Phenotypic Characterization and Comparative Genomics of the Melanin-Producing Yeast Exophiala lecanii-corni Reveals a Distinct Stress Tolerance Profile and Reduced Ribosomal Genetic Content
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of Fungi
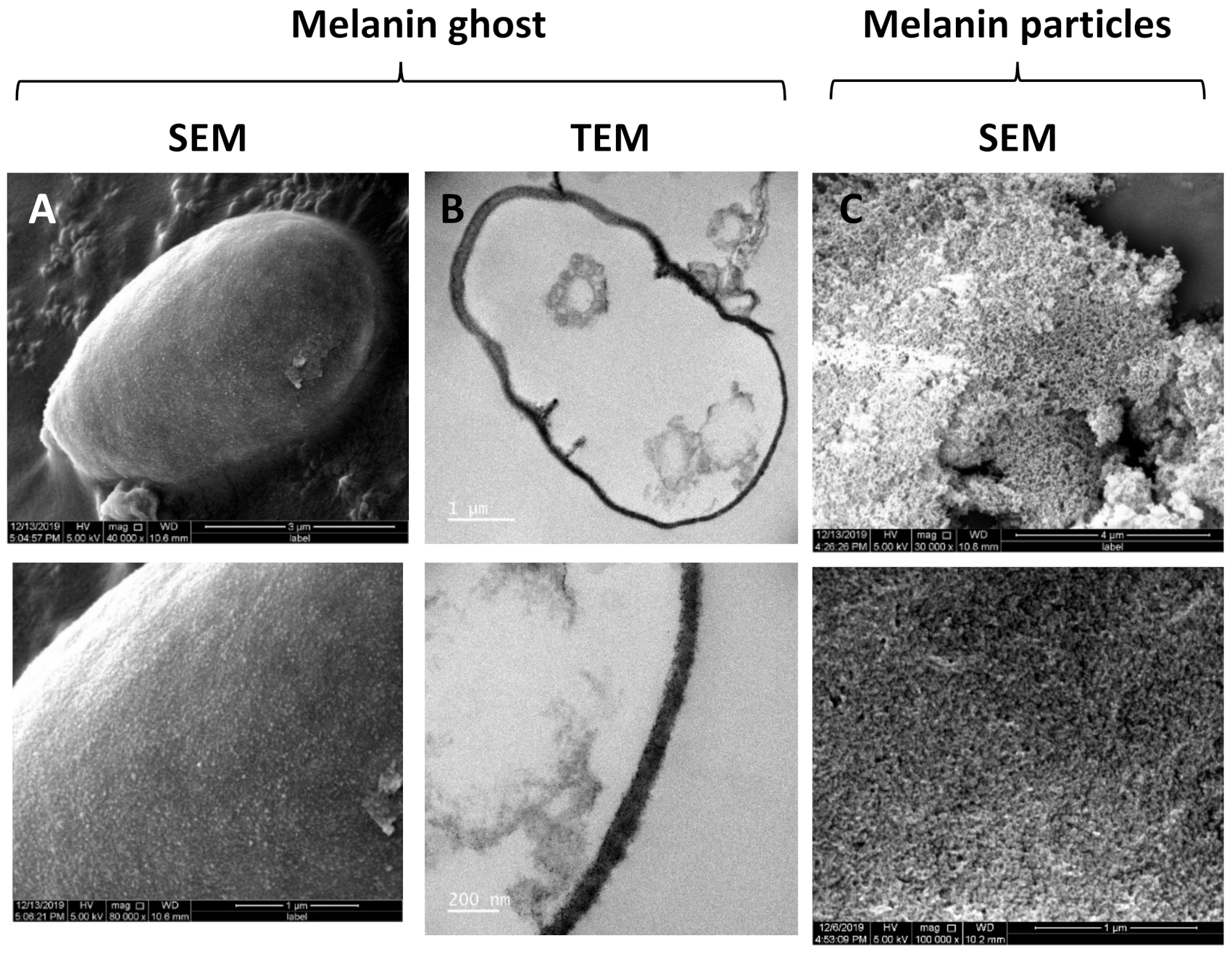
2.2. Isolation of Fungal Melanin
2.3. Genetic Manipulation
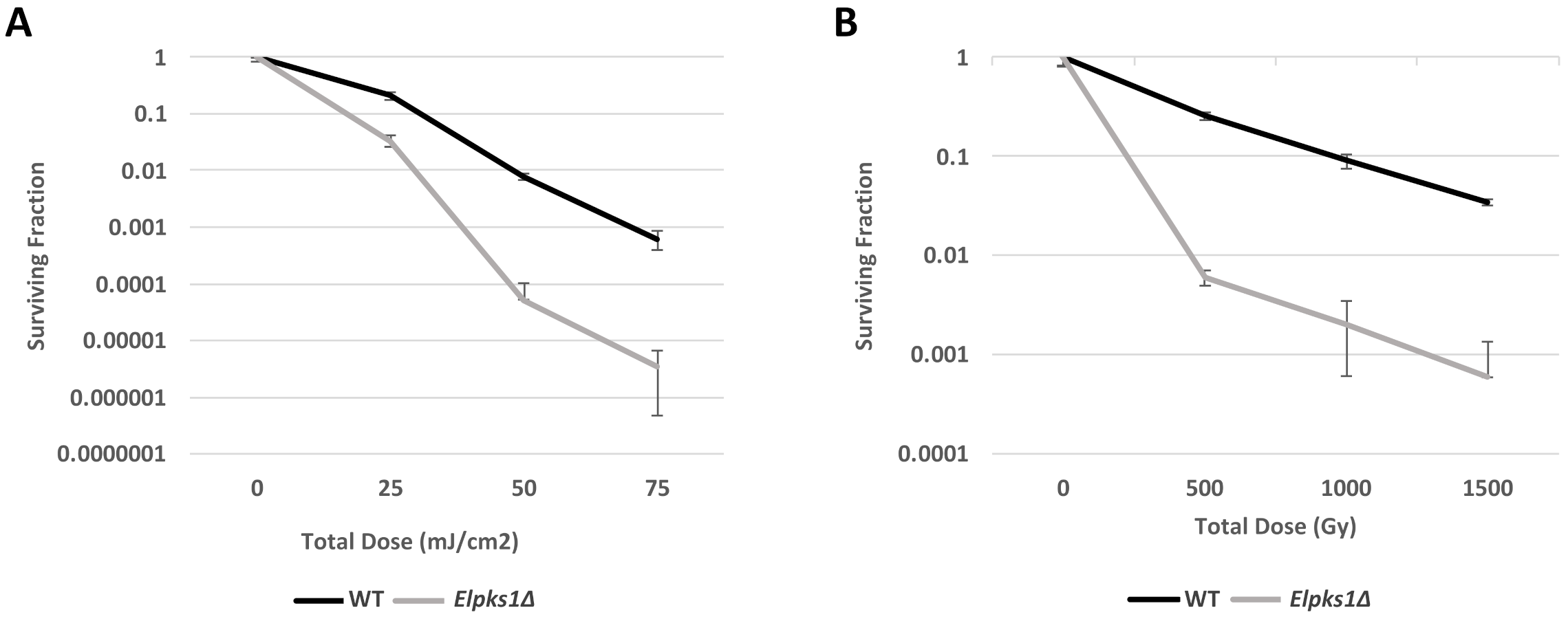
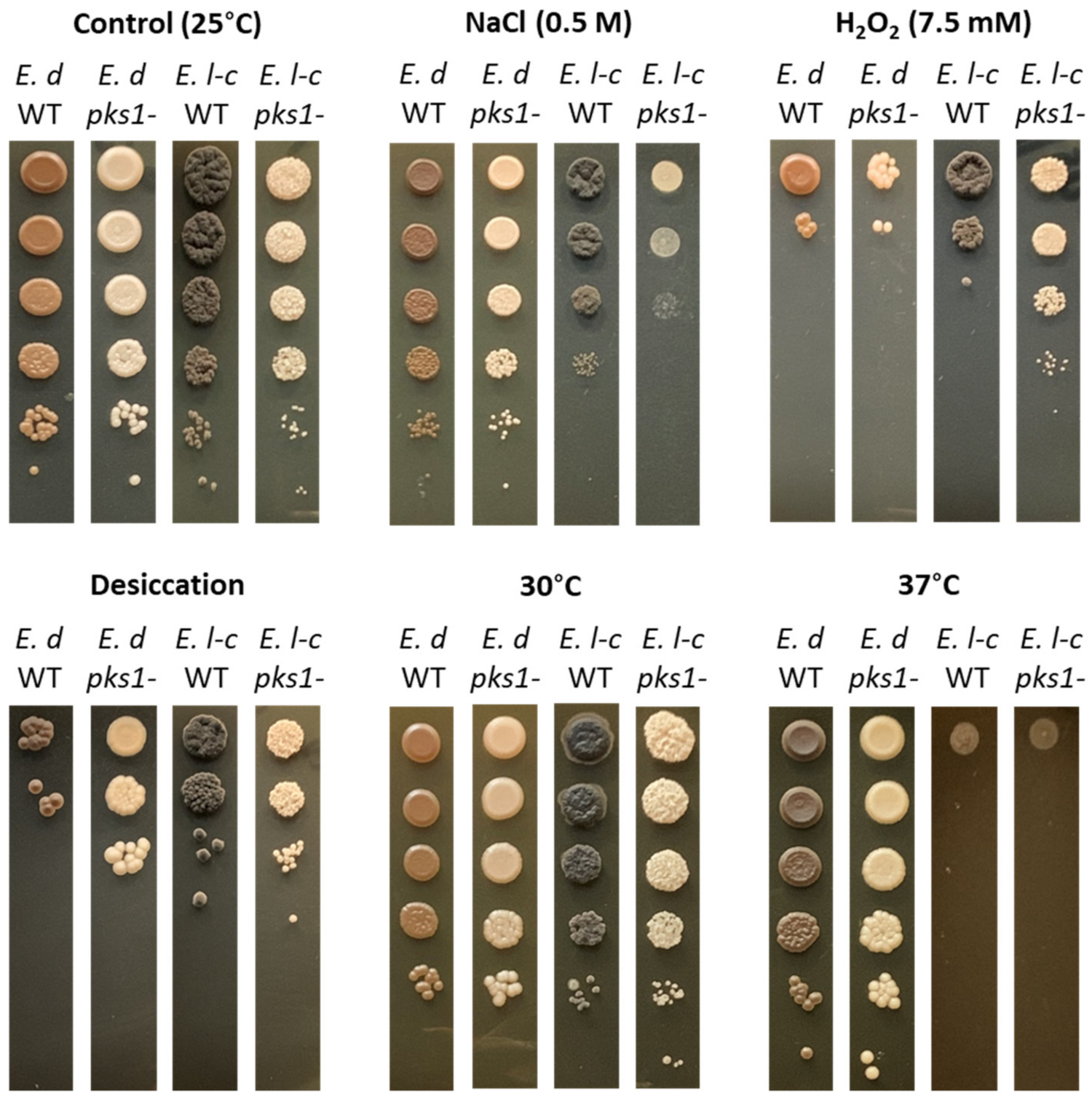
2.4. Cell Survival and Sensitivity Assays
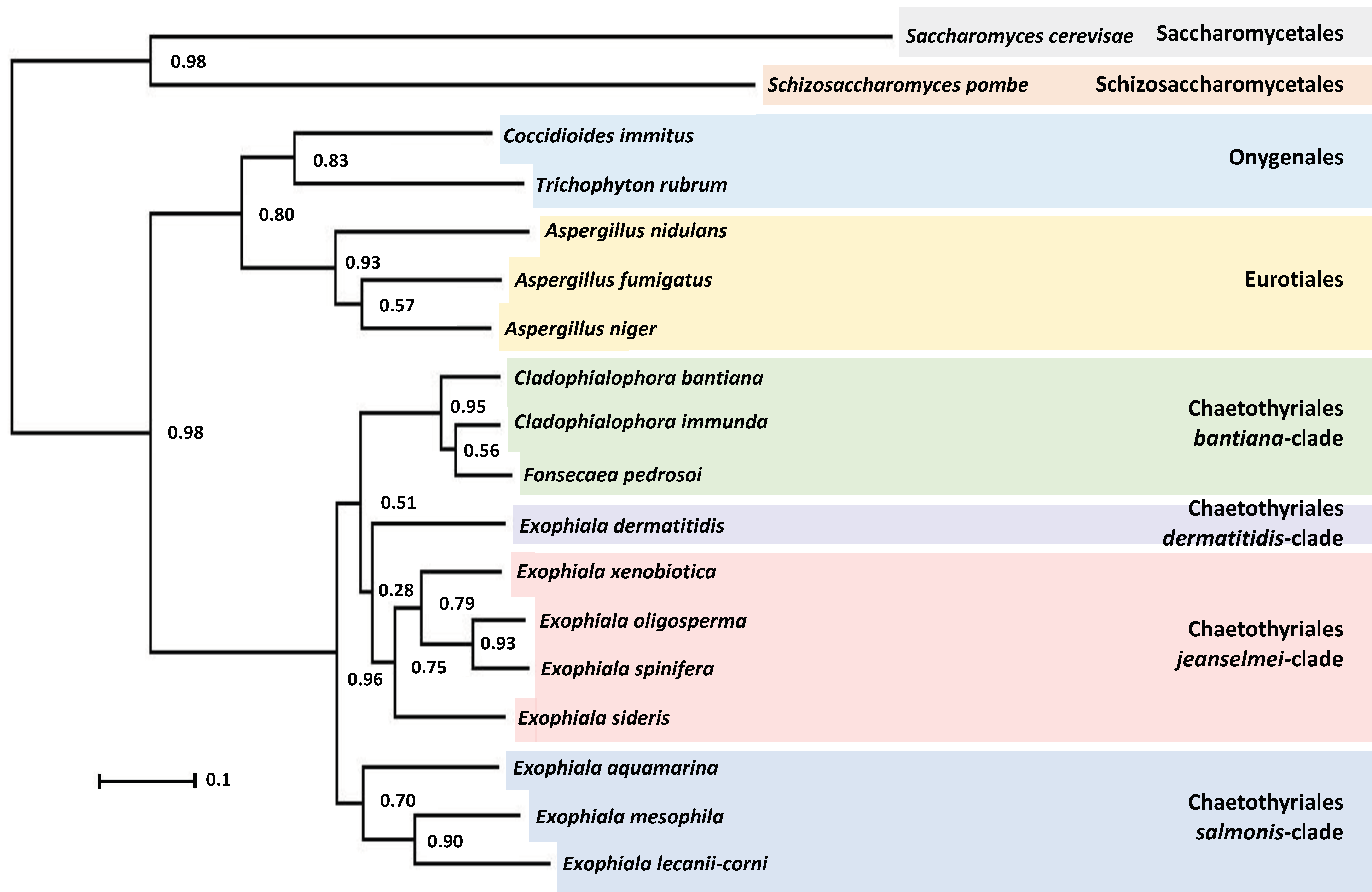
2.5. Phylogenetic and Comparative Genomic Analysis
3. Results
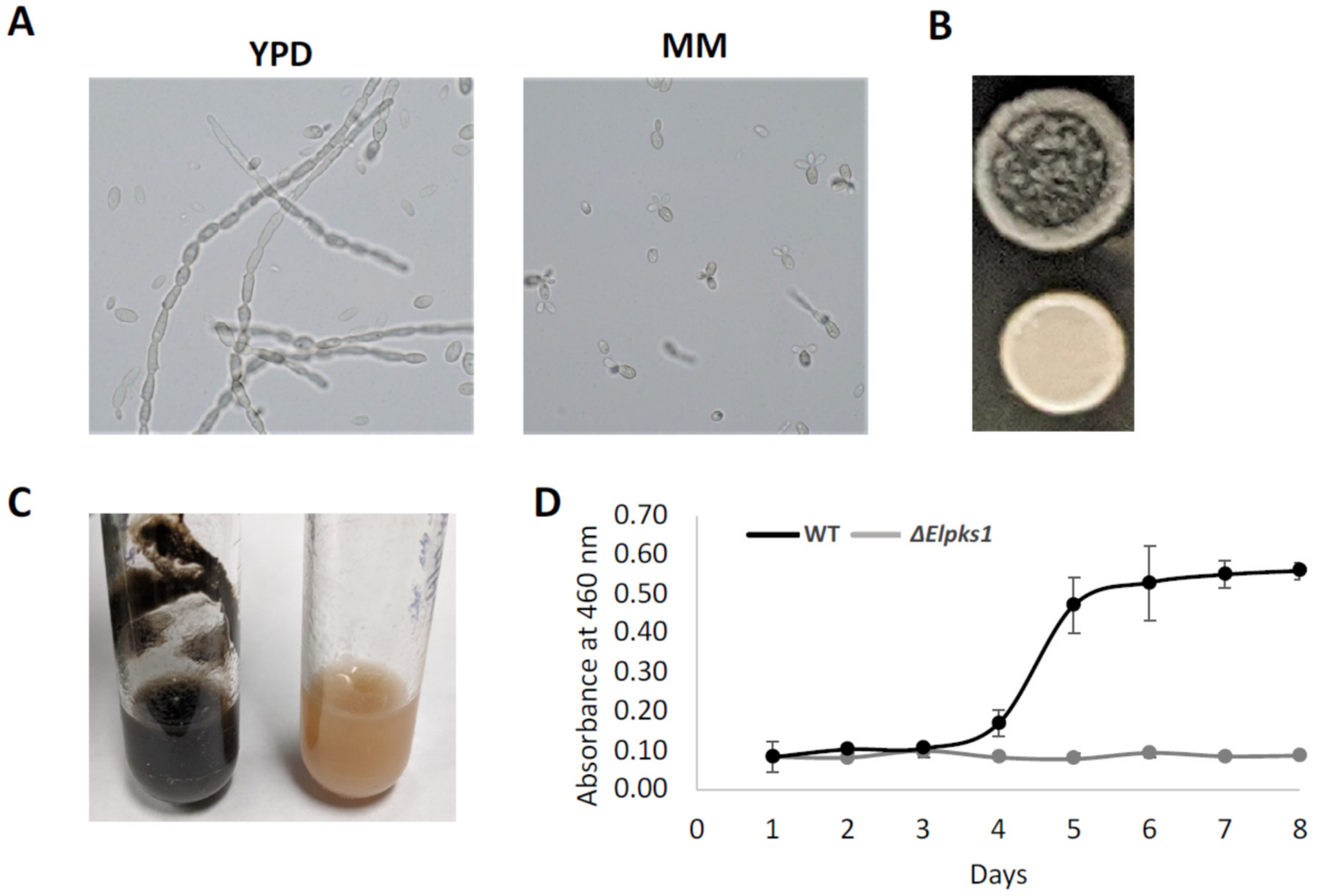
3.1. Phenotypic Analyses, Melanin Biosynthesis and Melanin Characterization in E. lecanii-corni
3.2. Effect of Melanin Production on Stress Resistance in E. lecanii-corni
3.3. Comparison of the E. lecanii-corni and E. dermatitidis Genomes
3.4. Phylogenetic Characterization of E. lecanii-corni
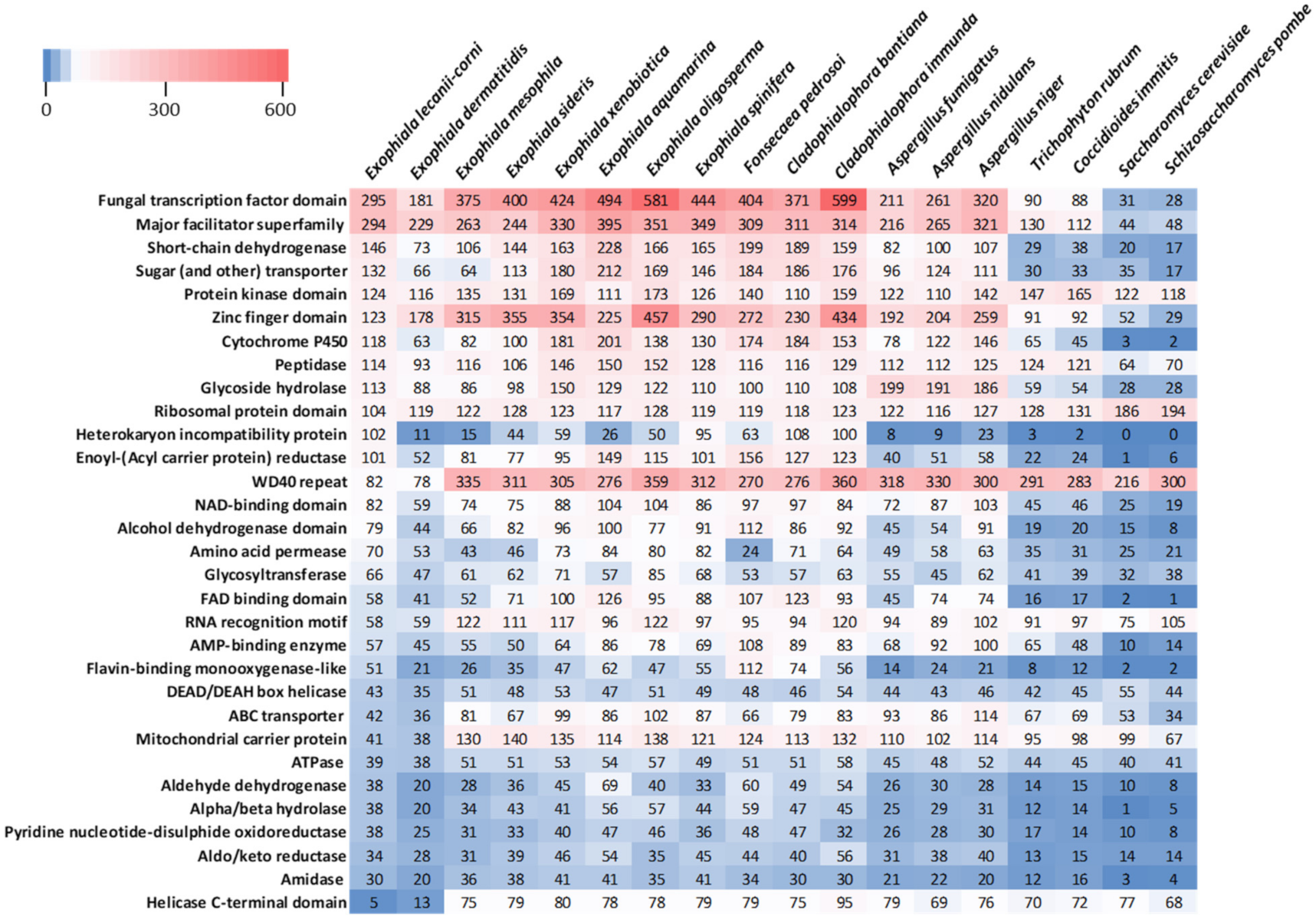
3.5. Comparative Analysis of Predicted Functional Content
3.6. Toluene Degradation Pathway Comparison
3.7. Melanin Biosynthesis and Regulation Pathway Comparison
3.8. Potential for Secondary Metabolite Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seyedmousavi, S.; Netea, M.G.; Mouton, J.W.; Melchers, W.J.G.; Verweij, P.E.; de Hoog, G.S. Black Yeasts and Their Filamentous Relatives: Principles of Pathogenesis and Host Defense. Clin. Microbiol. Rev. 2014, 27, 527–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onofri, S.; Barreca, D.; Selbmann, L.; Isola, D.; Rabbow, E.; Horneck, G.; de Vera, J.P.P.; Hatton, J.; Zucconi, L. Resistance of Antarctic Black Fungi and Cryptoendolithic Communities to Simulated Space and Martian Conditions. Stud. Mycol. 2008, 61, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Plemenitaš, A.; Vaupotič, T.; Lenassi, M.; Kogej, T.; Gunde-Cimerman, N. Adaptation of Extremely Halotolerant Black Yeast Hortaea Werneckii to Increased Osmolarity: A Molecular Perspective at a Glance. Stud. Mycol. 2008, 61, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; Zucconi, L.; Isola, D.; Onofri, S. Rock Black Fungi: Excellence in the Extremes, from the Antarctic to Space. Curr. Genet. 2015, 61, 335–345. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Moreno, L.F.; Stielow, B.J.; Muszewska, A.; Hainaut, M.; Gonzaga, L.; Abouelleil, A.; Patané, J.S.L.; Priest, M.; Souza, R.; et al. Exploring the Genomic Diversity of Black Yeasts and Relatives (Chaetothyriales, Ascomycota). Stud. Mycol. 