Additions to Lyophyllaceae s.l. from China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Sampling
2.2. Morphological Observation
2.3. DNA Extraction, PCR, Sequencing and Phylogenetic Analyses
2.4. Phylogenetic Analyses
3. Results
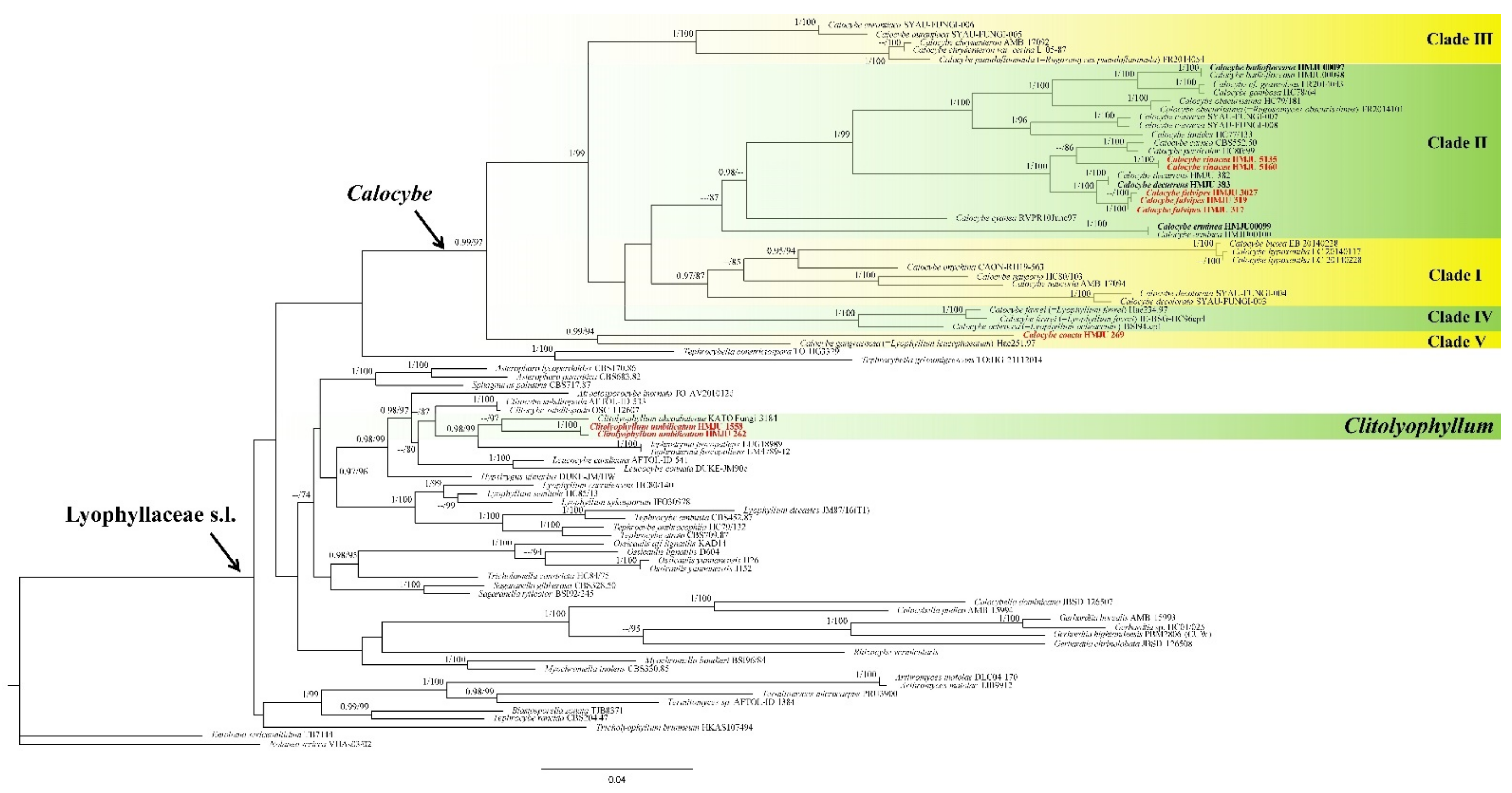
3.1. Phylogenetic Analyses
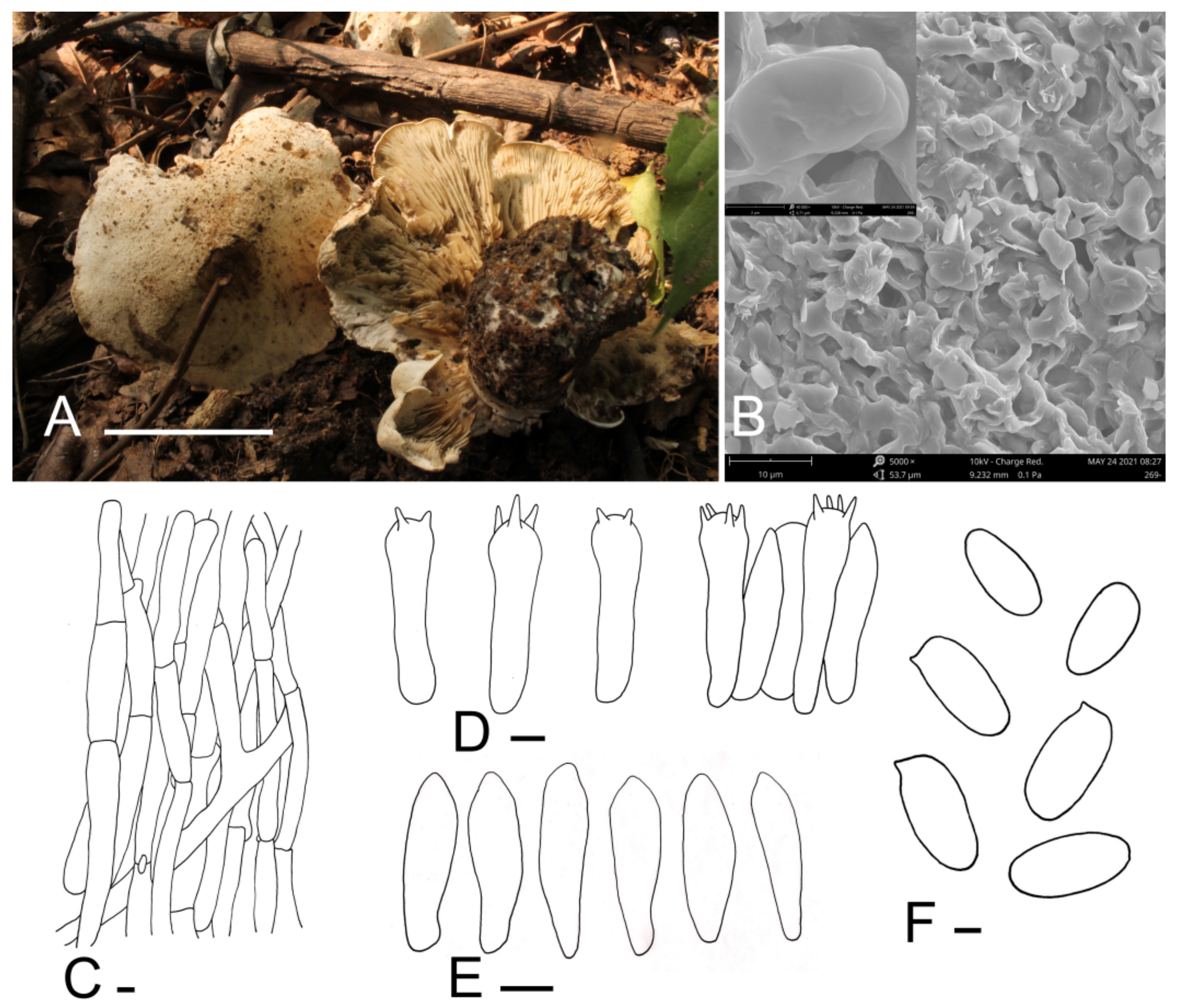
3.2. Taxonomy
4. Discussion
| 1. Basidiocarps medium to large, pileus usually more than 6 cm diam, stipe more than 3.5 cm long | 2 |
| 1’. Basidiocarps small, pileus usually less than 6 cm diam, stipe less than 3.5 cm long | 6 |
| 2. Lamellae sinuate | 3 |
| 2’. Lamellae not sinuate | 4 |
| 3. Pileus bruised blue, spore non-smooth | C. gangraenosa |
| 3’. Pileus bruised unchanged, spore smooth | C. gambosa |
| 4. Lamellae adnexed to slightly emarginate, lamellae less than 3 mm width | C. fulvipes |
| 4’. Lamellae decurrent, lamellae more than 3 mm width | 5 |
| 5. Stipe base broad, lamellae greyish orange when bruised, cystidia present | C. coacta |
| 5’. Stipe base narrow, lamellae unchanged when bruised, cystidia absent | C. decurrens |
| 6. Lamellae bruised blue | C. decolorata |
| 6’. Lamellae unchanged when bruised | 7 |
| 7. Stipe base with white pubescence | C. badiofloccosa |
| 7’. Stipe base without white pubescence | 8 |
| 8. Lamellae decurrent, pileus left plane and slightly depressed | 9 |
| 8’. Lamellae not decurrent, pileus left convex | 10 |
| 9. Pileus margin regular, stipe apex with whitish pruina | C. erminea |
| 9’. Pileus margin flexuous, stipe apex without whitish pruina | C. aurantiaca |
| 10. Lamellae sinuate, pileus with purple tones. | C. ionides |
| 10’. Lamellae adnate, pileus without purple tones | 11 |
| 11. Lamellae white | 12 |
| 11’. Lamellae not white | 13 |
| 12. Pileus yellow, pileus surface fibrillose, basidiospores less than 5 μm length | C. convexa |
| 12’. Pileus pink, pileus surface smooth, basidiospores more than 5 μm length | C. carnea |
| 13. Lamellae with yellow tones | 14 |
| 13’. Lamellae with taupe tones | C. obscurissima |
| 14. Pileus diam more than 5.5 cm, Stipe with pruinose at apex | C. chrysenteron |
| 14’. Pileus diam less than 5.5 cm, Stipe without pruinose at apex | 15 |
