Chitin Deacetylase, a Novel Target for the Design of Agricultural Fungicides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants, Fungi, Bacteria and Culture Conditions
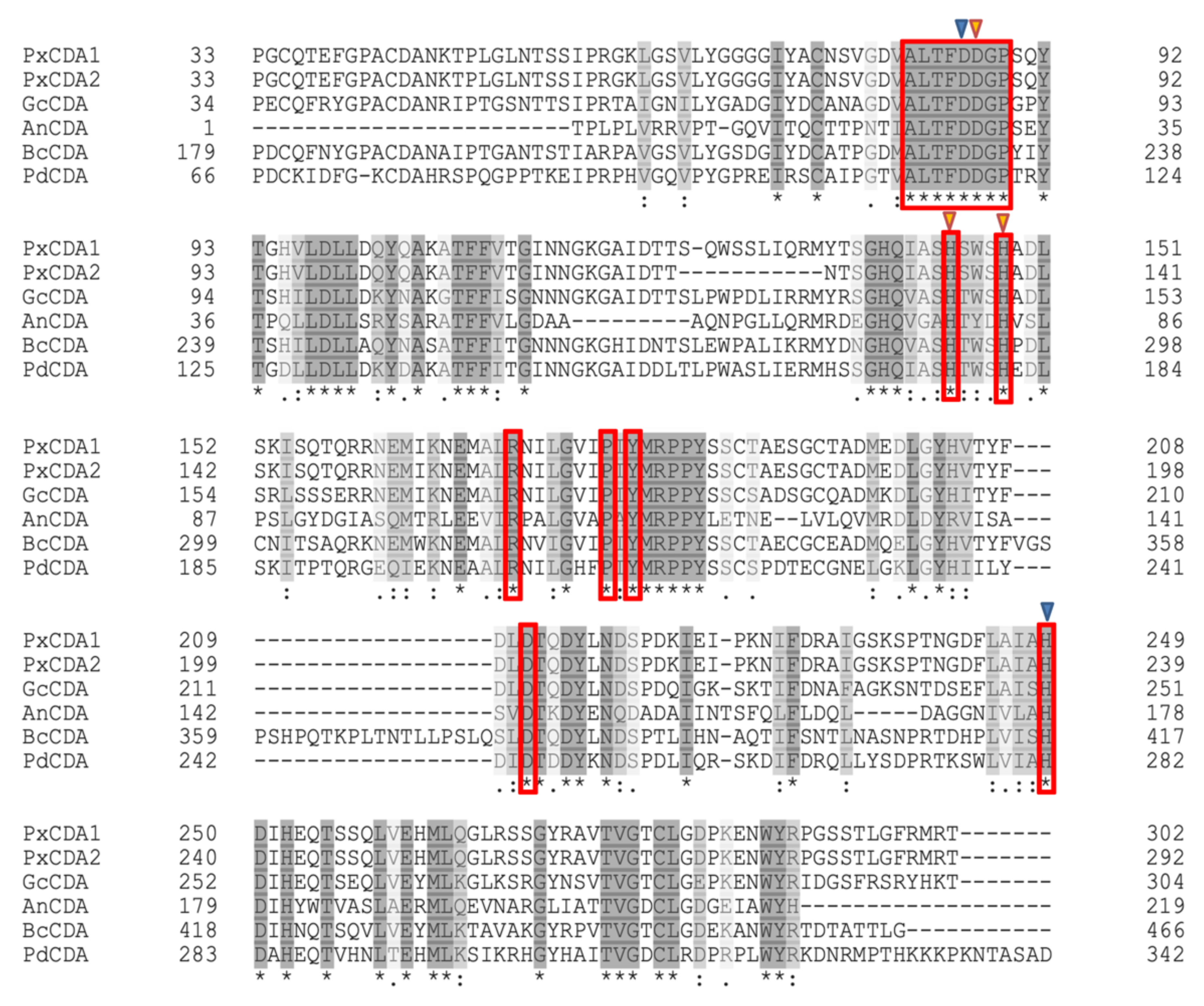
2.2. Sequence Analysis
2.3. RNA Isolation and cDNA Synthesis
2.4. Plasmid Construction
2.5. A. Tumefaciens-Mediated Host-Induced Gene Silencing (ATM−HIGS)
2.6. RT-qPCR
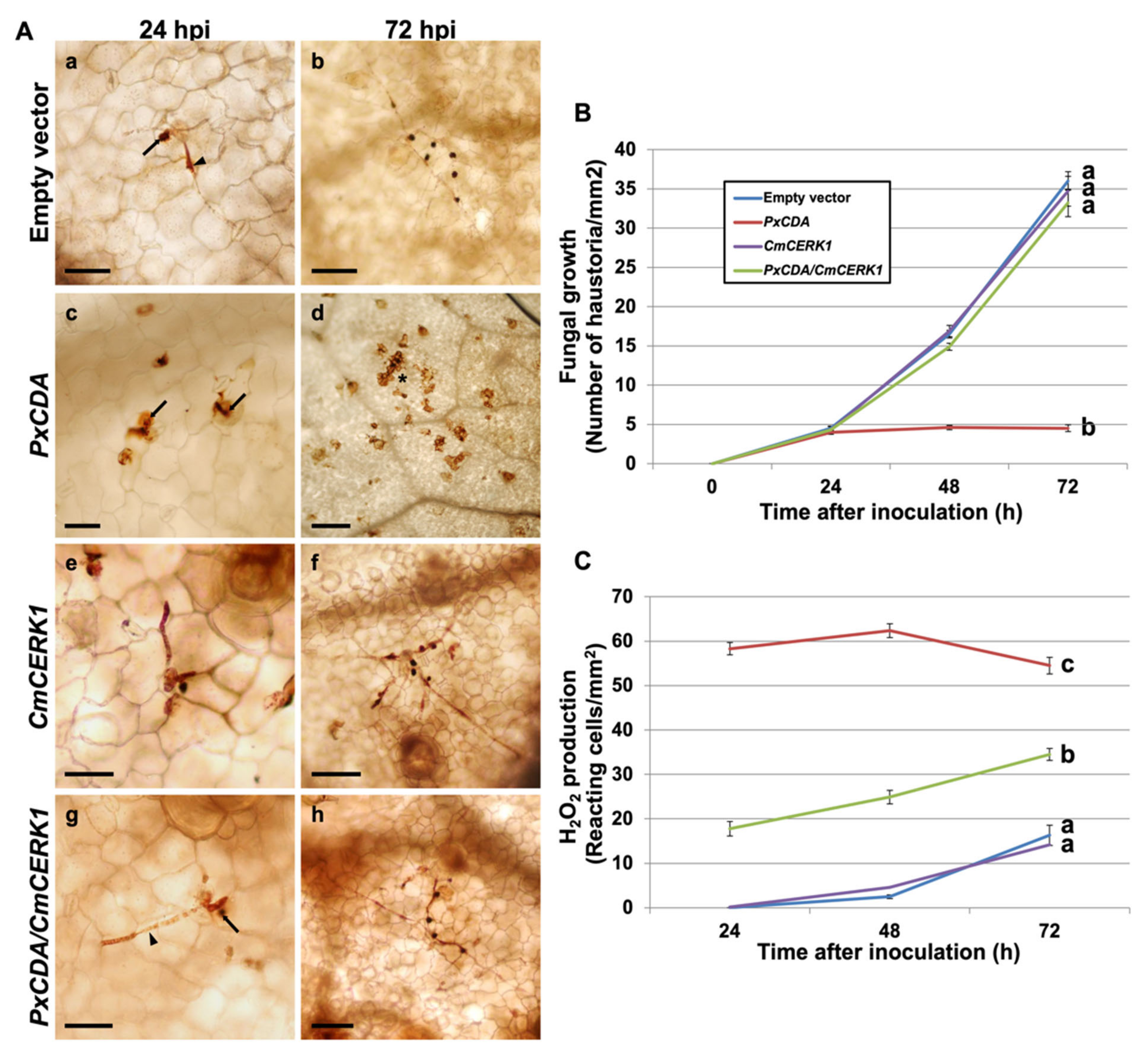
2.7. Hastorial Counts and Production of Hydrogen Peroxide
2.8. Leaf Disc and Plant Assays
2.9. Molecular Docking
2.10. Fruit Assays
2.11. Statistical Analysis
3. Results
3.1. Chitin Deacetylase Is an Essential Protein for P. xanthii Development
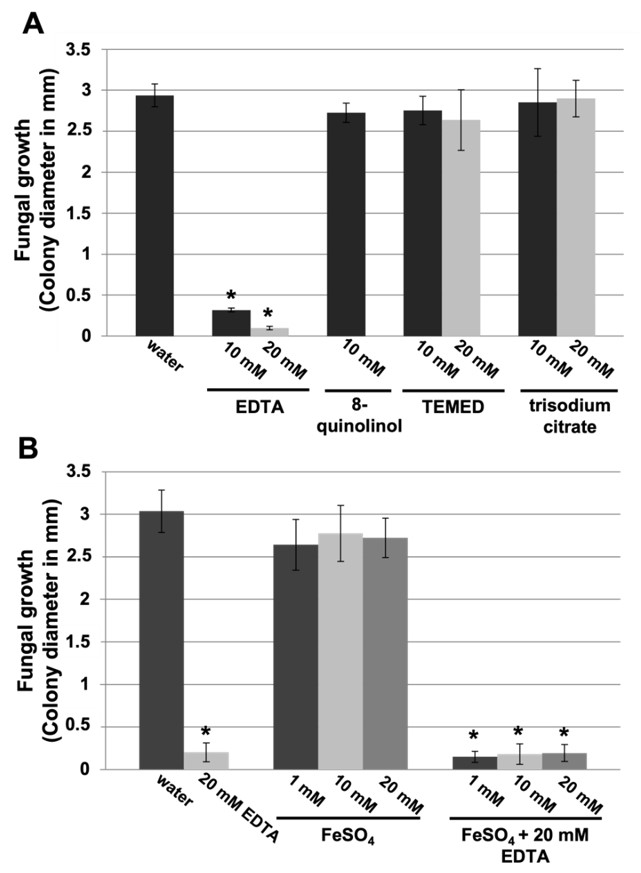
3.2. Small Carboxylic Acids Also Suppress Powdery Mildew Disease
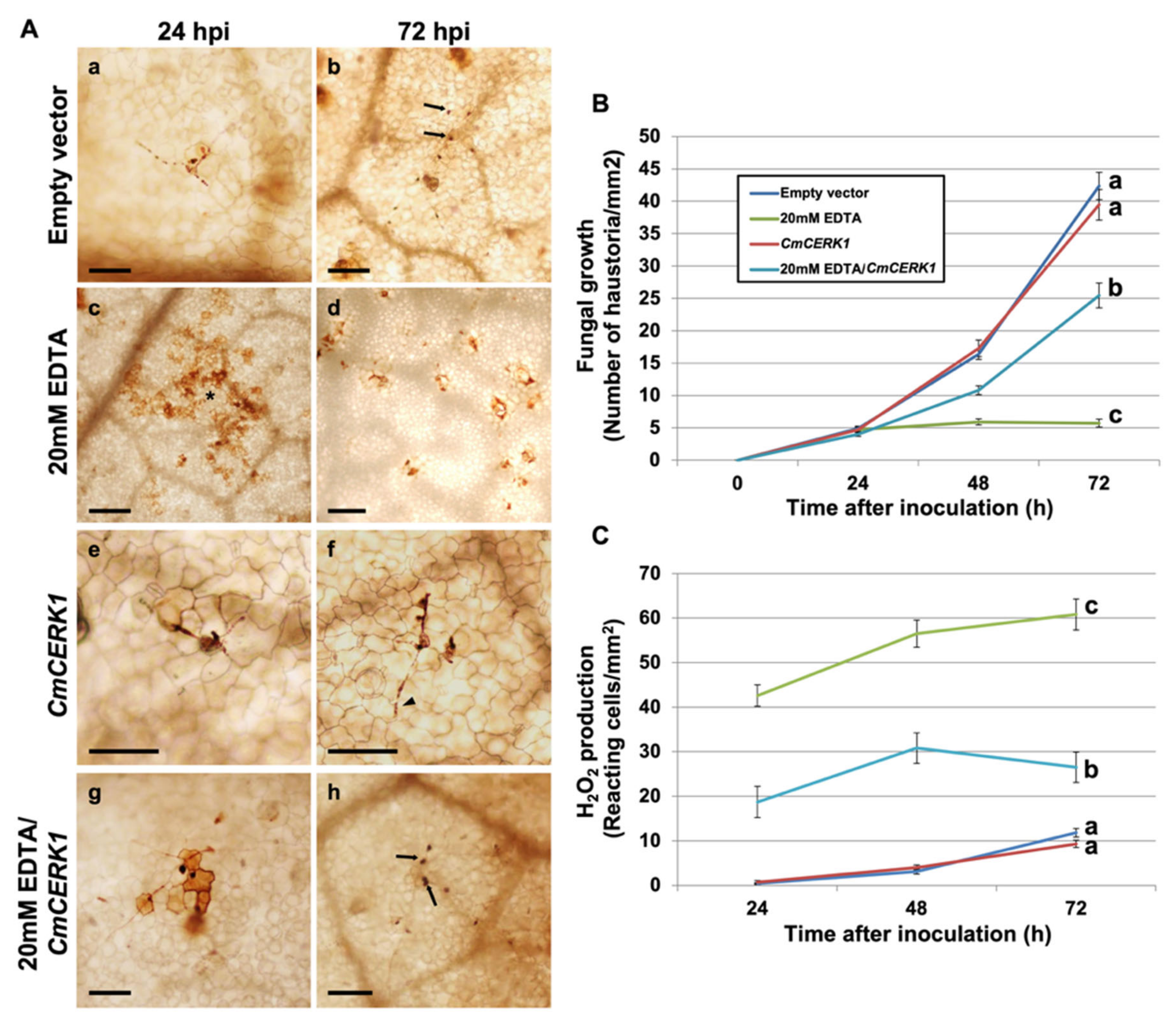
3.3. EDTA Inhibits P. xanthii Development by Targeting CDA and Eliciting Chitin-Triggered Immunity
3.4. EDTA Treatments Also Suppress Other Fungal Diseases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glawe, D.A. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phythopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef]
- Pérez-García, A.; Romero, D.; Fernández-Ortuño, D.; López-Ruiz, F.; De Vicente, A.; Torés, J.A. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 2009, 10, 153–160. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Pérez-García, A.; López-Ruiz, F.; Romero, D.; de Vicente, A.; Torés, J.A. Occurrence and distribution of resistance to QoI fungicides in populations of Podosphaera fusca in south central Spain. Eur. J. Plant Pathol. 2006, 115, 215–222. [Google Scholar] [CrossRef] [Green Version]
