Morphological and Phylogenetic Appraisal of Novel and Extant Taxa of Stictidaceae from Northern Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collections and Isolation
2.2. Morphological Studies
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Sequence Alignment and Phylogenetic Analyses
3. Results
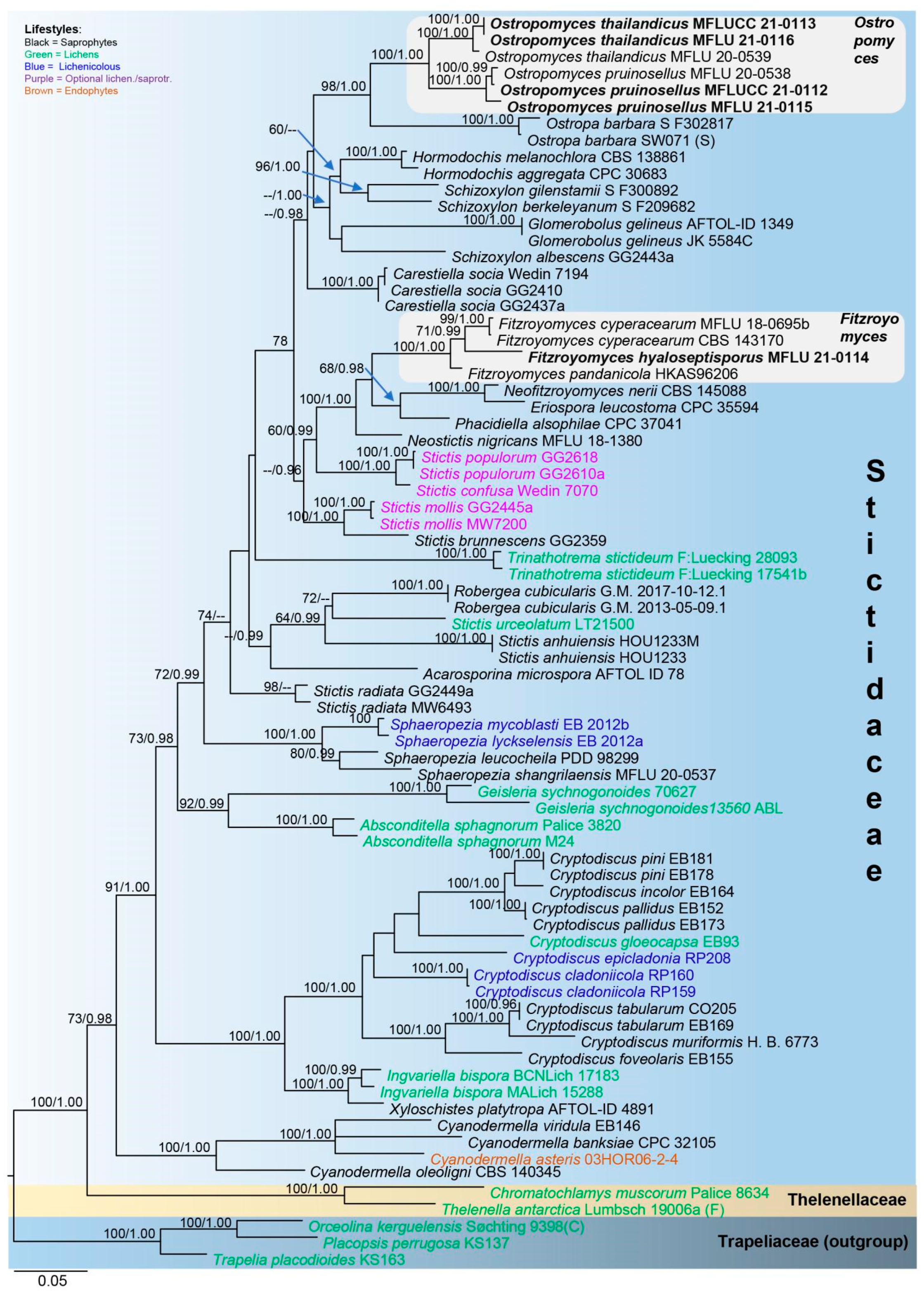
3.1. Phylogenetic Analysis
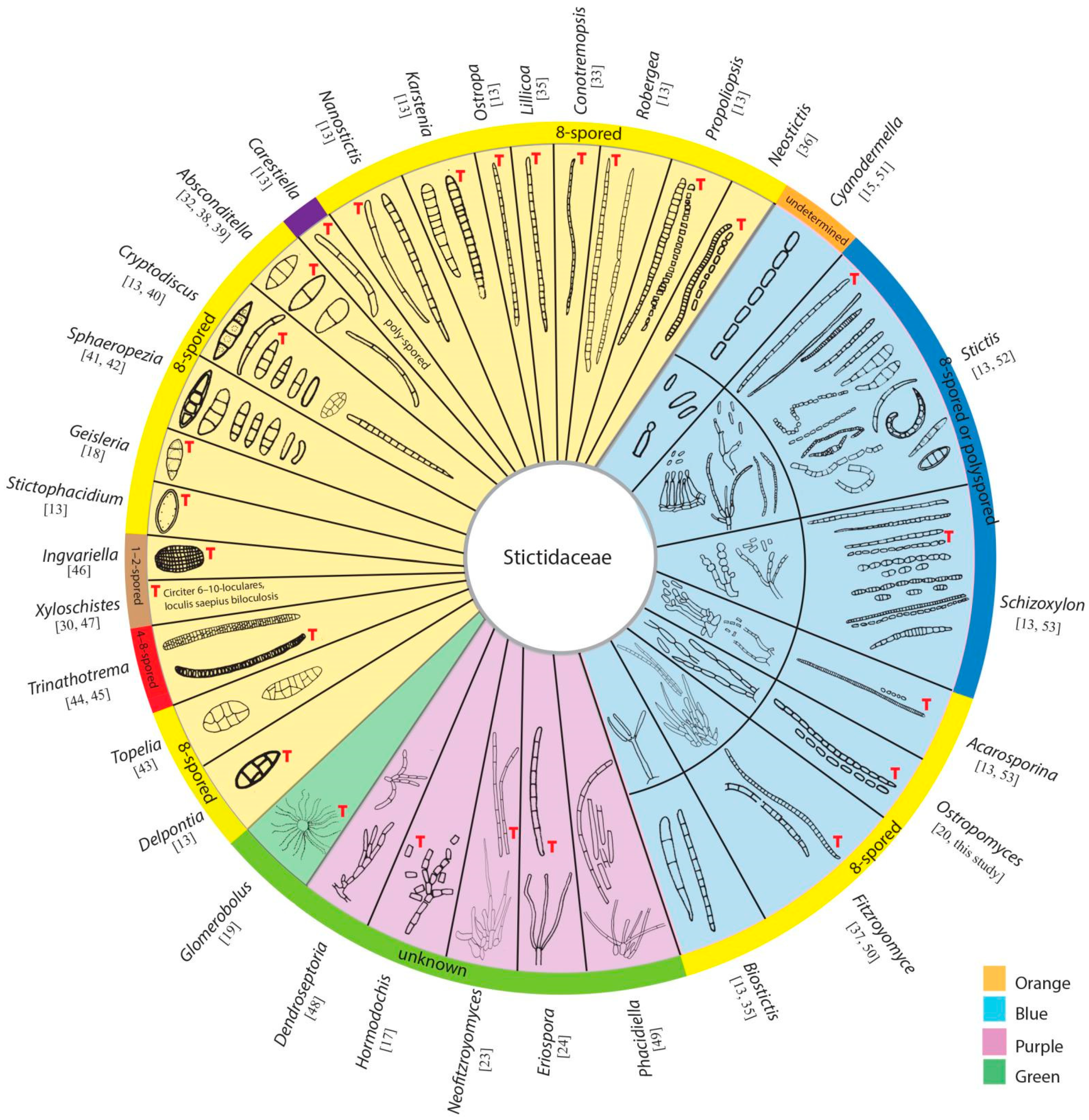
3.2. Taxonomy
3.2.1. Stictidaceae Fr. (as ‘Stictei’), Summa Veg. Scand., Sectio Post. (Stockholm): 345 (1849) Amend
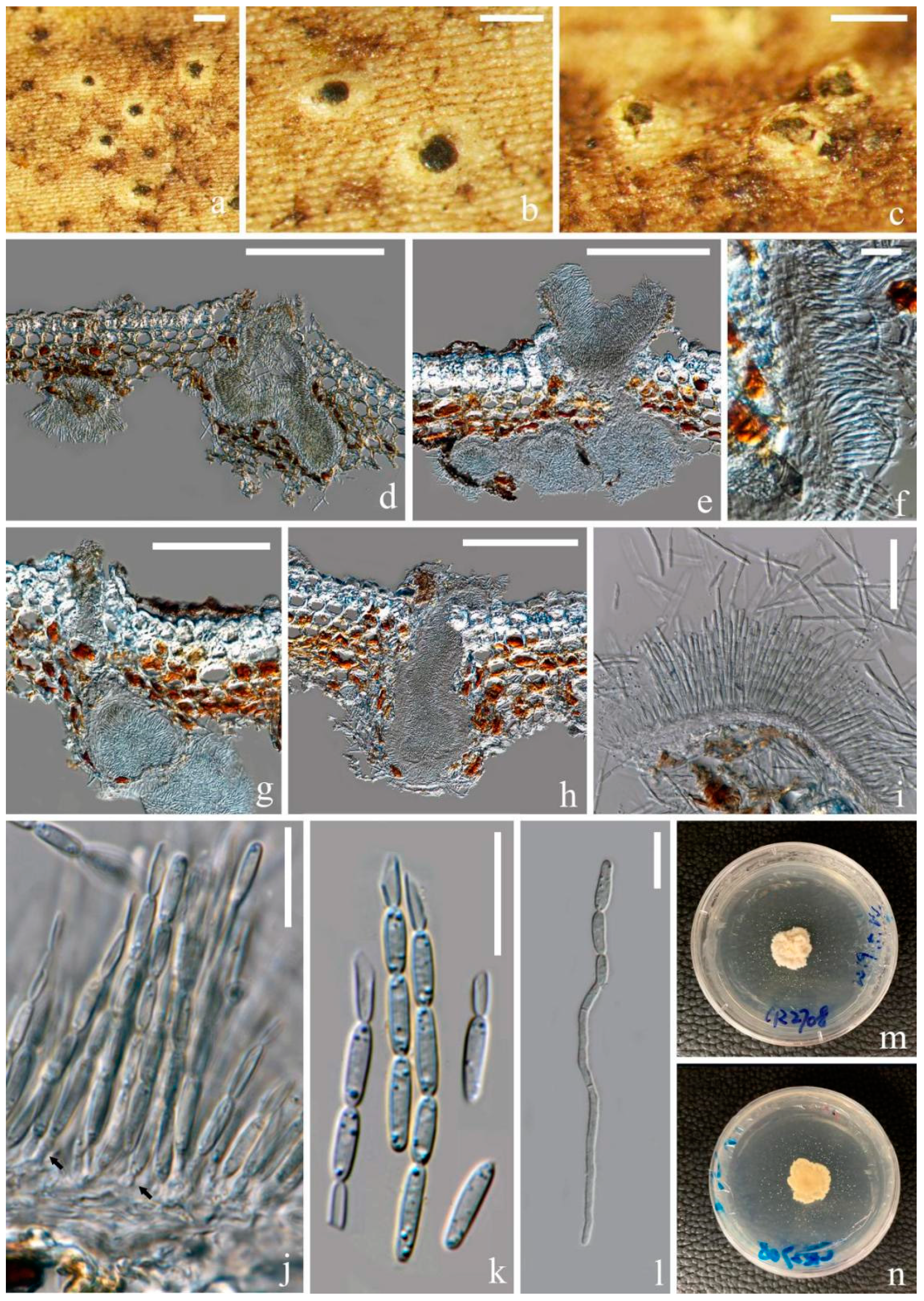
3.2.2. Fitzroyomyces Crous, Persoonia 39: 389 (2017)
3.2.3. Ostropomyces Thiyagaraja, Lücking, Ertz and K.D. Hyde, in Thiyagaraja, Lücking, Ertz, Karunarathna, Wanasinghe, Lumyong and Hyde, Journal of Fungi 7(no. 105): 11 (2021)
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nannfeldt, J.A. Studien iiber die Morphologie und Systematik der nichtlichenisierten inoperculaten Discomyceten. Nov. Acta Regiae Soc. Sci. Upsal. 1932, 8, 1–368. [Google Scholar]
- Baloch, E.; Lücking, R.; Lumbsch, H.T.; Wedin, M. Major clades and phylogenetic relationships between lichenized and non-lichenized lineages in Ostropales (Ascomycota: Lecanoromycetes). Taxon 2010, 59, 1483–1494. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Schmitt, I.; Lücking, R.; Wiklund, E.; Wedin, M. The phylogenetic placement of Ostropales within Lecanoromycetes (Ascomycota) revisited. Mycol. Res. 2007, 111, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Huhndorf, S.M. Outline of Ascomycota-2009. Myconet 2010, 14, 1–64. [Google Scholar]
- Lumbsch, H.T.; Huhndorf, S.M. Outline of Ascomycota-2007. Myconet 2007, 13, 1–58. [Google Scholar]
- Nelsen, M.P.; Lücking, R.; Andrew, C.J.; Aptroot, A.; Cáceres, M.E.S.; Mercado-Díaz, J.A.; Plata, E.R.; Lumbsch, H.T. Molecular phylogeny reveals the true colours of Myeloconidaceae (Ascomycota: Ostropales). Aust. Syst. Bot. 2014, 27, 38–47. [Google Scholar] [CrossRef]