| 15. Pileus surface pastel red to dullred, basidiospores more than 4 μm length | C. vinacea |
| 15’. Pileus surface reddish yellow, basidiospores less than 4 μm length | C. naucoria |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hofstetter, V.; Clémençon, H.; Vilgalys, R.; Moncalvo, J.-M. Phylogenetic analyses of the Lyophylleae (Agaricales, Basidiomycota) based on nuclear and mitochondrial rDNA sequences. Mycol. Res. 2002, 106, 104–1059. [Google Scholar] [CrossRef]
- Hofstetter, V.; Redhead, S.A.; Kauff, F.; Moncalvo, J.-M.; Matheny, P.B.; Vilgalys, R. Taxonomic Revision and Examination of Ecological Transitions of the Lyophyllaceae (Basidiomycota, Agaricales) Based on a Multigene Phylogeny. Cryptogam. Mycol. 2014, 35, 399–425. [Google Scholar] [CrossRef]
- Bellanger, J.-M.; Moreau, P.-A.; Corriol, G.; Bidaud, A.; Chalange, R.; Dudova, Z.; Richard, F. Plunging hands into the mushroom jar: A phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 2015, 143, 169–194. [Google Scholar] [CrossRef]
- Dentinger, B.T.M.; Gaya, E.; O’Brien, H.; Suz, L.M.; Lachlan, R.; Díaz-Valderrama, J.R.; Koch, R.A.; Aime, M.C. Tales from the crypt: Genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol. J. Linn. Soc. 2016, 117, 11–32. [Google Scholar] [CrossRef] [Green Version]
- Alvarado, P.; Moreau, P.-A.; Dima, B.; Vizzini, A.; Consiglio, G.; Moreno, G.; Setti, L.; Kekki, T.; Huhtinen, S.; Liimatainen, K.; et al. Pseudoclitocybaceae fam. nov. (Agaricales, Tricholomatineae), a new arrangement at family, genus and species level. Fungal Divers. 2018, 90, 109–133. [Google Scholar] [CrossRef]
- Vizzini, A.; Consiglio, G.; Setti, L.; Ercole, E. Calocybella, a new genus for Rugosomyces pudicus (Agaricales, Lyophyllaceae) and emendation of the genus Gerhardtia. IMA Fungus 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sesli, E.; Vizzini, A.; Ercole, E.; Contu, M. Clitolyophyllum akcaabatense gen. nov., sp. nov. (Agaricales, Tricholomatineae); a new fan-shaped clitocyboid agaric from Turkey. Botany 2016, 94, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Redhead, S.A. Nomenclatural novelties. Index Fungorum 2014, 202, 1. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Hernández-Restrepo, M.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; et al. Fungal Planet description sheets: 320–370. Persoonia 2015, 34, 167–266. [Google Scholar] [CrossRef]
