1. Introduction
The genera
Asproinocybe R. Heim (1970) and
Tricholosporum Guzmán (1975) are usually placed in Tricholomataceae due to their tricholomatoid basidioma [
1,
2,
3,
4,
5,
6].
Asproinocybe was originally described as
Leucinocybe Heim (1969) and typified by
Leucinocybe lactifera Heim (1969) [
7].
Leucinocybe is mainly characterized by indigo or violet basidiomata, hyaline and tuberculous spores, and the presence of laticifers [
7]. However,
Leucinocybe was used by Singer for accommodating
Mycena lenta Maire, meaning that
Leucinocybe Heim (1969) is invalid. Later, Heim (1970) proposed the new name
Asproinocybe, typified by
Asproinocybe lactifera [
8]. In the current sense, the genus is characterized by tricholomatoid; distinctive purplish, violaceous, or lilac-vinaceous basidioma; lamellae bruising reddish when damaged; spore hyaline and with irregularly tubercle; with laticifers present [
6,
7,
8,
9].
Tricholosporum was erected based on the combination of
Tricholoma goniospermum Bres. (as type) and
Tricholoma porphyrophyllum S. Imai [both from
Tricholoma section
Iorigida Singer (1945)] and the description of
Tricholosporum subporphyrophyllum Guzmán, due to its cruciform spores and lamella with lilac or purple pigments [
1,
10].
The segregation of the two genera was latter recognized, but the combinations were considered invalid because the original publications of the basionyms were not provided [
11]. Then,
Tricholosporum goniospermum and
T. porphyrophyllum, as well as
Tricholosporum atroviolaceum and
Tricholosporum pseudosordidum were added to
Tricholosporum [
11].
The independence of
Asproinocybe and
Tricholosporum was long debated. Singer recognized
Asproinocybe considered in tribe Tricholomateae [
12], but
Tricholosporum was considered synonym of
Tricholoma [
12,
13,
14,
15,
16]. On the other hand,
Asproinocybe and
Tricholosporum were also considered as independent entities [
2]. This opinion has been widely recognized [
3,
4,
6].
By now, eight species recognized in
Asproinocybe. Vicente et al. constructed a key for the species [
6,
7,
8,
9,
17]. With 14 species recognized in
Tricholosporum, Vicente et al. and Angelini et al. published a key for the species [
4,
5,
9].
Regarding the placement at the family level,
Asproinocybe was not indicated as belonging to a specific family when it was established: it was only compared with
Lyophyllum [
7,
8]. In 1977, Heinemann assigned
Asproinocybe russuloides to Tricholomataceae, later followed by Guzmán and Lebel et al. [
3,
6,
18].
Tricholosporum was established in Tricholomataceae [
1]. Thus,
Asproinocybe and
Tricholosporum were placed in Tricholomataceae for a long time based on morphological consideration.
However, more recently, phylogenetic studies are increasingly showing that they should not be placed in Tricholomataceae [
6,
19,
20]. The first phylogenetic approach recovered
Tricholosporum within Entolomataceae based on ITS dataset and within Tricholomataceae (s.l.) based on LSU [
19].
Later, Angelini et al. conducted a more comprehensive phylogenetic study on the relationships between
Tricholosporum and Tricholomatineae [
20]. They used ITS, LSU, SSU, and RPB2 DNA sequences to evaluate the phylogenetic position of
Tricholosporum within the clade of tricholomatoid fungi. Their analysis showed a weak relationship of
Tricholosporum in the clade of Tricholomataceae, and an isolated position of this genus within the Tricholomatineae. Their tree, based on SSU and RPB2 sequences, placed
Tricholosporum in the Entolomataceae/Lyophyllaceae, whereas the LSU and ITS trees placed
Tricholosporum within a group of morphologically heterogeneous species such as
Macrocybe gigantea,
Clitocybe fellea,
Pleurocollybia brunnescens, and
Callistosporium spp. The tree, combined RPB2-SSU-LSU sequences, showing a relationship of
Tricholosporum with the clade Entolomataceae, Lyophyllaceae, the Clytocybe/Lepista/Collybia, and the callistosporoid groups, but the relationship was poorly resolved and had weak bootstrap support [
20].
Both studies conducted a phylogenetic analysis of
Tricholosporum to find a suitable placement at the family level but failed. They confirmed that
Tricholosporum should not be placed in Tricholomataceae. However, the researchers only used a single or a few species of
Tricholosporum in the phylogenetic analysis. Heaton and Kropp postulated that using RPB1 would probably lead to a better understanding of the phylogenetic placement of
Tricholosporum [
19].
Only in 2020 were new species from
Asproinocybe found and included in phylogenetic analysis [
6]. Lebel et al. found two new species from
Asproinocybe and conducted a phylogenetic analysis based on ITS sequences only. They found that the species from
Asproinocybe and
Tricholosporum formed a clade, which suggested weak support for Biannulariaceae but strong support for a restricted Catathelasmataceae and for a clade with
Infundibulicybe,
Anupama,
Guyanagarika, Tricholomataceae sp.,
Asproinocybe, and
Tricholosporum as sisters to Catathelasmataceae [
6]. In a restricted multimarker analysis of a broad selection of taxa from Lyophyllaceae, Entolomataceae, and Tricholomatoid agarics, support for the placement of
Asproinocybe and
Tricholosporum in a broad Tricholomataceae was weak [
6].