- Kraichak, E.; Huang, J.-P.; Nelsen, M.; Leavitt, S.D.; Lumbsch, H.T. A revised classification of orders and families in the two major subclasses of Lecanoromycetes (Ascomycota) based on a temporal approach. Bot. J. Linn. Soc. 2018, 188, 233–249. [Google Scholar] [CrossRef]
- Lücking, R. Stop the abuse of time! Strict temporal banding is not the future of rank-based classifications in fungi (including lichens) and other organisms. CRC. Crit. Rev. Plant Sci. 2019, 38, 199–253. [Google Scholar] [CrossRef]
- Fries, E.M. Summa Vegetabilium Scandinaviae; Bonnier: Winter Park, FL, USA, 1849; Volume 2, pp. 259–572. [Google Scholar]
- Winka, K.; Ahlberg, C.; Eriksson, O.E. Are there lichenized Ostropales? Lichenologist 1998, 30, 455–462. [Google Scholar] [CrossRef]
- Sherwood, M.A. The ostropalean fungi. Mycotaxon 1977, 5, 1–277. [Google Scholar]
- Sherwood, M.A. The Ostropalean fungi II: Schizoxylon with notes on Stictis, Acarosporina, Coccopeziza, and Carestiella. Mycotaxon 1977, 6, 215–260. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.; Souza-Motta, C.M.; et al. Fungal Planet description sheets: 716–784. Persoonia 2018, 40, 240–393. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.-H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Wood, A.R.; Groenewald, J.Z. The Genera of Fungi–G5: Arthrinium, Ceratosphaeria, Dimerosporiopsis, Hormodochis, Lecanostictopsis, Lembosina, Neomelanconium, Phragmotrichum, Pseudomelanconium, Rutola, and Trullula. Fungal Syst. Evol. 2020, 5, 77–98. [Google Scholar] [CrossRef]
- Aptroot, A.; Parnmen, S.; Luecking, R.; Baloch, E.; Jungbluth, P.; Caceres, M.E.S.; Lumbsch, H.T. Molecular phylogeny resolves a taxonomic misunderstanding and places Geisleria close to Absconditella s. str.(Ostropales: Stictidaceae). Lichenologist 2014, 46, 115–128. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Volkmann-Kohlmeyer, B. Fungi on Juncus roemerianus. 6. Glomerobolus gen. nov., the first ballistic member of Agonomycetales. Mycologia 1996, 88, 328–337. [Google Scholar]