- Cai, Q.; LvLi, Y.-J.; Kost, G.; Yang, Z.-L. Tricholyophyllum brunneum gen. et. sp. nov. with bacilliform basidiospores in the family Lyophyllaceae. Mycosystema 2020, 39, 1–13. [Google Scholar]
- Kühner, R. Utilisation du carmin acétique dans la classification des agarics leucosporés. Bulletin Mensuel Societe Linneenne Lyon 1938, 7, 204–212. [Google Scholar] [CrossRef]
- Singer, R. The Agaricales in Modern Taxonomy, 4th ed.; Koeltz Scientific Books: Koenigstein, Germany, 1986; p. 981. [Google Scholar]
- Kühner, R.; Romagnesi, H. Flore Analytique des Champignons Supérieurs (Agarics, Boelts, Chanterelles); Masson: Paris, France, 1953; pp. 1–556. [Google Scholar]
- Arnolds, E. A confusing duo: Calocybe cerina and Callistosporium pinicola (Agaricales). Acta Mycol. 2013, 41, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-J.; Wu, S.-Y.; Yu, X.-D.; Zhang, S.-B.; Cao, D.-X. Three new species of Calocybe (Agaricales, Basidiomycota) from northeastern China are supported by morphological and molecular data. Mycologia 2017, 109, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yu, X.; Zhang, C.; Li, Y. Two new species of Calocybe (Lyophyllaceae) from northeast China. Phytotaxa 2019, 425, 219–232. [Google Scholar] [CrossRef]
- Xu, J.; Yu, X.; Zhang, C.; Li, Y. Morphological characteristics and phylogenetic analyses revealed a new Calocybe (Lyophyllaceae, Basidiomycota) species from northeast China. Phytotaxa 2021, 490, 203–210. [Google Scholar] [CrossRef]
- Consiglio, G.; Contu, M. Il genere Lyophyllum P. Karst. emend. Kühner, in Italia. Rivista di Micologia 2002, 45, 99–181. [Google Scholar]
- Kalamees, K. Palearctic Lyophyllaceae (Tricholomataceae) in northern and eastern Europe and Asia. Scr. Mycol. 2004, 18, 3–134. [Google Scholar]
- Yang, S.-D.; Huang, H.-Y.; Zhao, J.; Zeng, N.-K.; Tang, L.-P. Ossicaulis yunnanensis sp. nov. (Lyophyllaceae, Agaricales) from southwestern China. Mycoscience 2018, 59, 33–37. [Google Scholar] [CrossRef]