2017, 86, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Schultzhaus, Z.; Romsdahl, J.; Chen, A.; Tschirhart, T.; Kim, S.; Leary, D.; Wang, Z. The Response of the Melanized Yeast Exophiala Dermatitidis to Gamma Radiation Exposure. Environ. Microbiol. 2020, 22, 1310–1326. [Google Scholar] [CrossRef]
- Zeng, J.S.; Sutton, D.A.; Fothergill, A.W.; Rinaldi, M.G.; Harrak, M.J.; de Hoog, G.S. Spectrum of Clinically Relevant Exophiala Species in the United States. J. Clin. Microbiol. 2007, 45, 3713–3720. [Google Scholar] [CrossRef] [Green Version]
- Woertz, J.R.; Kinney, K.A.; McIntosh, N.D.P.; Szaniszlo, P.J. Removal of Toluene in a Vapor-Phase Bioreactor Containing a Strain of the Dimorphic Black YeastExophiala Lecanii-Corni. Biotechnol. Bioeng. 2001, 75, 550–558. [Google Scholar] [CrossRef]
- Qi, W.; Moe, K.; Kinney, B. Biodegradation of Volatile Organic Compounds by Five Fungal Species. Appl. Microbiol. Biotechnol. 2002, 58, 684–689. [Google Scholar] [CrossRef]
- Cheng, Q.; Kinney, K.A.; Whitman, C.P.; Szaniszlo, P.J. Characterization of Two Polyketide Synthase Genes in Exophiala Lecanii-Corni, a Melanized Fungus with Bioremediation Potential. Bioorg. Chem. 2004, 32, 92–108. [Google Scholar] [CrossRef]
- Fernandez, C.W.; Koide, R.T. The Function of Melanin in the Ectomycorrhizal Fungus Cenococcum Geophilum under Water Stress. Fungal Ecol. 2013, 6, 479–486. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Functions of Fungal Melanin beyond Virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Robert, V.; Cardinali, G.; Arinze, E.S.; Thon, S.M.; Casadevall, A. Impact of Yeast Pigmentation on Heat Capture and Latitudinal Distribution. Curr. Biol. 2018, 28, 2657–2664.e3. [Google Scholar] [CrossRef] [Green Version]
- Pihet, M.; Vandeputte, P.; Tronchin, G.; Renier, G.; Saulnier, P.; Georgeault, S.; Mallet, R.; Chabasse, D.; Symoens, F.; Bouchara, J.-P. Melanin Is an Essential Component for the Integrity of the Cell Wall of Aspergillus Fumigatus Conidia. BMC Microbiol. 2009, 9, 177. [Google Scholar] [CrossRef]
- Bashyal, B.M.; Chand, R.; Kushwaha, C.; Sen, D.; Prasad, L.C.; Joshi, A.K. Association of Melanin Content with Conidiogenesis in Bipolaris Sorokiniana of Barley (Hordeum Vulgare L.). World J. Microbiol. Biotechnol. 2010, 26, 309–316. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Casadevall, A. The Contribution of Melanin to Microbial Pathogenesis. Cell Microbiol. 2003, 5, 203–223. [Google Scholar] [CrossRef]