Lebel et al. demonstrated the phylogenetic relationship between Asproinocybe and Tricholosporum for the first time but could not solve the phylogenetic problems at the family level, and confirmed that the idea of Tricholosporum being distinct from Asproinocybe was problematic. All the abovementioned phylogenetic studies were either conducted using only a single marker or included only a single species, which prevented the determination of the relationship between Asproinocybe and Tricholosporum and of the placement at the family level. A more comprehensive sampling of Asproinocybe and Tricholosporum and involving multimarker data in the phylogenetic analysis may help to solve these problems.
The aim of this study is to determine the family-level placement of Asproinocybe and Tricholosporum and to further discuss the relationships between Asproinocybe and Tricholosporum from morphology and phylogeny perspectives. Two new species from Asproinocybe and Tricholosporum are described.
4. Taxonomy
Asproinocybaceae T.Bau et G.F.Mou, fam. nov.
Mycobank No: MB841852
Etymology: From the type genus Asproinocybe.
Description: Habit tricholomatoid. Basidiomata with distinctive purplish, violaceous, or lilac-vinaceous colors. Pileus broadly convex, subumbonate to flat-hemispherical, becoming plane to depressed with age, margin smooth or with light and short stripes, entire, incurved at first then straight, surface at first fibrillose-felted (due to very thin, white hairs) then finely velvety but smooth toward the center, nonviscid, or subviscidus; with varying degrees of purplish, violaceous, or lilac-vinaceous colors in surface, especially near the margin, center more or less yellowish, yellowish ochre, yellowish brown, brown to dark brown colors. Context firm, white or whitish, becoming greyish or cream yellowing. Lamellae adnate, adnexed, sinuate or emarginate to free, sometimes with small decurrent tooth; lamellulae exist; margins smooth or unevenly serrate; close to crowded or crowded; pale violet to deep violet or greyish violet, bruising reddish or pale brown when damaged. Stipe solid to fistulose-hollow, cylindric to slightly clavate, central, pale violet, violet, greyish violet to bluish violaceous when fresh, covered by white to pale violet flocculose pruina, bruising dull, fading to whitish with age. Base usually with white rhizomorphs. Odor not distinct or fragrant. Taste not distinct or bitter or sour. Spore-print white.
Basidiospores hyaline, colorless, inamyloid, thin-walled, cyanophilous or not, subglobose to subellipsoid, tuberculate to stellate (Asproinocybe), or cruciform to stauriform (Tricholosporum), usually with a single large oil-drop. Basidia cylindric to narrowly clavate, two sterigmate or four sterigmate, thin-walled, colorless. Cheilocystidia and pleurocystidia usually similar, present or absent, oblong to ellipsoid, utriform, ampullaceous, fusiform or clavate, with a swollen base and a neck, acute or mucronate at apex, thin-walled or occasionally thick-walled, colorless or golden brown, or sometimes with pinkish violet content or grey-violet pigment. Hymenophoral trama regular, inamyloid, not dextrinoid, thin-walled. Pileipellis consisting of a cutis of loosely interwoven, cylindric to clavate hyphae, smooth or with incrustation. Clamp connections present or absent. Laticifers present, both in Asproinocybe and Tricholosporum.
Type genus: Asproinocybe R. Heim, Revue Mycol., Paris 34(4): 343 (1970).