- Thiyagaraja, V.; Lücking, R.; Ertz, D.; Karunarathna, S.C.; Wanasinghe, D.N.; Lumyong, S.; Hyde, K.D. The evolution of life modes in Stictidaceae, with three novel taxa. J. Fungi 2021, 7, 105. [Google Scholar] [CrossRef]
- Ertz, D.; Sanderson, N.; Lebouvier, M. Thelopsis challenges the generic circumscription in the Gyalectaceae and brings new insights to the taxonomy of Ramonia. Lichenologist 2021, 53, 45–61. [Google Scholar] [CrossRef]
- Schoch, C.L.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; Tsui, C.K.M.; Spatafora, J.W. The halotolerant fungus Glomerobolus gelineus is a member of the Ostropales. Mycol. Res. 2006, 110, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Luangsa-Ard, J.J.; Wingfield, M.J.; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Akulov, A.; Bulgakov, T.S.; Carnegie, A.J.; Jurjević, Ž.; Decock, C.; Denman, S.; Lombard, L.; et al. New and Interesting Fungi. 3. Fungal Syst. Evol. 2020, 6, 157–231. [Google Scholar] [CrossRef] [PubMed]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar]
- Wedin, M.; Döring, H.; Koenberg, K.; Gilenstam, G. Generic delimitations in the family Stictidaceae (Ostropales, Ascomycota): The Stictis–Conotrema problem. Lichenologist 2005, 37, 67–75. [Google Scholar] [CrossRef]
- Baloch, E.; Gilenstam, G.; Wedin, M. Phylogeny and classification of Cryptodiscus, with a taxonomic synopsis of the Swedish species. Fungal Divers. 2009, 38, 51–68. [Google Scholar]
- Fernández-Brime, S.; Olariaga, I.; Baral, H.-O.; Friebes, G.; Jaklitsch, W.; Senn-Irlet, B.; Wedin, M. Cryptodiscus muriformis and Schizoxylon gilenstamii, two new species of Stictidaceae (Ascomycota). Mycol. Prog. 2018, 17, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.L.; Baral, H.-O.; Xu, X.; Liu, Y. Parakarstenia phyllostachydis, a new genus and species of non-lichenized Odontotremataceae (Ostropales, Ascomycota). Mycol. Prog. 2019, 18, 833–845. [Google Scholar] [CrossRef]
- Spribille, T.; Bjork, C.R. New records and range extensions in the North American lignicolous lichen flora. Mycotaxon 2008, 105, 455–468. [Google Scholar]