- Revankar, S.G.; Sutton, D.A. Melanized Fungi in Human Disease. CMR 2010, 23, 884–928. [Google Scholar] [CrossRef] [Green Version]
- d’Ischia, M.; Napolitano, A.; Ball, V.; Chen, C.-T.; Buehler, M.J. Polydopamine and Eumelanin: From Structure–Property Relationships to a Unified Tailoring Strategy. Acc. Chem. Res. 2014, 47, 3541–3550. [Google Scholar] [CrossRef]
- Ju, K.-Y.; Lee, Y.; Lee, S.; Park, S.B.; Lee, J.-K. Bioinspired Polymerization of Dopamine to Generate Melanin-Like Nanoparticles Having an Excellent Free-Radical-Scavenging Property. Biomacromolecules 2011, 12, 625–632. [Google Scholar] [CrossRef]
- Le Na, N.T.; Duc Loc, S.; Minh Tri, N.L.; Bich Loan, N.T.; Anh Son, H.; Linh Toan, N.; Phuong Thu, H.; My Nhung, H.T.; Lai Thanh, N.; Van Anh, N.T.; et al. Nanomelanin Potentially Protects the Spleen from Radiotherapy-Associated Damage and Enhances Immunoactivity in Tumor-Bearing Mice. Materials 2019, 12, 1725. [Google Scholar] [CrossRef] [Green Version]
- Meredith, P.; Sarna, T. The Physical and Chemical Properties of Eumelanin. Pigment Cell Res. 2006, 19, 572–594. [Google Scholar] [CrossRef]
- Bothma, J.P.; de Boor, J.; Divakar, U.; Schwenn, P.E.; Meredith, P. Device-Quality Electrically Conducting Melanin Thin Films. Adv. Mater. 2008, 20, 3539–3542. [Google Scholar] [CrossRef]
- Karlsson, O.; Lindquist, N.G. Melanin and Neuromelanin Binding of Drugs and Chemicals: Toxicological Implications. Arch. Toxicol. 2016, 90, 1883–1891. [Google Scholar] [CrossRef]
- Tran-Ly, A.N.; Ribera, J.; Schwarze, F.W.M.R.; Brunelli, M.; Fortunato, G. Fungal Melanin-Based Electrospun Membranes for Heavy Metal Detoxification of Water. Sustain. Mater. Technol. 2020, 23, e00146. [Google Scholar] [CrossRef]
- Pombeiro-Sponchiado, S.R.; Sousa, G.S.; Andrade, J.C.R.; Lisboa, H.F.; Gonçalves, R.C.R. Production of Melanin Pigment by Fungi and Its Biotechnological Applications. In Melanin; Blumenberg, M., Ed.; InTech: London, UK, 2017; ISBN 978-953-51-2979-0. [Google Scholar]
- Tran-Ly, A.N.; Reyes, C.; Schwarze, F.W.M.R.; Ribera, J. Microbial Production of Melanin and Its Various Applications. World J. Microbiol. Biotechnol. 2020, 36, 170. [Google Scholar] [CrossRef]