- López-Ruiz, F.J.; Pérez-García, A.; Fernández-Ortuño, D.; Romero, D.; García, E.; de Vicente, A.; Brown, J.K.; Torés, J.A. Sensitivities to DMI fungicides in populations of Podosphaera fusca in south central Spain. Pest Manag. Sci. 2010, 66, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Bellón-Gómez, D.; Vela-Corcía, D.; Pérez-García, A.; Torés, J.A. Sensitivity of Podosphaera xanthii populations to anti-powdery-mildew fungicides in Spain. Pest. Manag. Sci. 2015, 71, 1407–1413. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A.R. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujikawa, T.; Sakaguchi, A.; Nishizawa, Y.; Kouzai, Y.; Minami, E.; Yano, S.; Koga, H.; Meshi, T.; Nishimura, M. Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 2012, 8, e1002882. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M.F. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef] [Green Version]
- Christodoulidou, A.; Bouriotis, V.; Thireos, G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 31420–31425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Zhang, B.-S.; Zhao, J.-H.; Huang, J.-F.; Jia, P.-S.; Wang, S.; Zhang, J.; Zhou, J.-M.; Guo, H.-S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, Y.S.; Happel, P.; Lenz, S.; Mounashree, J.U.; Bonin, M.; Cord-Landwehr, S.; Singh, R.; Moerschbacher, B.M.; Kahmann, R. Chitosan and chitin deacetylase activity are necessary for development and virulence of Ustilago maydis. mBio 2021, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Brent, K.J.; Hollomon, D. Fungicide Resistance in Crop Pathogens. How Can It Be Managed? 2nd ed.; FRAC: Brussels, Belgium, 2007; pp. 9–13. [Google Scholar]
- Martínez-Cruz, J.M.; Romero, D.; de la Torre, F.N.; Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Pérez-García, A. The functional characterization of Podosphaera xanthii candidate effector genes reveals novel target functions for fungal pathogenicity. Mol. Plant-Microbe Interact. 2018, 31, 914–931. [Google Scholar] [CrossRef] [Green Version]