- Halda, J.P.; Oh, S.-O.; Liu, D.; Lee, B.G.; Kondratyuk, S.Y.; Lőkös, L.; Park, J.-S.; Woo, J.-J.; Hur, J.-S. Two New Lichen Species, Thelopsis ullungdoensis and Phylloblastia gyeongsangbukensis from Korea. Mycobiology 2020, 48, 443–449. [Google Scholar] [CrossRef]
- Van den Boom, P.; Brand, A.M.; Suija, A. A new species of Absconditella from western and central Europe with a key to the European members. Phytotaxa 2015, 238, 271–277. [Google Scholar] [CrossRef]
- Pino-Bodas, R.; Zhurbenko, M.P.; Stenroos, S. Phylogenetic placement within Lecanoromycetes of lichenicolous fungi associated with Cladonia and some other genera. Persoonia 2017, 39, 91–117. [Google Scholar] [CrossRef]
- Vězda, A. Flechtensystematische Studien X. Conotremopsis weberiana gen. novum et sp. nova, eine neue Flechte aus der Familie Ostropaceae. Folia Geobot. Phytotaxon. 1977, 12, 313–316. [Google Scholar]
- Sherwood, M.A. New Ostropales from the collections of the Farlow Herbarium. Occas. Pap. Farlow Herb. Cryptogam. Bot. 1978, 13, 39–47. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Hyde, K.D.; Jones, E.B.G.; Zhao, Q.; Bulgakov, T.S. New and known discolichens from Asia and eastern Europe. Asian J. Mycol. 2019, 2, 48–86. [Google Scholar] [CrossRef]
- Kantvilas, G. Two ephemeral species of the lichen genus Absconditella (Stictidaceae) new to Tasmania. Muelleria 2005, 21, 91–95. [Google Scholar]
- Kalb, K.; Aptroot, A. New lichen species from Brazil and Venezuela. Bryologist 2018, 121, 56–66. [Google Scholar] [CrossRef]
- Baloch, E.; Döring, H.; Spooner, B.M. The Genus Cryptodiscus in Great Britain. Field Mycol. 2010, 11, 26–32. [Google Scholar] [CrossRef]
- Hjortstam, K.; Ramos Bononi, V.L. A contribution to the knowledge of Corticiaceae sl (Aphyllophorales) in Brazil. Mycotaxon 1987, 28, 1–15. [Google Scholar]
