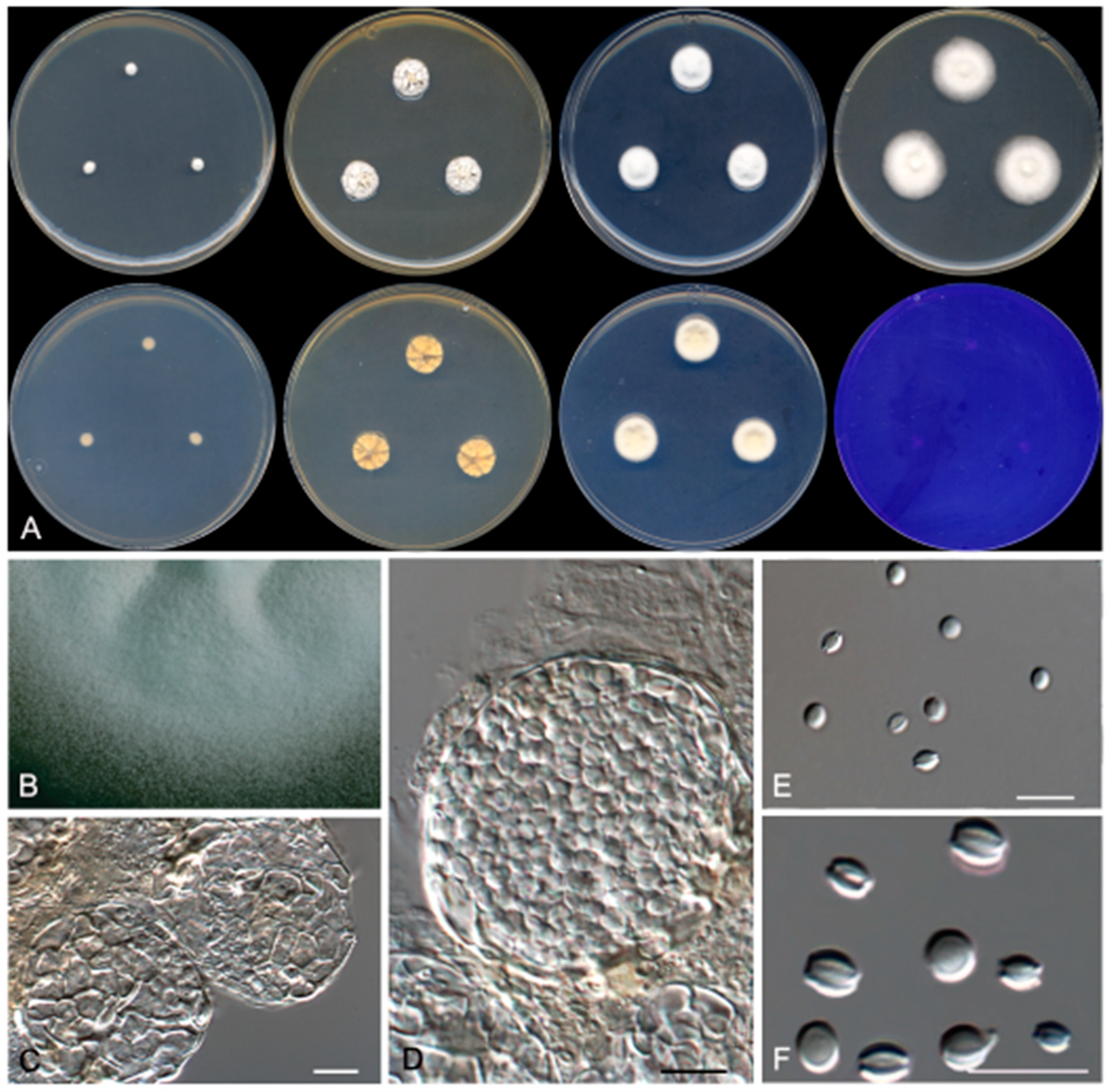
2.2. Phenotypic Study
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Phylogenetic Analysis
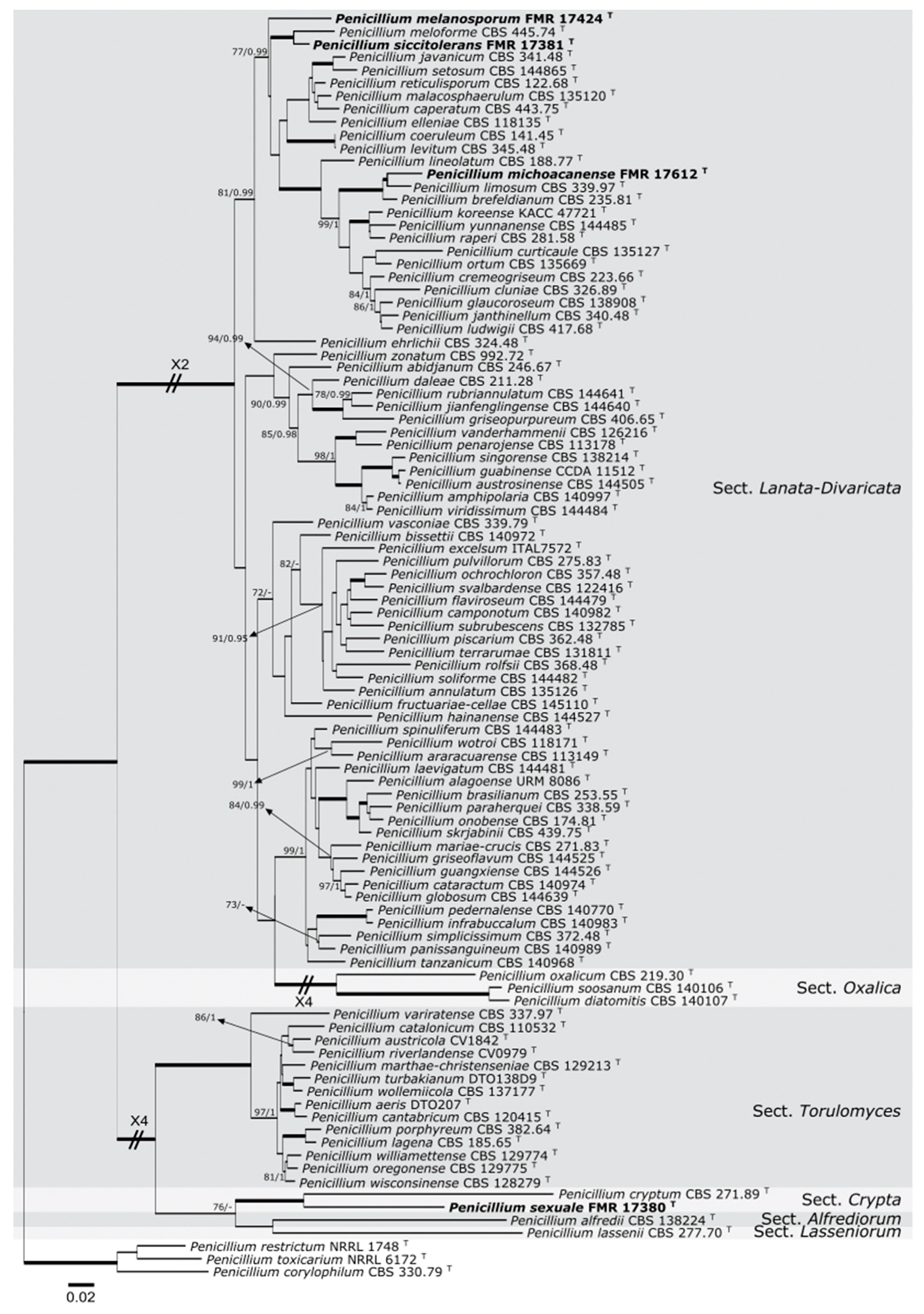
3. Results
3.1. Molecular Phylogeny
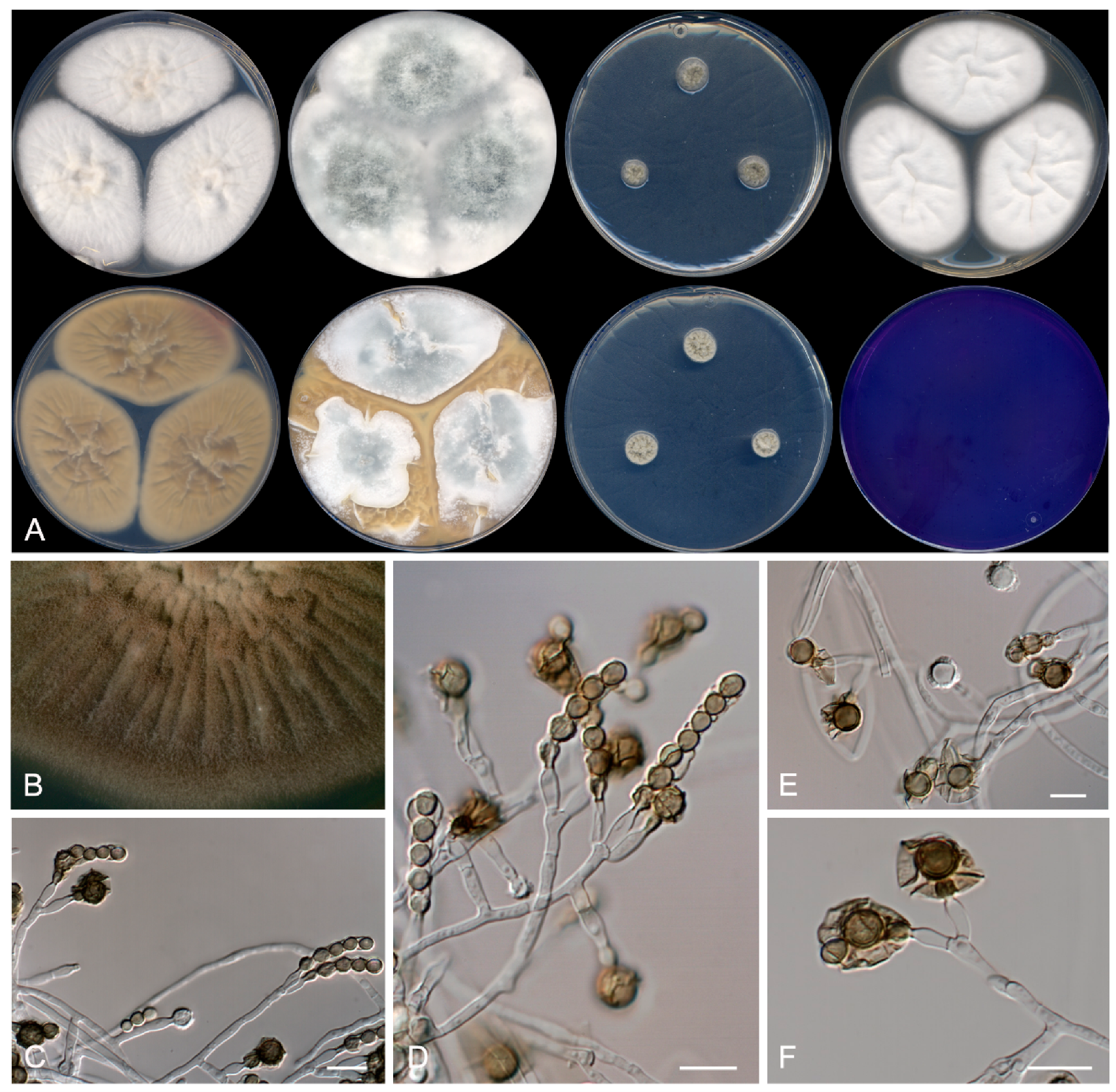
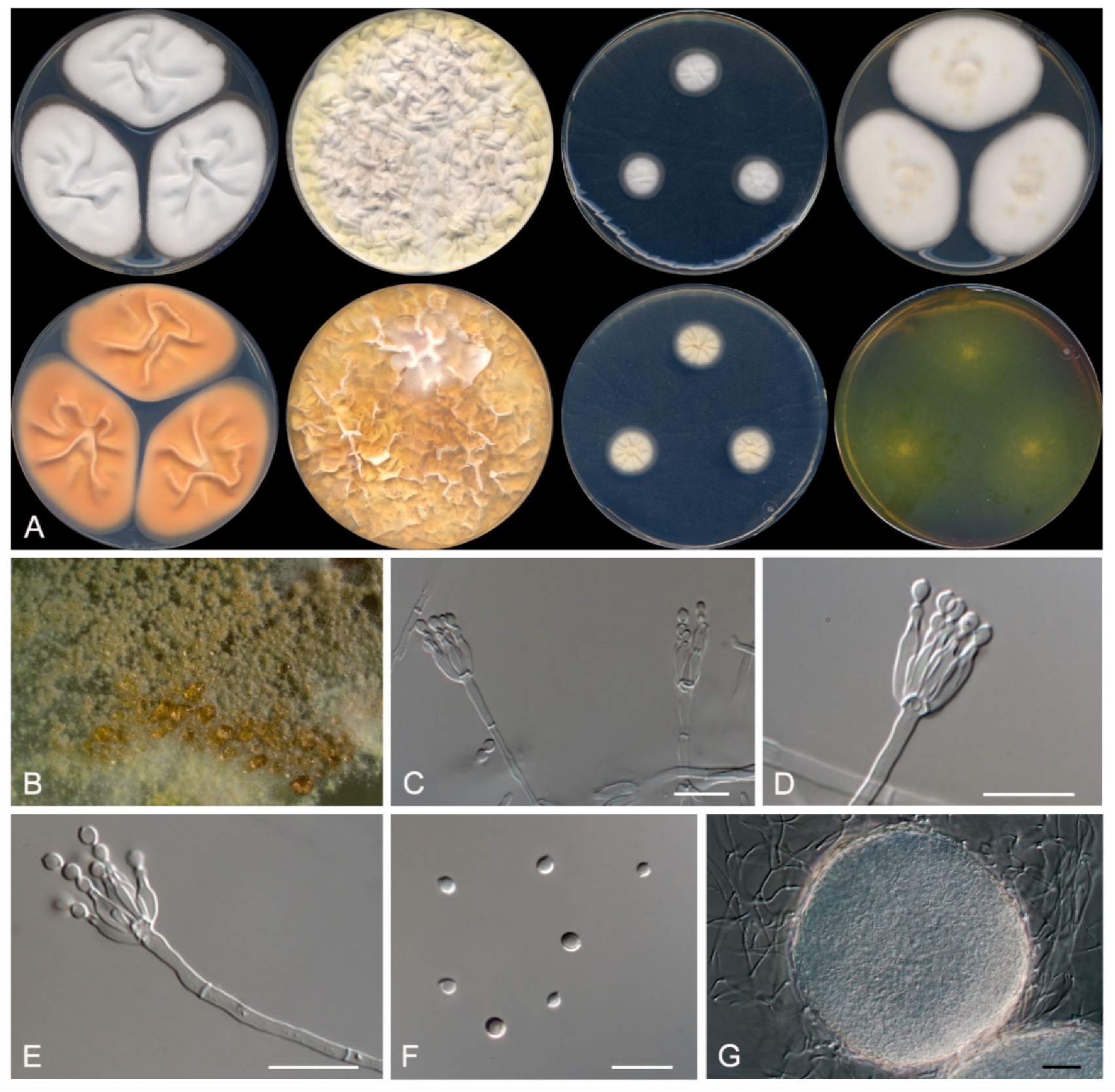
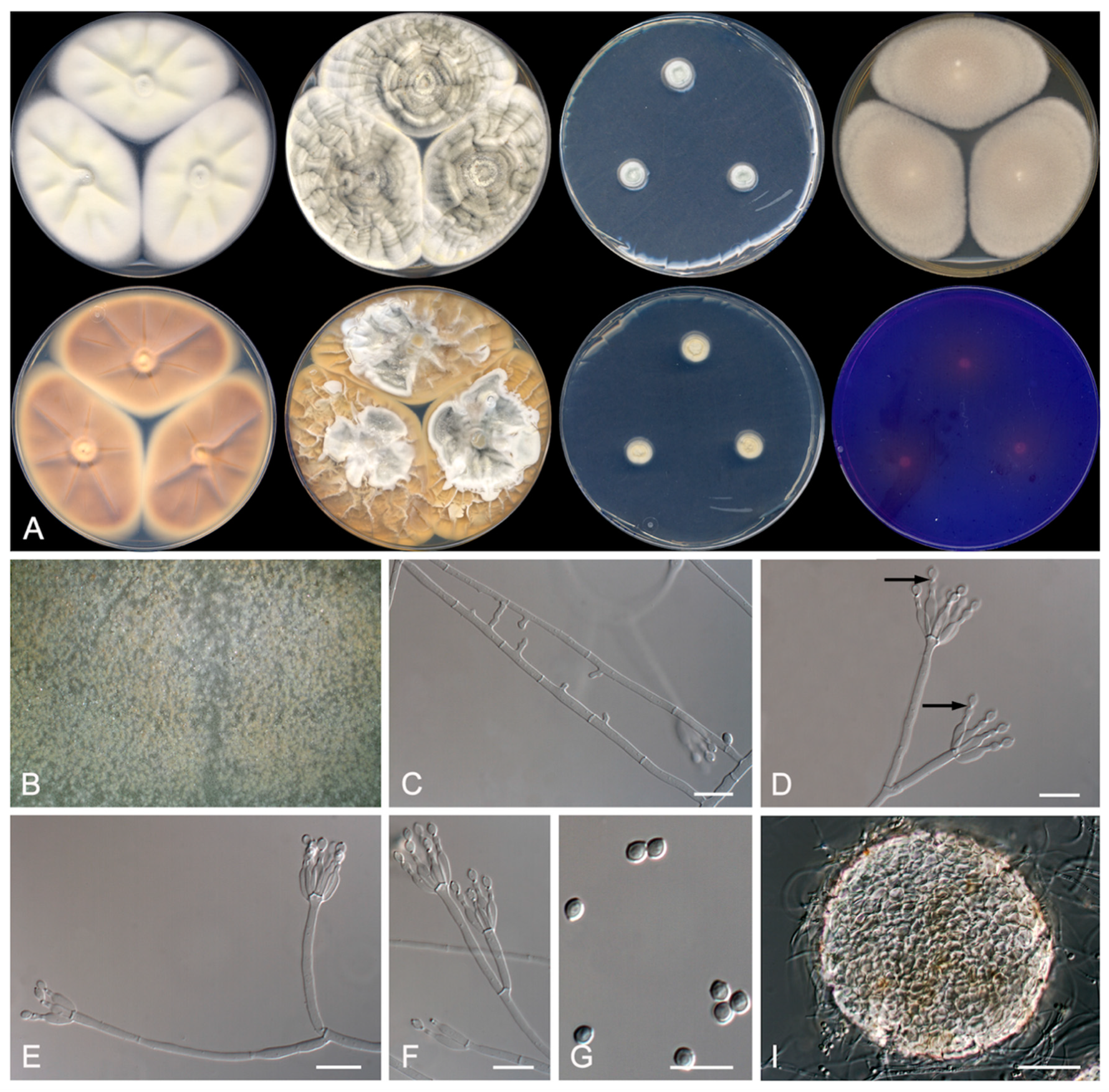
3.2. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garrett, P.W. Investigations of Pinus subsection strobi Loud. in Eastern United States. In Proceedings of the XVII IUFRO World Congress, Division 2, Kyoto, Japan, 6–17 September 1981; pp. 177–179. [Google Scholar]
- Carlile, M.J.; Watkinson, S.C. The Fungi; Academic Press: London, UK, 1994. [Google Scholar]
- Link, H.F. Observationes in ordines platarum naturales. Dissertatio 1ma. Mag. Ges. Naturf. Freunde Berl. 1809, 3, 3–42. [Google Scholar]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Robbins, E. Acidophilic and acid-tolerant fungi and yeasts. Hydrobiologia 2000, 433, 91–109. [Google Scholar] [CrossRef]