Habit: Scattered or gregarious on broad-leaved forests soil, usually found in summer or autumn.
Genera included: Asproinocybe R. Heim, Tricholosporum Guzmán
Distribution:
Asproinocybe mainly distributed in tropics, whereas
Tricholosporum is widespread [
45].
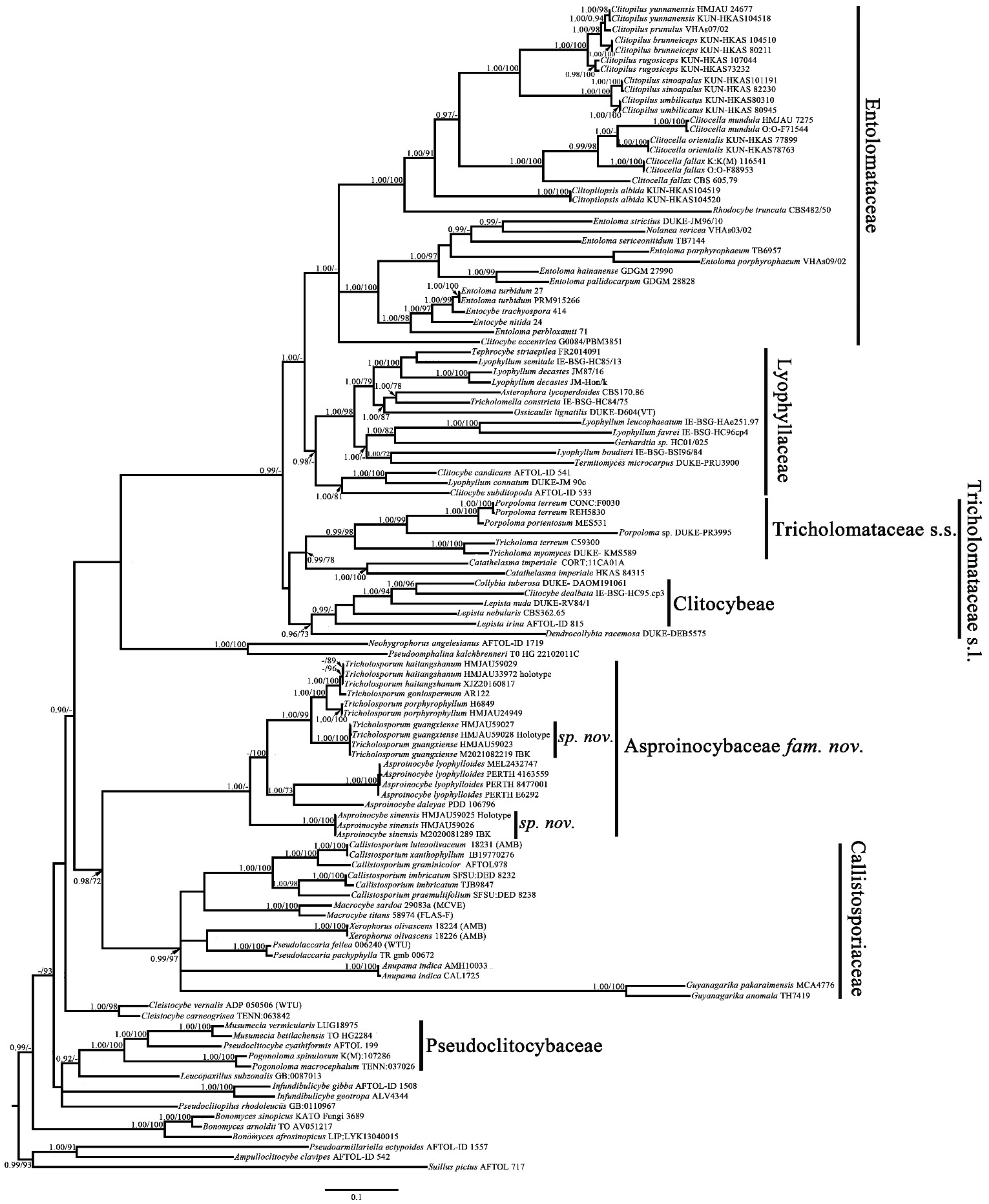
Notes: Our phylogenetic analysis results (based on partial ITS + nrLSU + RPB1 + RPB2 + nrSSU + TEF1-α) show that
Asproinocybe and
Tricholosporum form a single family-level clade and received strong statistical support (BPP = 0.98, BS = 72), and the clade is a sister to the Callistosporiaceae clade, which is in agreement with previously published phylogenetic results [
6,
20]. Taking all of the phylogenetic and morphological results into account, a new family,
Asproinocybaceae fam. nov., is proposed for the
Asproinocybe/
Tricholosporum clade.
The species of
Asproinocybe and
Tricholosporum are very similar in appearance: they can only be differentiated by the shape of the basidiospores. Some mycologists have discussed the split [
2,
6]. Our phylogenetic results (
Figure 1 and
Figure 2) show that the species of
Asproinocybe and
Tricholosporum always group together, but they do not form two single clades. However, when we removed the sequences from
Asproinocybe daleyae and
Asproinocybe lyophylloides and used the rest of the dataset in
Table S1 (in Supplementary Materials, the GenBank Accession numbers marked with asterisks) to reconstruct the phylogenetic tree, the species of
Asproinocybe and
Tricholosporum clearly formed two single clades (
Figure S1). However, when we removed the sequences of
Asproinocybe sinensis and used the rest of the dataset in
Table S1 (GenBank Accession numbers marked with asterisks) to reconstruct the phylogenetic tree, the species of
Asproinocybe or
Tricholosporum clearly formed two single clades (
Figure S2).
The morphological characteristics of our specimens (Asproinocybe sinensis) meet the definition of Asproinocybe; therefore, they must belong to Asproinocybe. Regarding why it did not form a single clade with Asproinocybe daleyae and Asproinocybe lyophylloides, we postulate that this may be due to the lack of sampling of species from Asproinocybe. When more species from Asproinocybe are included in the phylogenetic analysis, these questions may be able to be answered.
Thus, taking the results of
Figures S3 and S4 and the stable shape of spores into account, we still treat
Tricholosporum as being distinct from
Asproinocybe.
Asproinocybe R. Heim, Revue Mycol., Paris 34(4): 343 (1970).
Basionym: Leucinocybe Heim, Cah. de La maboké, VII, 2, 1969, p. 83.
Etymology: From the tuberculate basidiospores, similar to Inocybe but colorless.
Type species: Asproinocybe lactifera R. Heim 1970.
Basionym: Leucinocybe lactifera Heim, Cah. de La maboké, VII, 2, 1969, p. 83–85.
Ecology and distribution: Scattered or gregarious in broad-leaved forest soil, mainly distributed in the tropics.
Mycobank No: MB841850.
Diagnosis: Differs from other known species of this genus by the central pileus being dark brown, with larger basidia (33 × 10 µm on average); cheilocystidia (30–40 × 8–10 µm) and pleurocystidia (38 × 9 µm on average) present and the apex, not branched; hyphae of pileipellis with fine incrustation.
Etymology: sinensis (Lat.): The locality of the type specimen was China.
Type: China, Guangxi province, Baise city, Leye country, Yachang Orchidaceae National Nature Reserve, 24°50′51.48″ N, 106°24′55.43″ E, elevation 1053 m, 12 August 2020, Guang-fu Mou HMJAU59025 (Holotype HMJAU!).