- Ding, Y.; Tian, E.; Alvarado, P. A new species of Rhizocybe (tricholomatoid clade, Agaricales) from Yunnan, China. Phytotaxa 2017, 328, 267. [Google Scholar] [CrossRef]
- Mu, M.; Huang, H.-Y.; Huang, T.; Yang, S.-D.; Tang, L.-P. Gerhardtia yunnanensis (Agaricales, Lyophyllaceae), a new species from southwest China. Phytotaxa 2021, 484, 217–226. [Google Scholar] [CrossRef]
- Li, T.; Deng, W.; Wang, C.; Song, B. Gerhardtia sinensis (Agaricales, Lyophyllaceae), a new species and a newly recorded genus for China. Phytotaxa 2017, 332, 172. [Google Scholar] [CrossRef]
- Zhang, J.W.; Ma, S.Y.; Qi, L.L.; Li, Y. Macrofungal flora diversity in Liangshui Nature Reserve, Heilongjiang Province. J. Fungal Res. 2017, 15, 170–176. [Google Scholar]
- Wang, Y. Study on the Diversity of Macrofungi in Tanggou Greenstone Valley National Forest Park, Liaoning Province; Jilin Agricultural University: Changchun, China, 2019; pp. 1–79. [Google Scholar]
- Mao, X.L. The Macrofungi in China; Henan Science Technology Press: Zhengzhou, China, 2000; pp. 1–719. [Google Scholar]
- Li, Y.; Li, T.H.; Yang, Z.L.; Bau, T.; Dai, Y.C. Atlas of Chinese Macrofungal Resources; Central China Farmers Publishing House: Zhengzhou, China, 2015; pp. 1–1351. [Google Scholar]
- Kornerup, A.; Wanscher, J.H.K. The Methuen Handbook of Colour; Eyre Methuen: London, UK, 1978; pp. 1–252. [Google Scholar]
- Xu, J.; Yu, X.; Lu, M.; Hu, J.; Moodley, O.; Zhang, C.; Gong, L.; Li, Y. Phylogenetic Analyses of Some Melanoleuca Species (Agaricales, Tricholomataceae) in Northern China, With Descriptions of Two New Species and the Identification of Seven Species as a First Record. Front. Microbiol. 2019, 10, 2167. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Chethana, K.W.T.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.-Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhir, K.; Glen, S.; Koichiro, T. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml-vi-hpc: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floriani, M.; Vizzini, A. Calocybe pilosella sp. nov., a distinctive new lyophylloid agaric collected near Trento (Italy). Studi Trentini di Scienze Naturali 2016, 95, 17–24. [Google Scholar]