- Geiser, D.M.; Gueidan, C.; Miadlikowska, J.; Lutzoni, F.; Kauff, F. Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 2006, 98, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Dhakar, K.; Sharma, A.; Pandey, A. Cold, pH and salt tolerant Penicillium spp. inhabit the high altitude soils in Himalaya, India. World J. Microbiol. Biotechnol. 2014, 30, 1315–1324. [Google Scholar] [CrossRef]
- Houbraken, J.; Wang, L.; Lee, H.B.; Frisvad, J.C. New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 2016, 36, 299–314. [Google Scholar] [CrossRef]
- Barbosa, R.N.; Bezerra, J.D.; Souza-Motta, C.M.; Frisvad, J.C.; Samson, R.A.; Oliveira, N.T.; Houbraken, J. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek 2018, 111, 1883–1912. [Google Scholar] [CrossRef]
- Yong-Zhao, D.; Qian, C.; Xian-Zhi, J.; Houbraken, J.; Barbosa, R.N.; Cai, L.; Wu, W.P. Penicillium section Lanata-Divaricata from acidic soil. Cladistics 2018, 35, 514–549. [Google Scholar]
- Leong, S.L.; Pettersson, O.V.; Rice, T.; Hocking, A.D.; Schnurer, J. The extreme xerophilic mould Xeromyces bisporus-growth and competition at various water activities. Int. J. Food Microbiol. 2011, 145, 57–63. [Google Scholar] [CrossRef]