Description:
Basidiomata tricholomatoid
habit, solitary to gregarious (
Figure 3).
Pileus 35–55 mm in diam., broadly convex to subumbonate, becoming plane with age, margin smooth or with light short stripes, entire, incurved at first then straight, surface at first fibrillose-felted (due to very thin, white hairs), nonviscid; overall color from center to margin is dark brown (6E8), brown (6E6) to brownish orange (6C5), lilac grey (16C2) to violet (16C6).
Context 3–8.5 mm thick, firm, whitish becoming greyish.
Lamellae adnexed, close, 5 mm broad, with 1–2 tiers of lamellulae; margins smooth; lilac grey (16C2) to greyish violet (16C4) when immature, dull violet (16D4) to greyish violet (16D5) when mature, turning orange (6A7) to brownish orange (6C7) when damaged.
Stipe to 43 mm long × 5–11 mm in diam., stout, central, equal, dry, violet white (16A2) to light violet (16A5), covered by white (16A1) to violet white (16A2) flocculose pruina, bruising dull. Base with white rhizomorphs.
Odor not distinct.
Taste not recorded.
Spore-print white.
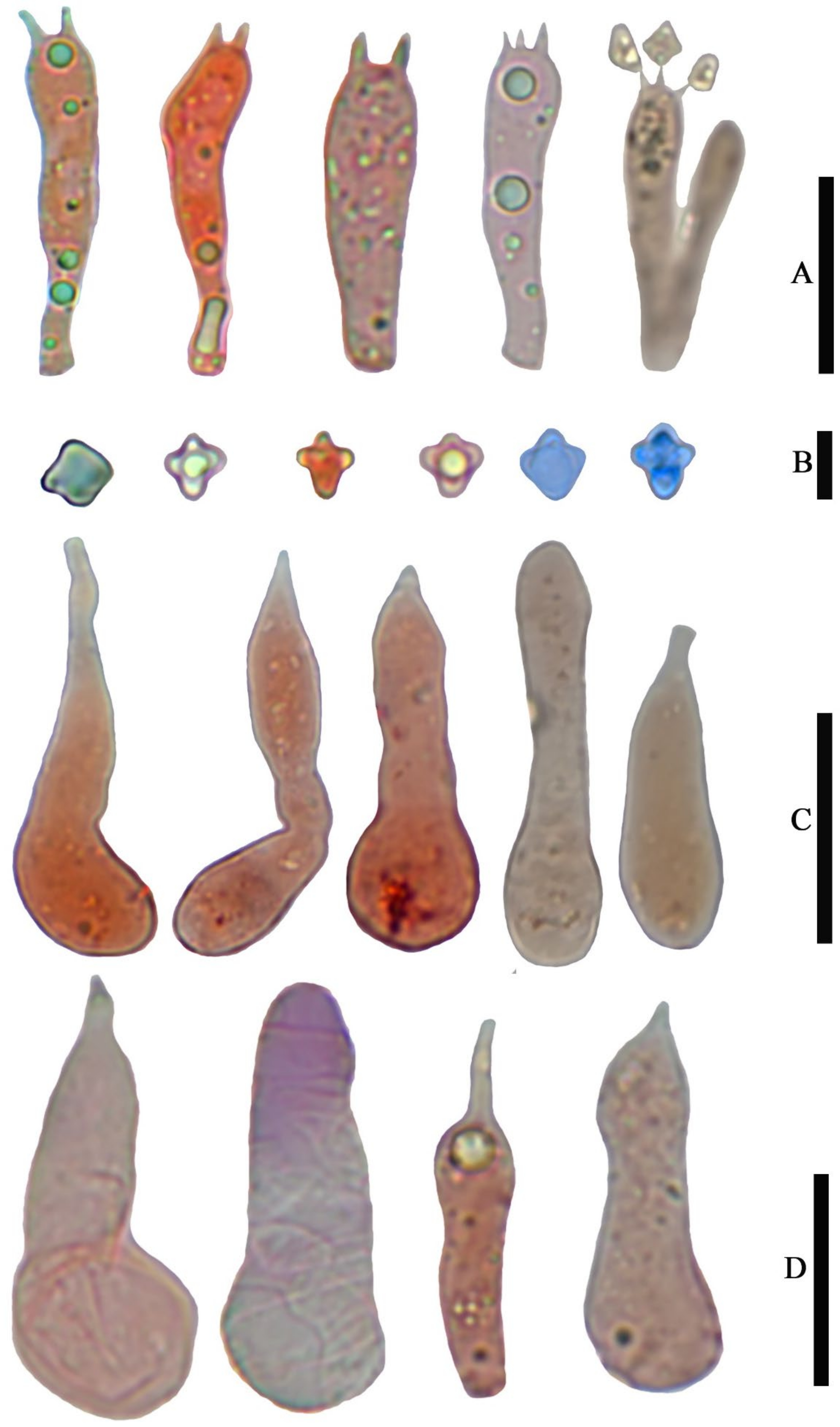
Basidiospores (6.5) 7.0–8.0 (9.0) × 4.8–6.0 (7.0) µm, 7.6 × 5.8 µm on average (Q = 1.1 − 1.5, Q
av = 1.3) [36/5/4], ornamentation not included), hyaline, colorless, inamyloid, thin-walled, densely tuberculate, ornamentation up to 1.0 µm high, sometimes with a single large oil-drop (
Figure 4 and
Figure 7).