- Baloch, E.; Gilenstam, G.; Wedin, M. The relationships of Odontotrema (Odontotremataceae) and the resurrected Sphaeropezia (Stictidaceae)—New combinations and three new Sphaeropezia species. Mycologia 2013, 105, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Aptroot, A.; de Oliveira, M.C.; Ferraro, L.I.; Cáceres, M.E.S. A world key to species of the genera Topelia and Thelopsis (Stictidaceae), with the description of three new species from Brazil and Argentina. Lichenologist 2014, 46, 801–807. [Google Scholar] [CrossRef]
- Kalb, J.; Kalb, K. New lichen species from Thailand, new combinations and new additions to the Thai lichen biota. Phytotaxa 2017, 332, 141–156. [Google Scholar] [CrossRef]
- Lücking, R.; Rivas Plata, E.; Mangold, A.; Sipman, H.J.M.; Aptroot, A.; Miranda González, R.; Kalb, K.; Chaves, J.L.; Ventura, N.; EsmeraldaEsquivel, R. Natural history of Nash’s Pore Lichens, Trinathotrema (Ascomycota: Lecanoromycetes: Ostropales: Stictidaceae). Bibl. Lichenol. 2011, 106, 183–206. [Google Scholar]
- Fernández-Brime, S.; Llimona, X.; Molnar, K.; Stenroos, S.; Hognabba, F.; Bjork, C.; Lutzoni, F.; Gaya, E. Expansion of the Stictidaceae by the addition of the saxicolous lichen-forming genus Ingvariella. Mycologia 2011, 103, 755–763. [Google Scholar] [CrossRef] [Green Version]
- Nylander, W. Addenda nova ad Lichenographiam Europaeam. Continuatio septima. Flora 1868, 51, 161–165. [Google Scholar]
- Koukol, O.; Hofmann, T.A.; Piepenbring, M. Dendroseptoria mucilaginosa: A new anamorphic fungus with stauroconidia and phylogenetic placement of Dendroseptoria. Mycol. Prog. 2017, 16, 1065–1070. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Summerell, B.A.; Giraldo, A.; Gené, J.; Guarro, J.; Wanasinghe, D.N.; Hyde, K.D.; Camporesi, E.; et al. Fungal Planet description sheets: 281–319. Persoonia 2014, 33, 212–289. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.J.; Smith, D.; Summerell, B.A.; Cano-Lira, J.F.; Guarro, J.; Houbraken, J.; et al. Fungal Planet description sheets: 625–715. Persoonia 2017, 39, 270–467. [Google Scholar] [CrossRef]
- Van Nieuwenhuijzen, E.J.; Miadlikowska, J.M.; Houbraken, J.A.M.P.; Adan, O.C.G.; Lutzoni, F.M.; Samson, R.A. Wood staining fungi revealed taxonomic novelties in Pezizomycotina: New order Superstratomycetales and new species Cyanodermella oleoligni. Stud. Mycol. 2016, 85, 107–124. [Google Scholar] [CrossRef]