- Stchigel, A.M.; Cano, J.; Guarro, J. A new Gelasinospora from Argentinian soil. Mycol. Res. 1998, 102, 1405–1408. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Kirk, P.M.; Sutton, B.C.; Pegler, D.N. Ainsworth & Bisby‘s Dictionary of the Fungi, 8th ed.; CAB International: Wallingford, UK, 1995. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010. [Google Scholar]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press: London, UK, 1979. [Google Scholar]
- Frisvad, J.C. Physiological criteria and mycotoxin production as aids in identification of common asymmetric penicillia. Appl. Environ. Microbiol. 1981, 41, 568–579. [Google Scholar] [CrossRef]
- Hocking, A.D.; Pitt, J.I. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl. Environ. Microbiol. 1980, 39, 488–492. [Google Scholar] [CrossRef]
- Pitt, J.I. An appraisal of identification methods for Penicillium species: Novel taxonomic criteria based on temperature and water relations. Mycologia 1973, 62, 1137–1157. [Google Scholar]
- Beuchat, L.R.; Hocking, A.D. Some considerations when analyzing foods for the presence of xerophilic fungi. J. Food Prot. 1990, 53, 984–989. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978. [Google Scholar]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- Marimon, R.J.; Gené, J.; Cano, J.; Trilles, L.; Dos Santos, L.M.; Guarro, J. Molecular phylogeny of Sporothrix schenckii. J. Clin. Microbiol. 2006, 44, 3251–3256. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldo, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.B.; Go, S.J.; Shin, H.D.; Frisvad, J.C.; Samsom, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationship among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Valenzuela-Lopez, N.; Cano-Lira, J.F.; Guarro, J.; Sutton, D.A.; Wiederhold, N.; Crous, P.W.; Stchigel, A.M. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2018, 90, 1–69. [Google Scholar] [CrossRef]
- Udagawa, S.I.; Horie, Y. Some Eupenicillium from soils of New Guinea. Trans. Mycol. Soc. Jpn. 1973, 14, 370–387. [Google Scholar]
- Ueda, S. A new species of Eupenicillium from marine sediment. Mycoscience 1995, 36, 451–454. [Google Scholar] [CrossRef]
- Gochenaur, S.E.; Cochrane, E. Eupenicillium cryptum sp. nov., a fungus with self-limiting growth and restricted carbon nutrition. Mycotaxon 1986, 26, 345–360. [Google Scholar]