Basidia (25) 30–40 (44) × (8) 9–12 (13) µm, 33 × 10 µm on average [48/4/4], cylindric to narrowly clavate, thin-walled, colorless, usually with one to multiple oil drops, two or four sterigmate (
Figure 4A).
Cheilocystidia 30–40 × 8–10 µm, mostly ampullaceous, with a swollen base and a neck, acute or mucronate at apex, thin-walled, colorless (
Figure 4D and
Figure 5A).
Pleurocystidia 29–44 (54) × 8–10 (13) µm, ampullaceous or clavate, with a swollen base and a neck, acute, mucronate or obtuse at apex, thin-walled, colorless (
Figure 4C).
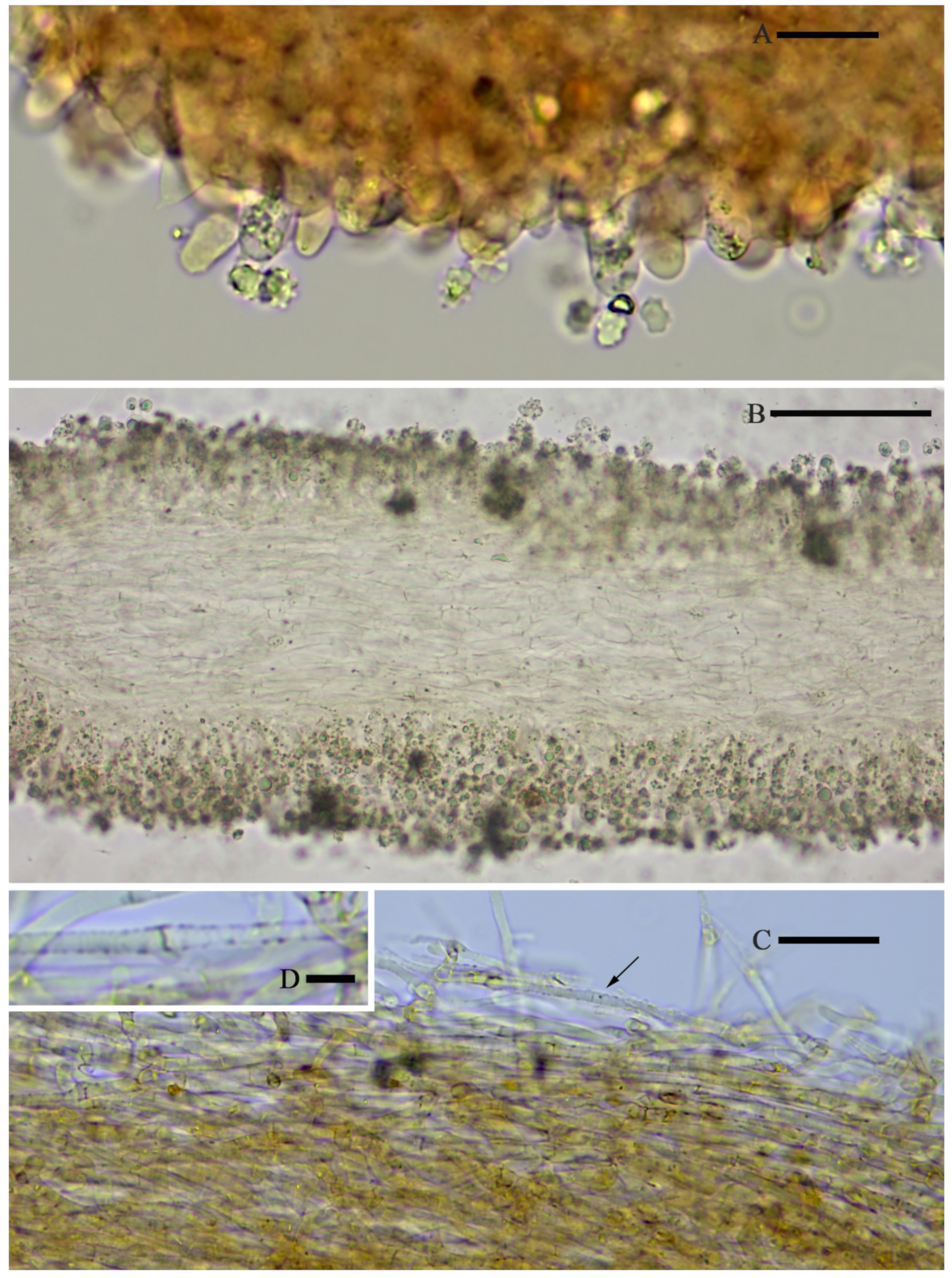
Hymenophoral trama regular, hyphae thin-walled (
Figure 5B).
Pileipellis an undifferentiated cutis, hyphae 3–5 µm in diam., light yellow (4A4–4A5, under water), some hyphae with fine incrustation on surface (
Figure 5C,D).
Laticifers present, pale yellow (4A3), thick-walled, branched, 5–7.5 µm in diam. (
Figure 6).
Clamp connections present (
Figure 5D).
Habitat: Scattered or gregarious in broad-leaved forest soil of karst areas, dominant tree species is Cyclobalanopsis myrsinifolia.
Known distribution: So far only known from Guangxi (China).