- Fries, E.M. Systema Mycologicum I: i–vii; Ex Officina Berlingiana: Lund, Sweden; Greifswald, Germany, 1821. [Google Scholar]
- Singer, R. Das System der Agaricales. III. Annales Mycologici 1943, 41, 1–189. [Google Scholar]
- Singer, R. New and interesting species of Basidiomycetes. II. Pap. Mich. Acad. Sci. 1948, 32, 103–150. [Google Scholar]
- Redhead, S.A.; Singer, R. On Calocybe names. Mycotaxon 1978, 6, 500–502. [Google Scholar]
- Singer, R. Keys for the identification of the species of Agaricales I. Sydowia 1978, 30, 192–279. [Google Scholar]



| Species | Collection | GenBank Accession Numbers | |
|---|---|---|---|
| ITS | 28S | ||
| Arthromyces matolae | TJB9912 | EU708338 | EU708335 |
| Arthromyces matolae | DLC04-170 | EU708339 | EU708336 |
| Asterophora lycoperdoides | CBS170.86 | AF357037 | AF223190 |
| Asterophora parasitica | CBS683.82 | AF357038 | AF223191 |
| Atractosporocybe inornata | TO AV201012d | KJ680993 | KJ681046 |
| Blastosporella zonata | TJB8371 | EU708340 | EU708337 |
| Calocybe aurantiaca | SYAU-FUNGI-005 | KU528828 | KU528833 |
| Calocybe aurantiaca | SYAU:FUNGI 006 | NR156304 | NG058937 |
| Calocybe badiofloccosa | HMJU00099 | OK649912 | OK649882 |
| Calocybe badiofloccosa | HMJU00100 | MN172331 | MN172333 |
| Calocybe buxea | EB 20140228 | KP885633 | KP885625 |
| Calocybe carnea | CBS552.50 | AF357028 | AF223178 |
| Calocybe cf. graveolens | FR2014043 | KP192609 | – |
| Calocybe chrysenteron | AMB 17092 | KP885639 | KP885628 |
| Calocybe chrysenteron var.cerina | L 05-87 | KP885640 | KP885629 |
| Calocybe coata | HMJU269 | OK649907 | OL687156 |
| Calocybe convexa | SYAU-FUNGI-007 | KU528826 | KU528830 |
| Calocybe convexa | SYAU:FUNGI 008 | NR156303 | NG058936 |
| Calocybe cyanea | RVPR10June97 | – | AF261400 |
| Calocybe decolorata | SYAU-FUNGI-003 | KU528824 | KU528834 |
| Calocybe decolorata | SYAU:FUNGI 004 | NR_156302 | NG_058938 |
| Calocybe decurrens | HMJU 382 | MT080028 | MW444857 |
| Calocybe decurrens | HMJU 383 | OK649913 | OK649883 |
| Calocybe erminea | HMJU00097 | OK649911 | OK649881 |
| Calocybe erminea | HMJU00098 | MN172332 | MN172334 |
| Calocybe fulvipes | HMJU 317 | MT071590 | OK649878 |
| Calocybe fulvipes | HMJU 319 | MW406907 | OK649879 |