- Álvarez, B.; Torés, J.A. Cultivo in vitro de Sphaerotheca fuliginea (Schlecht. ex Fr.), efecto de diferentes fuentes de carbono sobre su desarrollo. Bol. San. Veg. Plagas 1997, 23, 283–288. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; López, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornton, B.; Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011, 39, 145–154. [Google Scholar] [CrossRef]
- Polonio, Á.; Seoane, P.; Claros, M.G.; Pérez-García, A. The haustorial transcriptome of the cucurbit pathogen Podosphaera xanthii reveals new insights into the biotrophy and pathogenesis of powdery mildew fungi. BMC Genom. 2019, 20, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vela-Corcía, D.; Bellón-Gómez, D.; López-Ruiz, F.; Torés, J.A.; Perez-García, A. The Podosphaera fusca TUB2 gene, a molecular “Swiss Army knife” with multiple applications in powdery mildew research. Fungal Biol. 2014, 118, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; van Aalten, D.M.F.; Eijsink, V.G.H. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Ortuño, D.; Bryson, P.K.; Grabke, A.; Schnabel, G. First report of fludioxonil resistance in Botrytis cinerea from a strawberry field in Virginia. Plant Dis. 2013, 97, 848. [Google Scholar] [CrossRef] [PubMed]
- Holmes, G.J.; Eckert, J.W.; Pitt, J.I. Revised description of Penicillium ulaiense and its role as a pathogen of citrus fruits. Phytopathology 1994, 84, 719–727. [Google Scholar] [CrossRef]
- Vela-Corcía, D. Development of Molecular and Genomic Tools for Functional Analysis in the Cucurbit Powdery Mildew Fungus Podosphaera fusca. Ph.D. Thesis, University of Malaga, Malaga, Spain, 2014. [Google Scholar]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polonio, A.; Díaz-Martínez, L.; Fernández-Ortuño, D.; de Vicente, A.; Romero, D.; López-Ruíz, F.J.; Pérez-García, A. A hybrid genome assembly resource for Podosphaera xanthii, the main causal agent of powdery mildew disease in cucurbits. Mol. Plant-Microbe Interact. 