Additional material examined: China, Guangxi Province, Baise city, Leye country, Yachang Orchidaceae National Nature Reserve, 24°50′51.56″ N, 106°24′55.40″ E, elevation 1056 m, 12 August 2020, Guang-fu Mou HMJAU59026 (HMJAU!); China, Guangxi Province, Baise city, Leye country, Yachang Orchidaceae National Nature Reserve, 24°50′50.75″ N, 106°24′56.42″ E, elevation 1047 m, 12 August 2020, Guang-fu Mou M2020081289 (IBK!), China, Guangxi Province, Baise city, Leye country, Yachang Orchidaceae National Nature Reserve, 24°50′50.60″ N, 106°24′56.61″ E, elevation 1053 m, 12 August 2020, Guang-fu Mou M2020081292 (IBK!).
Notes:
Asproinocybe is a small genus, characterized by the tricholomatoid
basidiomata with distinctive purplish, violaceous, or lilac-vinaceous colors; spores with tuberculate ornamentation and present of the
laticifers. Our specimens present these features. In 2020, Lebel et al. described two new species of this genus [
6]. Our specimens are somewhat similar to
A. daleyae in appearance: they all present a dark brown pileus. However, our specimens had larger
basidia (33 × 10 µm vs. 20–30 × 5–7 µm), longer
cheilocystidia (30–40 × 8–10 µm vs. 25–30 × 8–12 µm), and
pleurocystidia (38 × 9 µm vs. 25–30 × 10–13 µm), and
hyphae of the
pileipellis had fine
incrustation. The dark brown
pileus, the presence of larger
cystidia, and
hyphae of
pileipellis with fine
incrustation can also be used to differentiate the rest of the known species. Our phylogenetic results (
Figure 1 and
Figure 2) agree with the morphological results.
Tricholosporum Guzmán, Boln. Soc. mex. Micol. 9: 61 (1975).
Etymology: From cruciform basidiospores.
Type species: Tricholosporum goniospermum (Bres.) Guzmán ex T.J. Baroni.
Ecology and distribution: Scattered or gregarious on broad-leaved forest soil, widespread.
Mycobank No: MB841851.
Diagnosis: Differs from other known species of this genus by these combined features: basidiomata medium in size, pileus no color spots, central with obvious yellowish-brownish color when mature; cheilocystidia and pleurocystidia unfurcate and sometimes with purplish pigment; spores cyanophilous, not exceeding 7 µm, 5.4 × 4.7 µm on average.
Etymology: guangxiense (Lat.): The type specimen was obtained in Guangxi, China.
Type: China, Guangxi province, Chongzuo city, Ningming country, Nongang National Nature Reserve, 22°14′31.54″ N, 107°03′50.14″ E, elevation 167 m, 22 August 2021, Guang-fu Mou HMJAU59028 (Holotype HMJAU!).
Description:
Basidiomata tricholomatoid, solitary to gregarious (
Figure 8).
Pileus 43–55 mm in diam., convex to flat-hemispherical, becoming plane to depressed with age, margin smooth or with light short stripes, entire, incurved at first then straight to slightly reflexed, surface at first fibrillose-felted (due to very thin, white hairs), nonviscid; greyish ruby (12D4–12D5) when young, light lilac (16A5) to greyish violet (16C5) near margin and central becoming light orange (5A5) to brownish yellow (16C5) with age.
Context up to 4 mm thick, firm, whitish.
Lamellae emarginate, with small decurrent tooth, close, 5 mm in broad, with 2–3 tiers of lamellulae; margins smooth; light violet (1AC5) to violet (17A7), turning orange (6A7) to brownish orange (6B7) when damaged.
Stipe 30 to 50 mm long × 5–8 mm in diam., stout, central, equal, dry, light violet (17A5) to violet (17A6), covered by violet white (16A2) to pale violet (16A3) pruina, bruising dull.
Basal with white rhizomorphs.
Odor not distinct.
Taste not recorded.
Spore-print white.
Basidiospores (4.0) 5.0–6.0 (7.0) × (3.6) 4.0–5.0 (5.4) µm, 5.4 × 4.7 µm on average (Q = 1.0–1.4, Q
av = 1.2) [38/5/5], cruciform, hyaline, colorless, inamyloid, cyanophilous (
Figure 9B 5–6), thin-walled, usually with a single large oil drop (
Figure 9B).
Basidia (21) 23–28 (32) × 5–7 µm [48/3/3], cylindric to narrowly clavate, thin-walled, colorless, usually with one to multiple oil drops, two or four sterigmate (
Figure 9A).
Cheilocystidia (23) 27–36 (40) × 6–13 (14) µm, ampullaceous or clavate, with a swollen base and a neck, acute, mucronate or obtuse at apex, thin-walled, sometimes with purplish pink (14A5) to greyish magenta pigment (14D5) (
Figure 9D and
Figure 10A).
Pleurocystidia (35) 40–50 (60) × (8) 9–13 (14) µm, ampullaceous or clavate, with a swollen base and a neck, acute, mucronate or obtuse at apex, sometimes curved, thin-walled, sometimes with grey-violet pigment (
Figure 9C).
Hymenophoral trama 148–243 µm broad, regular, hypha thin-walled (
Figure 10B,C).
Pileipellis an undifferentiated cutis, hyphae 4.6–5.8 µm in diam., colorless (
Figure 10D,E).
Laticifers present, pale yellow (4A3), thick-walled, branched, 5–10 µm in diam. (
Figure 10F).
Clamp connections present (
Figure 10E).