| Calocybe fulvipes | HMJU 3027 | OK649910 | OK649880 |
| Calocybe gambosa | HC78/64 | AF357027 | AF223177 |
| Calocybe gangraenosa | Hae251.97 | AF357032 | AF223202 |
| Calocybe hypoxantha | EC 20140117 | KP885634 | KP885626 |
| Calocybe hypoxantha | EC 20140228 | KP885635 | – |
| Calocybe ionides | HC77/133 | AF357029 | AF223179 |
| Calocybe naucoria | AMB 17094 | KP885642 | KP885630 |
| Calocybe naucoria | HC80/103 | AF357030 | AF223180 |
| Calocybe obscurissima | HC79/181 | AF357031 | AF223181 |
| Calocybe onychina | CAON-RH19-563 | MW084664 | MW084704 |
| Calocybe persicolor | HC80/99 | AF357026 | AF223176 |
| Calocybe vinacea | HMJU5135 | OK649908 | OK649876 |
| Calocybe vinacea | HMJU5160 | OK649909 | OK649877 |
| Calocybella dominicana | JBSD 126507 | KY363575 | KY363577 |
| Calocybella pudica | AMB 15994 | KP858000 | KP858005 |
| Clitocybe subditopoda | AFTOL-ID 533 | DQ202269 | AY691889 |
| Clitocybe subditopoda | OSC 112607 | EU697244 | EU852807 |
| Clitolyophyllum akcaabatense | KATO Fungi 3184 | KT934393 | KT934394 |
| Clitolyophyllum umbilicatum | HMJU 262 | OK649905 | OK649873 |
| Clitolyophyllum umbilicatum | HMJU 1558 | OK649906 | OK649874 |
| Entoloma sericeonitidum | TB7144 | EF421108 | AF261315 |
| Gerhardtia borealis | AMB 15993 | KP858004 | KP858009 |
| Gerhardtia citrinolobata | JBSD 126508 | KY363576 | KY363578 |
| Gerhardtia highlandensis | PBM2806 (CUW) | GU734744 | EF535275 |
| Gerhardtia sp. | HC01/025 | EF421103 | EF421091 |
| Hypsizygus ulmarius | DUKE-JM/HW | EF421105 | AF042584 |
| Leucocybe candicans | AFTOL-ID 541 | DQ202268 | AY645055 |
| Leucocybe connata | DUKE-JM90c | EF421104 | AF042590 |
| Lyophyllum caerulescens | HC80/140 | AF357052 | AF223209 |
| Lyophyllum decastes | JM87/16(T1) | AF357059 | AF042583 |
| Lyophyllum favrei | IE-BSG-HC96cp4 | EF421102 | AF223184 |
| Lyophyllum favrei | Hae234.97 | AF357034 | AF223183 |
| Lyophyllum leucophaeatum | Hae251.97 | AF357032 | AF223202 |
| Lyophyllum ochraceum | BSI94.cp1 | AF357033 | AF223185 |
| Lyophyllum semitale | HC85/13 | AF357049 | AF042581 |
| Lyophyllum sykosporum | IFO30978 | AF357050 | AF223208 |
| Myochromella boudieri | BSI96/84 | AF357047 | DQ825430 |