- Johnston, P.R. Stictis and its anamorphs in New Zealand. N.Z. J. Bot. 1983, 21, 249–279. [Google Scholar] [CrossRef]
- Johnston, P.R. Anamorphs of the ostropalean genera Schizoxylon and Acarosporina. Mycotaxon 1985, 24, 349–360. [Google Scholar]
- Diederich, P.; Lawrey, J.D.; Ertz, D. The 2018 classification and checklist of lichenicolous fungi, with 2000 non-lichenized, obligately lichenicolous taxa. Bryologist 2018, 121, 340–425. [Google Scholar] [CrossRef]
- Wedin, M.; Döring, H.; Gilenstam, G. Saprotrophy and lichenization as options for the same fungal species on different substrata: Environmental plasticity and fungal lifestyles in the Stictis–Conotrema complex. New Phytol. 2004, 164, 459–465. [Google Scholar] [CrossRef]
- Wedin, M.; Döring, H.; Gilenstam, G. Stictis s. lat. (Ostropales, Ascomycota) in northern Scandinavia, with a key and notes on morphological variation in relation to lifestyle. Mycol. Res. 2006, 110, 773–789. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayake, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.-C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. a guide to methods and applications. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Zoller, S.; Scheidegger, C.; Sperisen, C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 1999, 31, 511–516. [Google Scholar] [CrossRef]
- Clewley, J.P. Macintosh sequence analysis software. Mol. Biotechnol. 1995, 3, 221–224. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, R.; Serra, F.; Tárraga, J.; Medina, I.; Carbonell, J.; Pulido, L.; de María, A.; Capella-Gutíerrez, S.; Huerta-Cepas, J.; Gabaldón, T.; et al. Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011, 39, 470–474. [Google Scholar] [CrossRef] [Green Version]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. FigTree v1. 4.0. A Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 August 2021).