- Christensen, C.M.; Papavizas, G.C.; Benjamin, C.R. A new halophilic species of Eurotium. Mycologia 1959, 51, 636–640. [Google Scholar] [CrossRef]
- Kozakiewicz, Z. Aspergillus Species on Stored Products; C.A.B. International: Wallingford, UK, 1989. [Google Scholar]
- Hawsworth, D.L.; Pitt, J.L. A new taxonomy for Monascus species based on cultural and microscopial characters. Aus. J. Bot. 1983, 31, 51–61. [Google Scholar] [CrossRef]
- Hocking, A.D.; Pitt, J.L. Two new species of xerophilic and a further record of Eurotium halophilicum. Mycologia 1988, 80, 82–88. [Google Scholar] [CrossRef]
- Barbosa, R.N.; Leong, S.L.; Petterson, O.V.; Chen, A.J.; Souza-Motta, C.M.; Frisvad, J.C.; Samson, R.A.; Oliveira, N.T.; Houbraken, J. Phylogenetic analysis of Monascus and new species from honey, pollen and nests of stinglees bees. Stud. Mycol. 2018, 86, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I.; Lantz, H.; Petterson, O.V.; Leong, S.L. Xerochrysium gen. nov. and Bettsia, genera encompassing xerophilic species of Chrysosporium. IMA Fungus 2013, 4, 229–241. [Google Scholar] [CrossRef]