- Chen, Z.; Martinez, D.A.; Gujja, S.; Sykes, S.M.; Zeng, Q.; Szaniszlo, P.J.; Wang, Z.; Cuomo, C.A. Comparative Genomic and Transcriptomic Analysis of Wangiella Dermatitidis, A Major Cause of Phaeohyphomycosis and a Model Black Yeast Human Pathogen. G3 Genes|Genomes|Genet. 2014, 4, 561–578. [Google Scholar] [CrossRef] [Green Version]
- Romsdahl, J.; Schultzhaus, Z.; Chen, A.; Liu, J.; Ewing, A.; Hervey, J.; Wang, Z. Adaptive Evolution of a Melanized Fungus Reveals Robust Augmentation of Radiation Resistance by Abrogating Non-Homologous End-Joining. Environ. Microbiol. 2020, 23, 3627–3645. [Google Scholar] [CrossRef]
- Schultzhaus, Z.; Schultzhaus, J.N.; Romsdahl, J.; Chen, A.; Hervey IV, W.J.; Leary, D.H.; Wang, Z. Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala Dermatitidis. Genes 2020, 11, 1128. [Google Scholar] [CrossRef]
- Malo, M.E.; Schultzhaus, Z.; Frank, C.; Romsdahl, J.; Wang, Z.; Dadachova, E. Transcriptomic and Genomic Changes Associated with Radioadaptation in Exophiala Dermatitidis. Comput. Struct. Biotechnol. J. 2021, 19, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Dadachova, E.; Bryan, R.A.; Huang, X.; Moadel, T.; Schweitzer, A.D.; Aisen, P.; Nosanchuk, J.D.; Casadevall, A. Ionizing Radiation Changes the Electronic Properties of Melanin and Enhances the Growth of Melanized Fungi. PLoS ONE 2007, 2, e457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-Joint PCR: A PCR-Based Molecular Tool for Gene Manipulations in Filamentous Fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Schultzhaus, Z.; Cuomo, C.A.; Wang, Z. Genome Sequence of the Black Yeast Exophiala Lecanii-Corni. Microbiol. Resour. Announc. 2019, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoff, K.J.; Lange, S.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 2016, 32, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.J.; Lomsadze, A.; Borodovsky, M.; Stanke, M. Whole-Genome Annotation with BRAKER. In Gene Prediction; Kollmar, M., Ed.; Springer: New York, NY, USA, 2019; Volume 1962, pp. 65–95. ISBN 978-1-4939-9172-3. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Coleine, C.; Selbmann, L.; Masonjones, S.; Onofri, S.; Zucconi, L.; Stajich, J.E. Draft Genome Sequence of an Antarctic Isolate of the Black Yeast Fungus Exophiala Mesophila. Microbiol. Resour. Announc. 