2021, 34, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Ito, E. A pathway of chitosan formation in Mucor rouxii. Eur. J. Biochem. 1975, 55, 71–78. [Google Scholar] [CrossRef]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Park, R.D.; Muzzarelli, R.A.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Ann. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; Hierrezuelo, J.; Thon, M.; de Vicente, A.; Pérez-García, A. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar] [CrossRef] [PubMed]
- Polonio, A.; Fernández-Ortuño, D.; de Vicente, A.; Pérez-García, A. A haustorial-expressed lytic polysaccharide monooxygenase from the cucurbit powdery mildew pathogen Podosphaera xanthii contributes to the suppression of chitin-triggered immunity. Mol. Plant Pathol. 2021, 22, 580–601. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; Polonio, A.; Ruiz-Jiménez, L.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Resistance to the SDHI fungicides boscalid and fluopyram in Podospharea xanthii from comercial cucurbit fields in Spain. J. Fungi 2021, 7, 733. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. USA 1993, 90, 2564–2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christodoulidou, A.; Briza, P.; Ellinger, A.; Bouriotis, V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999, 460, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, N.; Trombotto, S.; David, L.; Shirai, K. Activity of chitin deacetylase from Colletotrichum gloeosporioides on chitinous substrates. Carbohydr. Polym. 2013, 96, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Kakiki, K.; Misato, T. Mechanism of action of the antifugal agent polyoxin D. J. Bacteriol. 