| Myochromella inolens | CBS330.85 | AF357045 | AF223201 |
| Nolanea sericea | VHAs03/02 | DQ367430 | DQ367423 |
| Ossicaulis aff. lignatilis | KAD14 | MG663236 | MT237454 |
| Ossicaulis lignatilis | D604 | DQ825426 | AF261397 |
| Ossicaulis yunnanensis | IJ152 | KY411962 | KY411960 |
| Ossicaulis yunnanensis | IH26 | KY411961 | KY411959 |
| Rugosomyces obscurissimus | FR2014101 | KP192650 | – |
| Rugosomyces pseudoflammulus | FR2014054 | KP192579 | – |
| Rhizocybe vermicularis | AH33078 | KJ681032 | – |
| Sagaranella gibberosa | CBS328.50 | AF357041 | AF223197 |
| Sagaranella tylicolor | BSI92/245 | AF357040 | AF223195 |
| Sphagnurus palustris | CBS717.87 | AF357044 | AF223200 |
| Tephrocybe ambusta | CBS452.87 | AF357057 | AF223216 |
| Tephrocybe anthracophila | HC79/132 | AF357055 | AF223212 |
| Tephrocybe atrata | CBS709.87 | AF357053 | AF223210 |
| Tephrocybe rancida | CBS204.47 | AF357025 | AF223203 |
| Tephrocybella constrictospora | TO HG3329 | MF614962 | MF614963 |
| Tephrocybella griseonigrescens | TO:HG 21112014 | NR137975 | KR476785 |
| Tephroderma fuscopallens | EM4789-12 | KJ701326 | KJ701332 |
| Tephroderma fuscopallens | LUG18989 | KJ701327 | KJ701333 |
| Termitomyces microcarpus | PRU3900 | AF357023 | AF042587 |
| Termitomyces sp. | AFTOL-ID 1384 | DQ494698 | DQ110875 |
| Tricholomella constricta | HC84/75 | AF357036 | AF223188 |
| Tricholyophyllum brunneum | HKAS107494 | MT705717 | MT734655 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Yu, X.; Suwannarach, N.; Jiang, Y.; Zhao, W.; Li, Y. Additions to Lyophyllaceae s.l. from China. J. Fungi 2021, 7, 1101. https://doi.org/10.3390/jof7121101
Xu J, Yu X, Suwannarach N, Jiang Y, Zhao W, Li Y. Additions to Lyophyllaceae s.l. from China. Journal of Fungi. 2021; 7(12):1101. https://doi.org/10.3390/jof7121101
Chicago/Turabian StyleXu, Jize, Xiaodong Yu, Nakarin Suwannarach, Yi Jiang, Wei Zhao, and Yu Li. 2021. "Additions to Lyophyllaceae s.l. from China" Journal of Fungi 7, no. 12: 1101. https://doi.org/10.3390/jof7121101
APA StyleXu, J., Yu, X., Suwannarach, N., Jiang, Y., Zhao, W., & Li, Y. (2021). Additions to Lyophyllaceae s.l. from China. Journal of Fungi, 7(12), 1101. https://doi.org/10.3390/jof7121101