2019, 8, e00142-19. [Google Scholar] [CrossRef] [Green Version]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic Sequence of the Pathogenic and Allergenic Filamentous Fungus Aspergillus Fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.-J.; Wortman, J.R.; Batzoglou, S.; Lee, S.-I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus Nidulans and Comparative Analysis with A. Fumigatus and A. Oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome Sequencing and Analysis of the Versatile Cell Factory Aspergillus Niger CBS 513.88. Nat. Biotechnol. 2007, 25, 11. [Google Scholar] [CrossRef] [Green Version]
- Martinez, D.A.; Oliver, B.G.; Gräser, Y.; Goldberg, J.M.; Li, W.; Martinez-Rossi, N.M.; Monod, M.; Shelest, E.; Barton, R.C.; Birch, E.; et al. Comparative Genome Analysis of Trichophyton Rubrum and Related Dermatophytes Reveals Candidate Genes Involved in Infection. mBio 2012, 3, e00259-12. [Google Scholar] [CrossRef] [Green Version]
- Sharpton, T.J.; Stajich, J.E.; Rounsley, S.D.; Gardner, M.J.; Wortman, J.R.; Jordar, V.S.; Maiti, R.; Kodira, C.D.; Neafsey, D.E.; Zeng, Q.; et al. Comparative Genomic Analyses of the Human Fungal Pathogens Coccidioides and Their Relatives. Genome Res. 2009, 19, 1722–1731. [Google Scholar] [CrossRef] [Green Version]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 Genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [Green Version]
- Wood, V.; Gwilliam, R.; Rajandream, M.-A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The Genome Sequence of Schizosaccharomyces Pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving Fundamental Biases in Whole Genome Comparisons Dramatically Improves Orthogroup Inference Accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- Kelly, S.; Maini, P.K. DendroBLAST: Approximate Phylogenetic Trees in the Absence of Multiple Sequence Alignments. PLoS ONE 2013, 8, e58537. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. STAG: Species Tree Inference from All Genes. BioRxiv 2018, 267914. [Google Scholar]
- Emms, D.M.; Kelly, S. STRIDE: Species Tree Root Inference from Gene Duplication Events. Mol. Biol. Evol. 2017, 34, 3267–3278. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatModeler Open-1.0 (2008–2015). Institute for Systems Biology: Seattle, WA, USA, 2015. Available online: www.repeatmasker.org (accessed on 20 November 2020).
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. 2015. Available online: www.repeatmasker.org (accessed on 20 November 2020).