Habitat: Scattered or gregarious on broad-leaved forest soil of karst areas; the associated tree species are Streblus tonkinensis, Wendlandia uvariifolia, Sterculia monosperma, Musa balbisiana, and Heptapleurum sp.
Known distribution: So far, only known in Guangxi (China).
Additional material examined: China, Guangxi Province, Chongzuo city, Ningming country, Nongang National Nature Reserve, 22°14′29.90″ N, 107°03′33.99″ E, elevation 263 m, 08 July 2018, Guang-fu Mou HMJAU59023 (HMJAU!); China, Guangxi Province, Chongzuo city, Ningming country, Nongang National Nature Reserve, 22°14′31.63″ N, 107°03′50.59″ E, elevation 166 m, 22 August 2021, Guang-fu Mou HMJAU59027 (HMJAU!). China, Guangxi Province, Chongzuo city, Ningming country, Nongang National Nature Reserve, 22°14′41.78″ N, 107°04′22.05″ E, elevation 145 m, 08 August 2021, Guang-fu Mou M2021082219 (IBK!); China, Guangxi Province, Chongzuo city, Ningming country, Nongang National Nature Reserve, 22°14′32.55″ N, 107°03′55.02″ E, elevation 181 m, 8 August 2021, Guang-fu Mou M2021082208 (IBK!).
Notes: Angelini et al., according to the size of the
basidiospores, divided the species of
Tricholosporum into two groups: group 1, the large-spored species with spores over 7 µm in length, and group 2, species with small spores, usually under 6 µm in length. According to the presence or absence of
hymenial cystidia and whether grey-violet pigmentation was shown, they further divided group 2. Our specimens should obviously be categorized into group 2, subgroup 2.3: species with pigmented
hymenial cystidia, which are grey-violet or brownish. Three species,
T. palmense, T. violaceum, and
T. caraibicum, were placed in this subgroup [
4].
Our specimens differ from
T. violaceum by their smaller and dry
pileus (4.3–5.5 vs. 8–13 cm), thicker
context (4 vs. 10–20 mm), narrower
lamellae (5 vs. 15 mm), shorter
stipe (5 vs. 5–11 cm), and larger
spores (5.4 × 4.7 vs. 4.5 × 3.5 µm, on average) [
4]; differ from
T. palmense by the unfurcate
cystidia; differ from
T. caraibicum by the
pileus lacking color spots, central with obvious yellowish-brownish color when mature, the larger
spores (5.4 × 4.7 vs. 4.1 × 3.8 µm, on average), and being cyanophilous [
4].
5. Discussion
The phylogenetic placement of the
Asproinocybe/
Tricholosporum clade has been discussed by Angelini et al. and Lebel et al. [
6,
20] but remains unresolved due to the poor sequencing of the species from this clade. Fortunately, we collected two new taxa from
Asproinocybe and
Tricholosporum and obtained another three specimens (one from the holotype) of the species
Tricholosporum haitangshanum. Thus, we had 12 specimens for this study. Finally, we successfully extracted the DNA from 10 specimens, and a total of 43 sequences (15 from
Asproinocybe and 28 from
Tricholosporum) were obtained, including ITS, nrLSU, RPB1, RPB2, nrSSU, and TEF1-α sequences (see
Table S1 in Supplementary Material, in bold).
The overall topology in
Figure 1 (the topology of the tree was obtained from Bayesian analysis) is consistent with the topologies published in previous studies [
22,
30,
31,
32,
33,
34,
35,
36,
37,
38], except for the positions of the genera
Bonomyces,
Catathelasma, and
Cleistocybe. Sánchez-García et al., Alvarado et al. and Raj et al. also reported results similar to those of the present study [
34,
35,
36,
37]. Vizzini et al. explained that this difference in arrangement is due to the taxon sampling within
Catathelasma,
Callistosporium, and
Macrocybe [
38]. In the additional analyses, we obtained the same results as Vizzini et al. when increasing the sampling within
Catathelasma,
Callistosporium, and
Macrocybe (not shown in the present study).
The relationships of the genera
Asproinocybe and
Tricholosporum have been discussed by many mycologists. Guzmán et al. and Baroni recognized
Tricholosporum is distinct from
Asproinocybe by the shape of spore; Lebel et al. believed that the relationship between
Tricholosporum and
Asproinocybe will remain problematic until further species of
Asproinocybe are sequenced; Singer, Bohus, Alessio, Hongo, and Bon and Braiotta recognized recognized
Tricholosporum is distinct from
Asproinocybe, but considered
Tricholosporum a synonym of
Tricholoma in the Section Iorigida [
2,
3,
4,
6,
11,
12,
13,
14,
15,
16]. Morphologically, they have many common features—key features used to tell them apart are the spore shapes and the presence of
laticifers [
2].
Laticifers are rarely recorded in
Tricholosporum and can even be considered as probably absent [
2]. However, we truly observed both in
Tricholosporum guangxiense (
Figure 10F) and
Tricholosporum haitangshanum (not shown in this study) but not so commonly as in
Asproinocybe. Moreover, based on our results in
Figure 1 and
Figure 2,
Asproinocybe and
Tricholosporum do not form two independent clades. Should we consider
Tricholosporum as a synonym of
Asproinocybe?
Tricholosporum cannot be a synonym of
Tricholoma based on our results. However, based on our results in
Figures S1 and S2, the species of
Asproinocybe and
Tricholosporum form two distinct clades, and based on the results in
Figure 1, the species of
Asproinocybe and
Tricholosporum do not cross over.