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Lutzoni, F.; Pagel, M.; Reeb, V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature 2001, 411, 937–940. [Google Scholar] [CrossRef]
- Johnston, P.R.; Park, D.; Renner, M.A.M. Sphaeropezia leucocheila sp. nov. (Stictidaceae): A liverwort pathogen from New Zealand. Phytotaxa 2019, 409, 222–226. [Google Scholar] [CrossRef] [Green Version]


| Species | Strain Number | LSU | ITS | mtSSU |
|---|---|---|---|---|
| Absconditella sphagnorum | M24 | EU940095 | - | EU940247 |
| Absconditella sphagnorum | Palice 3820 | AY300825 | - | AY300873 |
| Acarosporina microspora | AFTOL-ID 78 | AY584643 | DQ782834 | AY584612 |
| Carestiella socia | GG2410 | - | AY661687 | AY661677 |
| Carestiella socia | GG2437a | - | AY661682 | AY661678 |
| Carestiella socia | Wedin 7194 | - | - | JX266155 |
| Chromatochlamys muscorum | Palice 8634 | AY607731 | - | AY607743 |
| Cryptodiscus cladoniicola | RP160 | KY661653 | KY661620 | KY661675 |
| Cryptodiscus cladoniicola | RP159 | KY661652 | KY661619 | KY661674 |
| Cryptodiscus epicladonia | RP208 T | - | KY661628 | KY661680 |
| Cryptodiscus foveolaris | EB155 | - | FJ904673 | FJ904695 |
| Cryptodiscus gloeocapsa | EB93 | - | FJ904674 | FJ904696 |
| Cryptodiscus incolor | EB164 | - | FJ904675 | FJ904697 |
| Cryptodiscus muriformis | H.B. 6773 | MG281963 | MG281963 | MG281973 |
| Cryptodiscus pallidus | EB173 | - | FJ904680 | FJ904702 |
| Cryptodiscus pallidus | EB152 | - | FJ904679 | FJ904701 |
| Cryptodiscus pini | EB181 | - | FJ904684 | FJ904706 |
| Cryptodiscus pini | EB178 | - | FJ904683 | FJ904705 |
| Cryptodiscus tabularum | CO205 | - | FJ904690 | FJ904712 |
| Cryptodiscus tabularum | EB169 | - | FJ904689 | FJ904711 |
| Cyanodermella asteris | 03HOR06-2-4 T | KT758843 | KT758843 | |
| Cyanodermella banksiae | CPC:32105 T | NG_064548 | NR_159835 | - |
| Cyanodermella oleoligni | CBS 140345 T | NG_058973 | NR_153930 | KX999144 |
| Cyanodermella viridula | EB146 | HM244763 | - | HM244739 |
| Eriospora leucostoma | CPC:35594 | MT223890 | MT223795 | - |
| Fitzroyomyces cyperacearum | MFLU 18-0695b | MK499361 | MK499349 | - |
| Fitzroyomyces cyperacearum | CBS 143170 T | NG_058513 | NR_156387 | - |
| Fitzroyomyces hyaloseptisporus | MFLUCC 21-0111 T | MZ868921 | MZ868916 | MZ868911 |
| Stictis pandanicola | HKAS 96206 T | MH260319 | MH275085 | - |
| Geisleria sychnogonoides | Caceres and Aptroot 13560 (ABL) | KC689752 | - | KC689751 |
| Geisleria sychnogonoides | 70627 | KF220304 | - | KF220306 |
| Glomerobolus gelineus | JK 5584C | DQ247798 | - | DQ247783 |
| Glomerobolus gelineus | AFTOL-ID 1349 | - | DQ247782 | DQ247784 |
| Hormodochis aggregata | CPC 30683T | MN317280 | NR_166307 | - |
| Hormodochis melanochlora | CBS 138861 T | NG_070381 | NR_165507 | - |
| Ingvariella bispora | MALich 15288 | HQ659184 | - | HQ659173 |
| Ingvariella bispora | BCNLich 17183 | HQ659185 | - | HQ659174 |
| Neofitzroyomyces nerii | CBS 145088 T | NG_068278 | NR_161144 | - |
| Neostictis nigricans | MFLU 18-1380 T | MT214610 | MT310654 | - |
| Orceolina kerguelensis | Søchting 9398(C) | AY212830 | AY212814 | AY212853 |
| Ostropa barbara | SW071 (S) | HM244773 | HM244773 | HM244752 |
| Ostropa barbara | S F302817 | MG281965 | MG281965 | MG281974 |
| Ostropomyces pruinosellus | MFLU 20-0538 T | MW400966 | MW400964 | |
| Ostropomyces pruinosellus | MFLUCC 21-0112 | MZ868917 | MZ868912 | MZ868907 |
| Ostropomyces pruinosellus | MFLU 21-0115 | MZ868918 | MZ868913 | MZ868908 |
| Ostropomyces thailandicus | MFLU 20-0539 T | MW397060 | MW400967 | - |
| Ostropomyces thailandicus | MFLUCC 21-0113 | MZ868919 | MZ868914 | MZ868909 |