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a Database of Repetitive Elements in Eukaryotic Genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A Web Server for Whole-Genome Comparison and Annotation of Orthologous Clusters across Multiple Species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A Comprehensive Online Resource for Systematic Analysis of Gene Lists from Fungal Species. Bioinformatics 2015, 31, 445–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenman, H.C.; Casadevall, A. Synthesis and Assembly of Fungal Melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Aisen, P.; Casadevall, A. Melanin, Melanin “Ghosts,” and Melanin Composition in Cryptococcus Neoformans. Infect. Immun. 1996, 64, 2420–2424. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Kang, M.; Tschirhart, T.; Malo, M.; Dadachova, E.; Cao, G.; Yin, J.-J.; Bentley, W.E.; Wang, Z.; Payne, G.F. Spectroelectrochemical Reverse Engineering DemonstratesThat Melanin’s Redox and Radical Scavenging Activities Are Linked. Biomacromolecules 2017, 18, 4084–4098. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Decreased Susceptibility of Melanized Cryptococcus Neoformans to UV Light. Appl. Environ. Microbiol. 1994, 60, 3864–3866. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Duan, Z.; Huang, W.; Gao, Q.; Wang, C. Improving UV Resistance and Virulence of Beauveria Bassiana by Genetic Engineering with an Exogenous Tyrosinase Gene. J. Invertebr. Pathol. 2012, 109, 105–109. [Google Scholar] [CrossRef]
- Blachowicz, A.; Raffa, N.; Bok, J.W.; Choera, T.; Knox, B.; Lim, F.Y.; Huttenlocher, A.; Wang, C.C.C.; Venkateswaran, K.; Keller, N.P. Contributions of Spore Secondary Metabolites to UV-C Protection and Virulence Vary in Different Aspergillus Fumigatus Strains. mBio 2020, 11, e03415-19. [Google Scholar] [CrossRef] [Green Version]
- Cortesão, M.; de Haas, A.; Unterbusch, R.; Fujimori, A.; Schütze, T.; Meyer, V.; Moeller, R. Aspergillus Niger Spores Are Highly Resistant to Space Radiation. Front. Microbiol. 2020, 11, 560. [Google Scholar] [CrossRef] [Green Version]
- Maity, J.P.; Kar, S.; Banerjee, S.; Sudershan, M.; Chakraborty, A.; Santra, S.C. Effects of Gamma Radiation on Fungi Infected Rice (in Vitro). Int. J. Radiat. Biol. 2011, 87, 1097–1102. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P. Sex or No Sex: Evolutionary Adaptation Occurs Regardless. Bioessays 2014, 36, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable Elements Contribute to Fungal Genes and Impact Fungal Lifestyle. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Vicente, V.A.; Najafzadeh, M.J.; Harrak, M.J.; Badali, H.; Seyedmousavi, S. Waterborne Exophiala Species Causing Disease in Cold-Blooded Animals. Pers. Mol. Phylogeny Evol. Fungi 2011, 27, 46–72. [Google Scholar] [CrossRef] [Green Version]
- Blasi, B.; Poyntner, C.; Rudavsky, T.; Prenafeta-Boldú, F.X.; de Hoog, S.; Tafer, H.; Sterflinger, K. Pathogenic Yet Environmentally Friendly? Black Fungal Candidates for Bioremediation of Pollutants. Geomicrobiol. J. 2016, 33, 308–317. [Google Scholar] [CrossRef]
- Parales, R.E.; Parales, J.V.; Pelletier, D.A.; Ditty, J.L. Chapter 1 Diversity of Microbial Toluene Degradation Pathways. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 64, pp. 1–73. ISBN 978-0-12-374338-1. [Google Scholar]
- Blasi, B.; Tafer, H.; Kustor, C.; Poyntner, C.; Lopandic, K.; Sterflinger, K. Genomic and Transcriptomic Analysis of the Toluene Degrading Black Yeast Cladophialophora Immunda. Sci. Rep. 2017, 7, 11436. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Toledo, A.V.; Franco, M.E.E.; Yanil Lopez, S.M.; Troncozo, M.I.; Saparrat, M.C.N.; Balatti, P.A. Melanins in Fungi: Types, Localization and Putative Biological Roles. Physiol. Mol. Plant Pathol. 2017, 99, 2–6. [Google Scholar] [CrossRef]
- Bell, A.A.; Wheeler, M.H. Biosynthesis and Functions of Fungal Melanins. Annu. Rev. Phytopathol. 1986, 24, 411–451. [Google Scholar] [CrossRef]
- Langfelder, K.; Streibel, M.; Jahn, B.; Haase, G.; Brakhage, A.A. Biosynthesis of Fungal Melanins and Their Importance for Human Pathogenic Fungi. Fungal Genet. Biol. 2003, 38, 143–158. [Google Scholar] [CrossRef]
- Feng, B.; Wang, X.; Hauser, M.; Kaufmann, S.; Jentsch, S.; Haase, G.; Becker, J.M.; Szaniszlo, P.J. Molecular Cloning and Characterization of WdPKS1, a Gene Involved in Dihydroxynaphthalene Melanin Biosynthesis and Virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 2001, 69, 1781–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, M.H.; Abramczyk, D.; Puckhaber, L.S.; Naruse, M.; Ebizuka, Y.; Fujii, I.; Szaniszlo, P.J. New Biosynthetic Step in the Melanin Pathway of Wangiella (Exophiala) dermatitidis: Evidence for 2-Acetyl-1,3,6,8-Tetrahydroxynaphthalene as a Novel Precursor. Eukaryot. Cell 2008, 7, 1699–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, S.; Chi, Z.; Chen, L.; Liu, G.-L.; Hu, Z.; Chi, Z.-M. Molecular Evolution and Regulation of DHN Melanin-Related Gene Clusters Are Closely Related to Adaptation of Different Melanin-Producing Fungi. Genomics 2021, 113, 1962–1975. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shan, L.; Yang, S.; Ma, A. Identification and Antioxidant Activity of Melanin Isolated FromHypoxylon Archeri, a Companion Fungus OfTremella Fuciformis. J. Basic Microbiol. 2008, 48, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.C.R.; Lisboa, H.C.F.; Pombeiro-Sponchiado, S.R. Characterization of Melanin Pigment Produced by Aspergillus Nidulans. World J. Microbiol. Biotechnol. 2012, 28, 1467–1474. [Google Scholar] [CrossRef]
- Liu, D.; Wei, L.; Guo, T.; Tan, W. Detection of DOPA-Melanin in the Dimorphic Fungal Pathogen Penicillium Marneffei and Its Effect on Macrophage Phagocytosis In Vitro. PLoS ONE 2014, 9, e92610. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Paes, R.; Figueiredo-Carvalho, M.H.G.; Brito-Santos, F.; Almeida-Silva, F.; Oliveira, M.M.E.; Zancopé-Oliveira, R.M. Melanins Protect Sporothrix Brasiliensis and Sporothrix Schenckii from the Antifungal Effects of Terbinafine. PLoS ONE 2016, 11, e0152796. [Google Scholar] [CrossRef]