Based on our results shown in
Figure 1 and
Figure 2, the species of
Asproinocybe lyophylloides,
Asproinocybe daleyae, or
Asproinocybe sinensis form a monophyletic clade with the taxa of
Tricholosporum. Should we treat them as independent genera? If so, no morphological delimitation is shown between the independent clades abovementioned. If we treat
Tricholosporum as being distinct from
Asproinocybe, it seems more reasonable. Thus, the
Tricholosporum clade is a monophyletic clade with clear a morphological basis (from cruciform to stauriform spores). The taxa of
Asproinocybe do not form a monophyletic clade in
Figure 1 and
Figure 2 but instead form a monophyletic clade in
Figures S1 and S2 with a clear morphological basis (the tuberculate to stellate spores). A possible explanation for the
Asproinocybe clades is that the present phylogenetic tree lacks of sampling between
Asproinocybe sinensis,
A. daleyae, and
A. lyophylloides. Stronger evidence is needed to prove that
Tricholosporum is a synonym of
Asproinocybe; as such, we maintain the opinion that
Tricholosporum is distinct from
Asproinocybe due to the spore’s shape and the not-so-abundant
laticifers.
We also noticed that Tricholosporum haitangshanum was close to Tricholosporum goniospermum in terms of both morphological and phylogenetic features. We will not analyze it until more specimens of Tricholosporum goniospermum have been studied.
At the family level, the clades of
Asproinocybe and
Tricholosporum were commonly placed Tricholomataceae s.l., Lyophyllaceae, and Entolomataceae [
1,
2,
3,
4,
5,
6,
7,
8,
12,
19,
20]. Morphologically, those taxa have the tricholomatoid habit (especially in Tricholomataceae s.l. and Lyophyllaceae) and tuberculate spores. However, the species of
Asproinocybe and
Tricholosporum are always distinctive purplish, violaceous, or lilac-vinaceous colors, and the tuberculate spores are more remarkable. Some species in
Cortinarius and
Inocybe also have purplish basidiomata and tuberculate spores. However, their spores are brownish, and the results of Heaton and Kropp refute the possible relationships [
19]. Another possible group is the Clitocybeae, which includes the genus
Lepista, which could be similar to the species of
Asproinocybe and
Tricholosporum. Our phylogenetic analysis included these species: they were clearly separated and could be easily discriminated under a microscope. Another important feature indicating
Asproinocybe and
Tricholosporum as a new family is that they have
laticifers.
Morphologically, the species of
Asproinocybe and
Tricholosporum are somewhat similar to those of Callistosporiaceae. They have the same features: tricholomatoid
habit,
veils absent;
lamellae adnate, adnexed, sinuate, emarginated to decurrent;
spore print white,
spores cyanophilous or acyanophilous, thin-walled;
hymenophoral trama regular; and
pileipellis arranged as a cutis [
38]. All the species of
Asproinocybe and
Tricholosporum are more or less purplish, violaceous, or lilac-vinaceous; the species of Callistosporiaceae can also have similar coloration, such as
Callistosporium elegans.
The species of Callistosporiaceae grow in soil or rotten wood and aresaprotrophic or ectomycorrhizal [
38]. The species of
Asproinocybe and
Tricholosporum also grow in soil. We do not know if they form mycorrhizal relationships with plants, but they usually have white rhizomorphs. Recently,
Asproinocybe lactifera was reported as an ectomycorrhizae fungus [
46]. This is worthy of further study, but finding species of Asproinocybaceae is challenging.
Compared to Callistosporiaceae,
Asproinocybe and
Tricholosporum have some unique features, such as the basidiomata being distinctively purplish, violaceous, or lilac-vinaceous, spores tuberculate to stellate (
Asproinocybe) or cruciform to stauriform (
Tricholosporum),
laticifers present, and the
lamellae bruising reddish or pale brown when damaged [
1,
2,
3,
4,
5,
6,
7,
8].
Asproinocybaceae was an important lineage in the evolution of agarics. The presence of
laticifers,
lamellae bruising reddish, and
spores with ornamentation and ectomycorrhizae [
46] led to us link it with Russulaceae,
Lactarius. The species of
Lactarius also have
basidiomata shapes similar to those of species of Asproinocybaceae, but the spores of
Lactarius are amyloid. The relationship of the spore shapes between
Tricholosporum and Entolomataceae was discussed by Angelini et al. [
20]. According to the results reported by David et al. [
31], the spore walls forming the ornamentation of Entolomataceae may not be homologous to those of other tricholomatoid species with bumped spores [
20]. Our study confirms that
Tricholosporum is included in a new clade that is different from the tricholomatoid species previously known. Thus, we may have to reconsider the homology of spores between Asproinocybaceae and Entolomataceae.
Species from Callistosporiaceae are saprotrophic or ectomycorrhizal [
38]; as the sister family, species from Asproinocybaceae may be ectomycorrhizal [
46]. The new species proposed here were collected from karst areas, where the soil is thin and infertile, where stony desertification is common, and where it is difficult for the vegetation to recover. If the species from Asproinocybaceae are ectomycorrhizal, they could help with vegetation recovery in areas suffering from stony desertification using mycorrhizal techniques. Species from Callistosporiaceae and Asproinocybaceae may provide suitable study material for explaining the evolution of mycorrhizal and nonmycorrhizal fungi.