| Ostropomyces thailandicus | MFLU 21-0116 | MZ868920 | MZ868915 | MZ868910 |
| Phacidiella alsophilae | CPC:37041 T | MT373344 | MT373361 | - |
| Placopsis perrugosa | KS137 | KU844613 | KU844737 | KU844549 |
| Robergea cubicularis | G.M. 2017-10-12.1 | MN833317 | MN833317 | - |
| Robergea cubicularis | G.M. 2013-05-09.1 | KY611899 | KY611899 | - |
| Schizoxylon albescens | GG2443a | AY661690 | AY661690 | AY661681 |
| Schizoxylon berkeleyanum | S F209682 | MG281966 | MG281966 | MG281975 |
| Schizoxylon gilenstamii | S F300892 | MG281968 | MG281968 | MG281977 |
| Sphaeropezia leucocheila | PDD:98299 T | MK547099 | MK547090 | MK547101 |
| Sphaeropezia lyckselensis | EB-2012a | JX266158 | - | JX266156 |
| Sphaeropezia mycoblasti | EB-2012b | JX266159 | - | JX266157 |
| Sphaeropezia shangrilaensis | MFLU 20-0537 T | MW400965 | MW400955 | MW400962 |
| Stictis anhuiensis | HOU1233M | KX447623 | - | KX447625 |
| Stictis anhuiensis | HOU1233 | KX447622 | - | KX447624 |
| Stictis brunnescens | GG2359 | AY661688 | AY661688 | AY661679 |
| Stictis confusa | Wedin 7070 | - | NR_121318 | DQ401141 |
| Stictis mollis | MW7200 | AY527313 | AY527313 | - |
| Stictis mollis | GG2445a | AY527318 | AY527318 | - |
| Stictis populorum | GG2610a | AY527327 | AY527327 | AY527356 |
| Stictis populorum | GG2618 T | AY527331 | AY527331 | AY527360 |
| Stictis radiata | GG2449a | AY527308 | AY527308 | - |
| Stictis radiata | MW6493 | AY527309 | AY527309 | - |
| Stictis urceolatum | LT21500 | AY661686 | AY661686 | AY661676 |
| Thelenella antarctica | Lumbsch 19006a (F) | AY607739 | - | AY607749 |
| Trapelia placodioides | KS163 | KU844623 | KU844758 | KU844568 |
| Trinathotrema stictideum | F:Luecking 17541b | - | - | GU380288 |
| Trinathotrema stictideum | F:Luecking 28093 | - | - | GU380287 |
| Xyloschistes platytropa | AFTOL-ID 4891 | KJ766680 | - | KJ766517 |
| Species | Strain | Apothecia (μm) | Exciple (μm) | Paraphyses (μm) | Asci (μm) | Ascospores (μm) | Septation | Reference |
|---|---|---|---|---|---|---|---|---|
| F. cyperacearum | MFLU 17–1480 | 201–260 × 210–310 | 17–70, textura intricata | 1.3–3, aseptate | 110–150 × 10–20 | 100–145 × 2.5–3.5, eguttulate | 17–21 | [36] |
| F. hyaloseptisporus | MFLU 21–0114 | 140–200 × 150–200 | 6–20, textura angularis | 1–3, septate | 165–200 × 10–25 | 150–200 × 3.5–6, finely guttulate | up to 58 | this study |
| F. pandanicola | HKAS 96206 | 350–410 × 520–650 | 27–46, textura epidermoidea | 0.8–1.1, aseptate | 160–240 × 7.5–23 | 190–265 × 4–5, eguttulate | up to 40 | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, D.-P.; Wanasinghe, D.N.; Gentekaki, E.; Thiyagaraja, V.; Lumyong, S.; Hyde, K.D. Morphological and Phylogenetic Appraisal of Novel and Extant Taxa of Stictidaceae from Northern Thailand. J. Fungi 2021, 7, 880. https://doi.org/10.3390/jof7100880
Wei D-P, Wanasinghe DN, Gentekaki E, Thiyagaraja V, Lumyong S, Hyde KD. Morphological and Phylogenetic Appraisal of Novel and Extant Taxa of Stictidaceae from Northern Thailand. Journal of Fungi. 2021; 7(10):880. https://doi.org/10.3390/jof7100880
Chicago/Turabian StyleWei, De-Ping, Dhanushka N. Wanasinghe, Eleni Gentekaki, Vinodhini Thiyagaraja, Saisamorn Lumyong, and Kevin D. Hyde. 2021. "Morphological and Phylogenetic Appraisal of Novel and Extant Taxa of Stictidaceae from Northern Thailand" Journal of Fungi 7, no. 10: 880. https://doi.org/10.3390/jof7100880
APA StyleWei, D.-P., Wanasinghe, D. N., Gentekaki, E., Thiyagaraja, V., Lumyong, S., & Hyde, K. D. (2021). Morphological and Phylogenetic Appraisal of Novel and Extant Taxa of Stictidaceae from Northern Thailand. Journal of Fungi, 7(10), 880. https://doi.org/10.3390/jof7100880