- Prados-Rosales, R.; Toriola, S.; Nakouzi, A.; Chatterjee, S.; Stark, R.; Gerfen, G.; Tumpowsky, P.; Dadachova, E.; Casadevall, A. Structural Characterization of Melanin Pigments from Commercial Preparations of the Edible Mushroom Auricularia Auricula. J. Agric. Food Chem. 2015, 63, 7326–7332. [Google Scholar] [CrossRef] [Green Version]
- Schmaler-Ripcke, J.; Sugareva, V.; Gebhardt, P.; Winkler, R.; Kniemeyer, O.; Heinekamp, T.; Brakhage, A.A. Production of Pyomelanin, a Second Type of Melanin, via the Tyrosine Degradation Pathway in Aspergillus Fumigatus. AEM 2009, 75, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Vasanthakumar, A.; DeAraujo, M.; DeAraujo, A.; Schilling, M.; Mitchell, R. Pyomelanin Production in Penicillium Chrysogenum Is Stimulated by L-Tyrosine. Microbiology 2015, 161, 1211–1218. [Google Scholar] [CrossRef]
- Demain, A.; Fang, A. The Natural Functions of Secondary Metabolites. In History of Modern Biotechnology I; Springer: Singapore, 2000; pp. 1–39. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Narsing Rao, M.P.; Xiao, M.; Li, W.-J. Fungal and Bacterial Pigments: Secondary Metabolites with Wide Applications. Front. Microbiol. 2017, 8, 1113. [Google Scholar] [CrossRef]
- Romsdahl, J.; Wang, C.C.C. Recent Advances in the Genome Mining of Aspergillus Secondary Metabolites (Covering 2012–2018). Med. Chem. Commun. 2019, 10, 840–866. [Google Scholar] [CrossRef]
- Dadachova, E.; Bryan, R.A.; Howell, R.C.; Schweitzer, A.D.; Aisen, P.; Nosanchuk, J.D.; Casadevall, A. The Radioprotective Properties of Fungal Melanin Are a Function of Its Chemical Composition, Stable Radical Presence and Spatial Arrangement: Radioprotective Properties of Fungal Melanin. Pigment Cell Melanoma Res. 2007, 21, 192–199. [Google Scholar] [CrossRef]
- Pacelli, C.; Bryan, R.A.; Onofri, S.; Selbmann, L.; Shuryak, I.; Dadachova, E. Melanin Is Effective in Protecting Fast and Slow Growing Fungi from Various Types of Ionizing Radiation. Environ. Microbiol. 2017, 19, 1612–1624. [Google Scholar] [CrossRef]
- Schultzhaus, Z.; Chen, A.; Kim, S.; Shuryak, I.; Chang, M.; Wang, Z. Transcriptomic Analysis Reveals the Relationship of Melanization to Growth and Resistance to Gamma Radiation in Cryptococcus Neoformans. Environ. Microbiol. 2019, 21, 2613–2628. [Google Scholar] [CrossRef]
- Robertson, K.L.; Mostaghim, A.; Cuomo, C.A.; Soto, C.M.; Lebedev, N.; Bailey, R.F.; Wang, Z. Adaptation of the Black Yeast Wangiella Dermatitidis to Ionizing Radiation: Molecular and Cellular Mechanisms. PLoS ONE 2012, 7, e48674. [Google Scholar] [CrossRef] [Green Version]
- Robert, V.A.; Casadevall, A. Vertebrate Endothermy Restricts Most Fungi as Potential Pathogens. J. Infect. Dis. 2009, 200, 1623–1626. [Google Scholar] [CrossRef]
- Elias, P.M. The Skin Barrier as an Innate Immune Element. Semin. Immunopathol. 2007, 29, 3–14. [Google Scholar] [CrossRef]
- Dantas, A.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative Stress Responses in the Human Fungal Pathogen, Candida Albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef] [Green Version]
- Nomura, M. Regulation of Ribosome Biosynthesis in Escherichia Coli and Saccharomyces Cerevisiae: Diversity and Common Principles. J. Bacteriol. 1999, 181, 6857–6864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brauer, M.J.; Huttenhower, C.; Airoldi, E.M.; Rosenstein, R.; Matese, J.C.; Gresham, D.; Boer, V.M.; Troyanskaya, O.G.; Botstein, D. Coordination of Growth Rate, Cell Cycle, Stress Response, and Metabolic Activity in Yeast. MBoC 2008, 19, 352–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wool, I.G. Extraribosomal Functions of Ribosomal Proteins. Trends Biochem. Sci. 1996, 21, 164–165. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How Common Are Extraribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-D.; Xie, L.; Wei, Y.; Zhou, X.; Jia, B.; Liu, J.; Zhang, S. Abiotic Stress Resistance, a Novel Moonlighting Function of Ribosomal Protein RPL44 in the Halophilic Fungus Aspergillus Glaucus. Appl. Environ. Microbiol. 2014, 80, 4294–4300. [Google Scholar] [CrossRef] [Green Version]
- Prenafeta-Boldú, F.X.; Kuhn, A.; Luykx, D.M.A.M.; Anke, H.; van Groenestijn, J.W.; de Bont, J.A.M. Isolation and Characterisation of Fungi Growing on Volatile Aromatic Hydrocarbons as Their Sole Carbon and Energy Source. Mycol. Res. 2001, 105, 477–484. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Summerbell, R.; Sybren de Hoog, G. Fungi Growing on Aromatic Hydrocarbons: Biotechnology’s Unexpected Encounter with Biohazard? FEMS Microbiol. Rev. 2006, 30, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Pavan, M.E.; López, N.I.; Pettinari, M.J. Melanin Biosynthesis in Bacteria, Regulation and Production Perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 1357–1370. [Google Scholar] [CrossRef]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [Green Version]


| Genome Feature | Exophiala lecanii-corni | Exophiala mesophila | Exophiala dermatitidis |
|---|---|---|---|
| CBS 102400 | CCFEE 6314 [38] | NIH/UT8656 | |
| Size (Mb) | 34.42 | 30.43 | 26.38 |
| GC content (%) | 48.94 | 50 | 51.47 |
| Scaffolds | 13 | 207 | 11 |
| N50 (Mb) | 2.95 | 0.43 | 3.62 |
| tRNA (#) | 64 | 38 | 73 |
| Protein-coding genes (#) | 11,005 | 10,355 | 9562 |
| Exophiala lecanii-corni | Exophiala dermatitidis | ||||||
|---|---|---|---|---|---|---|---|
| Number of Elements | % of Genome Sequences | % of Repeat Sequences | Number of Elements | % of Genome Sequences | % of Repeat Sequences | ||
| Retro- elements | LINEs | 0 | 0% | 0% | 14 | 0.21% | 5.8% |
| Ty1/Copia (LTR) | 76 | 0.78% | 20.8% | 53 | 0.30% | 8.4% | |
| Gypsy/DIRS1 (LTR) | 59 | 0.45% | 12.0% | 128 | 1.09% | 30.4% | |
| Total | 135 | 1.23% | 32.8% | 195 | 1.60% | 44.7% | |
| DNA transposons | Tcl-IS630-Pogo | 103 | 0.12% | 3.2% | 30 | 0.12% | 3.3% |
| PiggyBac | 39 | 0.22% | 5.90% | 0 | 0.00% | 0.00% | |
| Total | 142 | 0.34% | 9.0% | 30 | 0.12% | 3.3% | |
| Unclassified | 383 | 1.13% | 30.0% | 811 | 0.95% | 26.7% | |
| Total | 660 | 2.70% | 71.70% | 1036 | 2.67% | 74.70% | |
| Gene Annotation | E. lecanii-corni | C. immunda CBS 110551 | C. immunda CBS 834.96 | BLAST Score (Bits) | E-Value |
|---|---|---|---|---|---|
| Cytochrome P450 | EXLC_010870T0 | CLAIMM_07379 | XP_016251043.1 | 723 | 0.0 |