1970, 104, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanni, R.; Galvez-Llompart, M.; García-Domenech, R.; Galvez, J. What place does molecular topology have in today’s drug discovery? Expert. Opin. Drug Discov. 2020, 15, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Zanni, R.; Galvez-Llompart, M.; García-Domenech, R.; Galvez, J. Latest advances in molecular topology applications for drug discovery. Expert. Opin. Drug Discov. 2015, 10, 945–957. [Google Scholar] [CrossRef] [PubMed]



| Compound | Affinity (kcal mol−1) | Residues | Hydrogen Bonds (Number) | Distance (Å) |
|---|---|---|---|---|
| Chitotriose 1 | −6.1 | Tyr138 | 1 | 1.73 |
| His196 | 1 | 1.86 | ||
| EDTA | −6.8 | His97 | 1 | 1.76 |
| Tyr138 | 1 | 1.80 | ||
| Tyr166 | 1 | 1.77 | ||
| His199 | 1 | 1.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cruz, J.M.; Polonio, Á.; Zanni, R.; Romero, D.; Gálvez, J.; Fernández-Ortuño, D.; Pérez-García, A. Chitin Deacetylase, a Novel Target for the Design of Agricultural Fungicides. J. Fungi 2021, 7, 1009. https://doi.org/10.3390/jof7121009
Martínez-Cruz JM, Polonio Á, Zanni R, Romero D, Gálvez J, Fernández-Ortuño D, Pérez-García A. Chitin Deacetylase, a Novel Target for the Design of Agricultural Fungicides. Journal of Fungi. 2021; 7(12):1009. https://doi.org/10.3390/jof7121009
Chicago/Turabian StyleMartínez-Cruz, Jesús M., Álvaro Polonio, Riccardo Zanni, Diego Romero, Jorge Gálvez, Dolores Fernández-Ortuño, and Alejandro Pérez-García. 2021. "Chitin Deacetylase, a Novel Target for the Design of Agricultural Fungicides" Journal of Fungi 7, no. 12: 1009. https://doi.org/10.3390/jof7121009
APA StyleMartínez-Cruz, J. M., Polonio, Á., Zanni, R., Romero, D., Gálvez, J., Fernández-Ortuño, D., & Pérez-García, A. (2021). Chitin Deacetylase, a Novel Target for the Design of Agricultural Fungicides. Journal of Fungi, 7(12), 1009. https://doi.org/10.3390/jof7121009