- Stchigel, A.M. Xeromyces: The Most Extreme Xerophilic Fungus. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press (Elsevier): London, UK, 2014; pp. 818–822. [Google Scholar]
- Hocking, A.D.; Pitt, J.I. Water relations of some Penicillium species at 25 °C. Trans. Br. Mycol. Soc. 1979, 73, 141–145. [Google Scholar] [CrossRef]
- Abellana, M.; Sanchis, V.; Ramos, A.J. Effect of water activity and temperature on growth of three Penicillium species and Aspergillus flavus on a sponge cake analogue. Int. J. Food Microbiol. 2001, 71, 151–157. [Google Scholar] [CrossRef]
- Fontana, A.J. Appendix D: Minimum water activity limits for growth of microorganisms. In Water Activity in Foods: Fundamentals and Applications; Barbosa-Cánovas, G.V., Fontana, A.J., Schmidt, S.J., Labuza, T.P., Eds.; Blackwell Publishing Ldt: Hoboken, NJ, USA, 2008. [Google Scholar]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Pettersson, O.V.; Leong, S.L. Fungal xerophiles (osmophiles). In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar]
- Kalai, S.; Anzala, L.; Bensoussan, M.; Dantigny, P. Modelling the effect of temperature, pH, water activity, and organic acids on the germination time of Penicillium camemberti and Penicillium roqueforti conidia. Int. J. Food Microbiol. 2017, 240, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Visagie, C.M.; Meijer, M.; Frisvad, J.; Busby, P.E.; Pitt, J.I.; Seifert, K.A.; Louis-Seize, G.; Demirel, R.; Yilmaz, N.; et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud. Mycol. 2014, 78, 373–451. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Andrade, E.; Stchigel, A.M.; Cano-Lira, J.F. New Xerophilic Species of Penicillium from Soil. J. Fungi 2021, 7, 126. https://doi.org/10.3390/jof7020126
Rodríguez-Andrade E, Stchigel AM, Cano-Lira JF. New Xerophilic Species of Penicillium from Soil. Journal of Fungi. 2021; 7(2):126. https://doi.org/10.3390/jof7020126
Chicago/Turabian StyleRodríguez-Andrade, Ernesto, Alberto M. Stchigel, and José F. Cano-Lira. 2021. "New Xerophilic Species of Penicillium from Soil" Journal of Fungi 7, no. 2: 126. https://doi.org/10.3390/jof7020126
APA StyleRodríguez-Andrade, E., Stchigel, A. M., & Cano-Lira, J. F. (2021). New Xerophilic Species of Penicillium from Soil. Journal of Fungi, 7(2), 126. https://doi.org/10.3390/jof7020126