| Cytochrome P450 | EXLC_003659T0 | CLAIMM_09044 | XP_016247671.1 | 565 | 0.0 |
| Cytochrome P450 | EXLC_001047T0 | CLAIMM_11603 | 519 | 0.0 | |
| Cytochrome P450 | EXLC_007222T0 | CLAIMM_10796 | XP_016250278.1 | 668 | 0.0 |
| Benzylalcohol dehydrogenase (BADH) | EXLC_009515T0 | CLAIMM_14566 | 317 | 1 × 10−70 | |
| Benzylalcohol dehydrogenase (BADH) | EXLC_000716T0 | CLAIMM_12646 | 290 | 4 × 10−96 | |
| Benzylalcohol dehydrogenase (BADH) | EXLC_010898T0 | CLAIMM_02069 | 350 | 2 × 10−119 | |
| Benzaldehyde dehydrogenase (BZDH) | EXLC_003682T0 | CLAIMM_14573 | XP_016255229.1 | 536 | 0.0 |
| Benzaldehyde dehydrogenase (BZDH) | EXLC_007394T0 | CLAIMM_12645 | 410 | 4 × 10−140 | |
| Benzaldehyde dehydrogenase (BZDH) | EXLC_003083T0 | CLAIMM_07604 | 310 | 4 × 10−101 | |
| Benzoate hydroxylase (BH) | EXLC_000444T0 | CLAIMM_00094 | XP_016243286.1 | 936 | 0.0 |
| p-hydroxybenzoate hydroxylase (PHBH) | EXLC_002510T0 | CLAIMM_03385 | 1021 | 0.0 | |
| Decarboxylase (PCAD) | EXLC_008460T0 | CLAIMM_02205 | 339 | 2 × 10−111 | |
| Dioxygenase (P34O) | EXLC_010602T0 | CLAIMM_02325 | XP_016246009.1 | 517 | 0.0 |
| Catechol 1,2-dioxygenase (C12O) | EXLC_006633T0 | CLAIMM_07423 | 234 | 1 × 10−75 | |
| Catechol 1,2-dioxygenase (C12O) | EXLC_005860T0 | CLAIMM_03467 | XP_016253603.1 | 336 | 1 × 10−115 |
| β-carboxy-muconate lactonizing enzyme (CMLE) | EXLC_001488T0 | CLAIMM_03466 | 285 | 6 × 10−96 | |
| β-carboxy-muconate lactonizing enzyme (CMLE) | EXLC_010962T0 | CLAIMM_08374 | 112 | 1 × 10−32 | |
| Carboxy-muconolactone decarboxylase (CMD) | EXLC_001487T0 | CLAIMM_03468 | XP_016253602.1 | 203 | 2 × 10−67 |
| Carboxy-muconolactone decarboxylase (CMD) | EXLC_004832T0 | CLAIMM_07415 | XP_016251077.1 | 73.9 | 6 × 10−15 |
| Muconate lactonizing enzyme (MLE) | EXLC_006601T0 | CLAIMM_14446 | XP_016251156.1 | 449 | 6 × 10−157 |
| β-ketoadipate-enol-actone hydrolase (ELH) | EXLC_008892T0 | CLAIMM_05933 | 417 | 1 × 10−149 | |
| β-ketoadipate succinyl-CoA transferase (TR) | EXLC_006182T0 | CLAIMM_07426 | 645 | 0.0 | |
| β-ketoadipyl-CoA thiolase (TH) | EXLC_000315T0 | CLAIMM_07421 | 427 | 2 × 10−148 |
| Gene | Annotation | Exophiala lecanii-corni | Exophiala dermatitidis |
|---|---|---|---|
| DHN-melanin biosynthetic pathway | |||
| Pks1 | Polyketide synthase | EXLC_004613T0, EXLC_009455T0 | HMPREF1120_03173 |
| Arp1 | Scytalone dehydratase | EXLC_004054T0 | HMPREF1120_07724 |
| Arp2 | 1,3,6,8-tetrahydroxynaphthalene reductase | EXLC_001201T0, EXLC_006778T0 | HMPREF1120_05939 |
| Ayg1 | Alpha/beta hydrolase | EXLC_008577T0 | HMPREF1120_00377 |
| Alpha/beta hydrolase | EXLC_003727T0 | HMPREF1120_02312 | |
| Abr2 | Laccase | EXLC_004610T0, EXLC_010027T0 | HMPREF1120_02828, HMPREF1120_05645 |
| Abr1 | Ferrooxidoreductase | EXLC_006706T0 | HMPREF1120_00173, HMPREF1120_01590, HMPREF1120_04510 |
| L-ascorbate oxidase | EXLC_003729T0, EXLC_006667T0 | HMPREF1120_03706, HMPREF1120_04536 | |
| DOPA-melanin biosynthetic pathway | |||
| MelC2 | Tyrosinase | EXLC_006713T0, EXLC_009460T0, EXLC_010702T0 | HMPREF1120_03345, HMPREF1120_04514, HMPREF1120_05316 |
| MelO | Tyrosinase | EXLC_000450T0, EXLC_009626T0 | HMPREF1120_07692 |
| MelO | Multicopper oxidase | EXLC_005401T0, EXLC_010629T0 | HMPREF1120_05865 |
| MelO | Multicopper oxidase | EXLC_001274T0 | HMPREF1120_00199 |
| MelO | Multicopper oxidase | EXLC_002340T0, EXLC_008951T0 | HMPREF1120_08116 |
| MelO | Multicopper oxidase | EXLC_009732T0, EXLC_009741T0 | HMPREF1120_02754, HMPREF1120_04578, HMPREF1120_08564 |
| L-tyrosine degradation pathway | |||
| Tat | Tyrosine aminotransferase | EXLC_000526T0 | HMPREF1120_02164 |
| HppD | 4-Hydroxyphenylpyruvate dioxygenase | EXLC_000706T0 | HMPREF1120_05584 |
| HmgA | Homogentisate dioxygenase | EXLC_000997T0, EXLC_002037T0, EXLC_006635T0, EXLC_007332T0, EXLC_008087T0, EXLC_010843T0 | HMPREF1120_03827 |
| FahA | Fumarylacetoacetate hydrolase | EXLC_000996T0, EXLC_002038T0, EXLC_007334T0 | HMPREF1120_03825 |
| MaiA | Maleylacetoacetate isomerase | EXLC_009316T0 | HMPREF1120_03438 |
| PKS/PKS-Like | NRPS/NRPS-Like | TC | Total | |
|---|---|---|---|---|
| Aspergillus niger | 46 | 35 | 7 | 88 |
| Aspergillus nidulans | 33 | 25 | 2 | 60 |
| Coccidioides immitis | 10 | 11 | 4 | 25 |
| Exophiala aquamarina | 7 | 12 | 6 | 25 |
| Fonsecaea pedrosoi | 4 | 11 | 5 | 20 |
| Cladophialophora bantiana | 4 | 10 | 5 | 19 |
| Exophiala spinifera | 5 | 11 | 3 | 19 |
| Exophiala dermatitidis | 3 | 8 | 4 | 15 |
| Exophiala lecanii-corni | 5 | 7 | 3 | 15 |
| Exophiala mesophila | 2 | 7 | 4 | 13 |
| Schizosaccharomyces pombe | 0 | 3 | 1 | 4 |
| Saccharomyces cerevisiae | 0 | 1 | 1 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romsdahl, J.; Schultzhaus, Z.; Cuomo, C.A.; Dong, H.; Abeyratne-Perera, H.; Hervey, W.J., IV; Wang, Z. Phenotypic Characterization and Comparative Genomics of the Melanin-Producing Yeast Exophiala lecanii-corni Reveals a Distinct Stress Tolerance Profile and Reduced Ribosomal Genetic Content. J. Fungi 2021, 7, 1078. https://doi.org/10.3390/jof7121078
Romsdahl J, Schultzhaus Z, Cuomo CA, Dong H, Abeyratne-Perera H, Hervey WJ IV, Wang Z. Phenotypic Characterization and Comparative Genomics of the Melanin-Producing Yeast Exophiala lecanii-corni Reveals a Distinct Stress Tolerance Profile and Reduced Ribosomal Genetic Content. Journal of Fungi. 2021; 7(12):1078. https://doi.org/10.3390/jof7121078
Chicago/Turabian StyleRomsdahl, Jillian, Zachary Schultzhaus, Christina A. Cuomo, Hong Dong, Hashanthi Abeyratne-Perera, W. Judson Hervey, IV, and Zheng Wang. 2021. "Phenotypic Characterization and Comparative Genomics of the Melanin-Producing Yeast Exophiala lecanii-corni Reveals a Distinct Stress Tolerance Profile and Reduced Ribosomal Genetic Content" Journal of Fungi 7, no. 12: 1078. https://doi.org/10.3390/jof7121078
APA StyleRomsdahl, J., Schultzhaus, Z., Cuomo, C. A., Dong, H., Abeyratne-Perera, H., Hervey, W. J., IV, & Wang, Z. (2021). Phenotypic Characterization and Comparative Genomics of the Melanin-Producing Yeast Exophiala lecanii-corni Reveals a Distinct Stress Tolerance Profile and Reduced Ribosomal Genetic Content. Journal of Fungi, 7(12), 1078. https://doi.org/10.3390/jof7121078







