Drk1, a Dimorphism Histidine Kinase, Contributes to Morphology, Virulence, and Stress Adaptation in Paracoccidioides brasiliensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. P. brasiliensis Isolate and Culture Conditions
2.2. Vector Construction, Heterologous Expression of PbDrk1 Recombinant (rPbDrk1), and Purification
2.3. Mass Spectrometric Analysis of rPbDrk1
2.4. Antibody Production
2.5. Protein Extraction, SDS-PAGE, and Western Blotting
2.6. Protein Localization: Indirect Immunofluorescence and Immunogold
2.7. P. brasiliensis Viability after the Anti-PbDrk1 Treatment
2.8. Inhibition Assays with Anti-PbDrk11 Antibody and Consequences to P. brasiliensis-Pneumocytes Interaction by Flow Cytometry
2.9. Effect of the P. brasiliensis Anti-PbDrk1 Antibody Treatment on G. mellonella Infection
2.10. PbDRK1 Gene Silencing with Antisense Molecules and Agrobacterium Tumefaciens Transformation (ATMT)
2.11. PCR Detection of Hygromycin Resistance Gene (hph) and Analysis of PbDRK1 Transcript Levels by qRT-PCR (Quantitative Real Time PCR)
2.12. Phenotypic Effects of PbDRK1 Silencing
2.12.1. Growth Curve and Cell Viability
2.12.2. Morphological Analysis
2.12.3. Virulence Assay
2.12.4. Chitin Level
2.12.5. Susceptibility to Antifungals
2.12.6. Susceptibility to Cell Wall Stresses
2.13. Statistical Analysis
3. Results
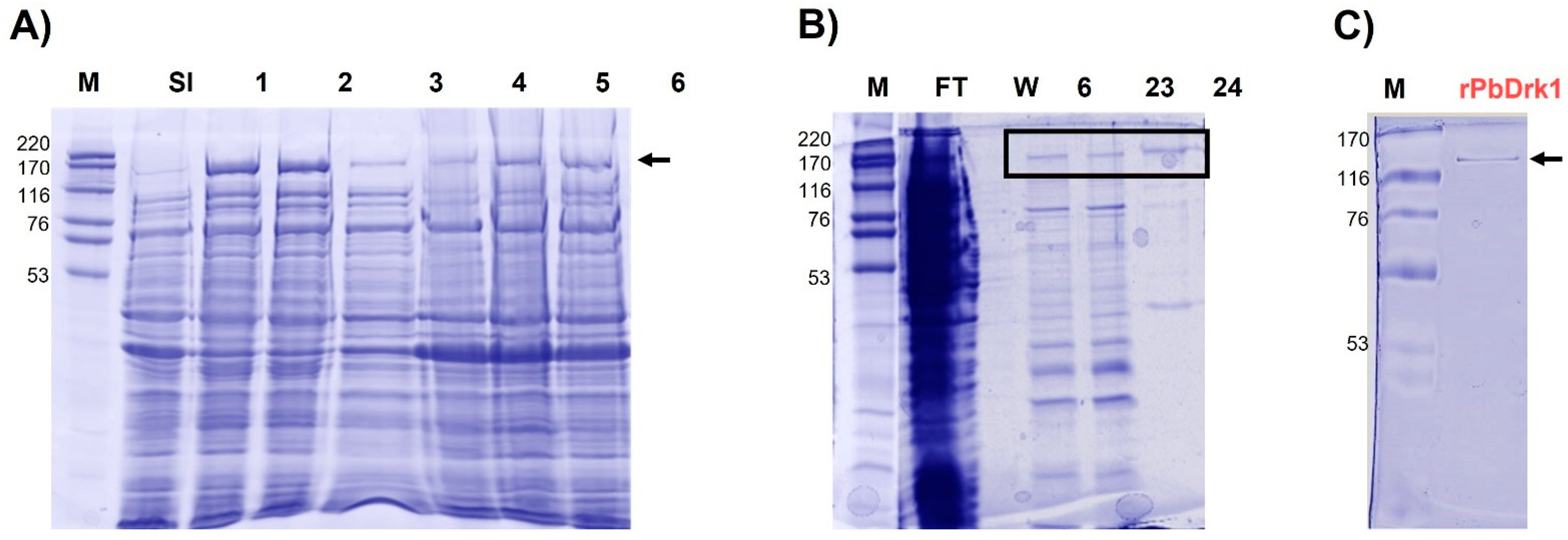
3.1. Molecular Cloning of cDNA Encoding a DRK1 of P. brasiliensis, Expression and Recombinant Protein Purification
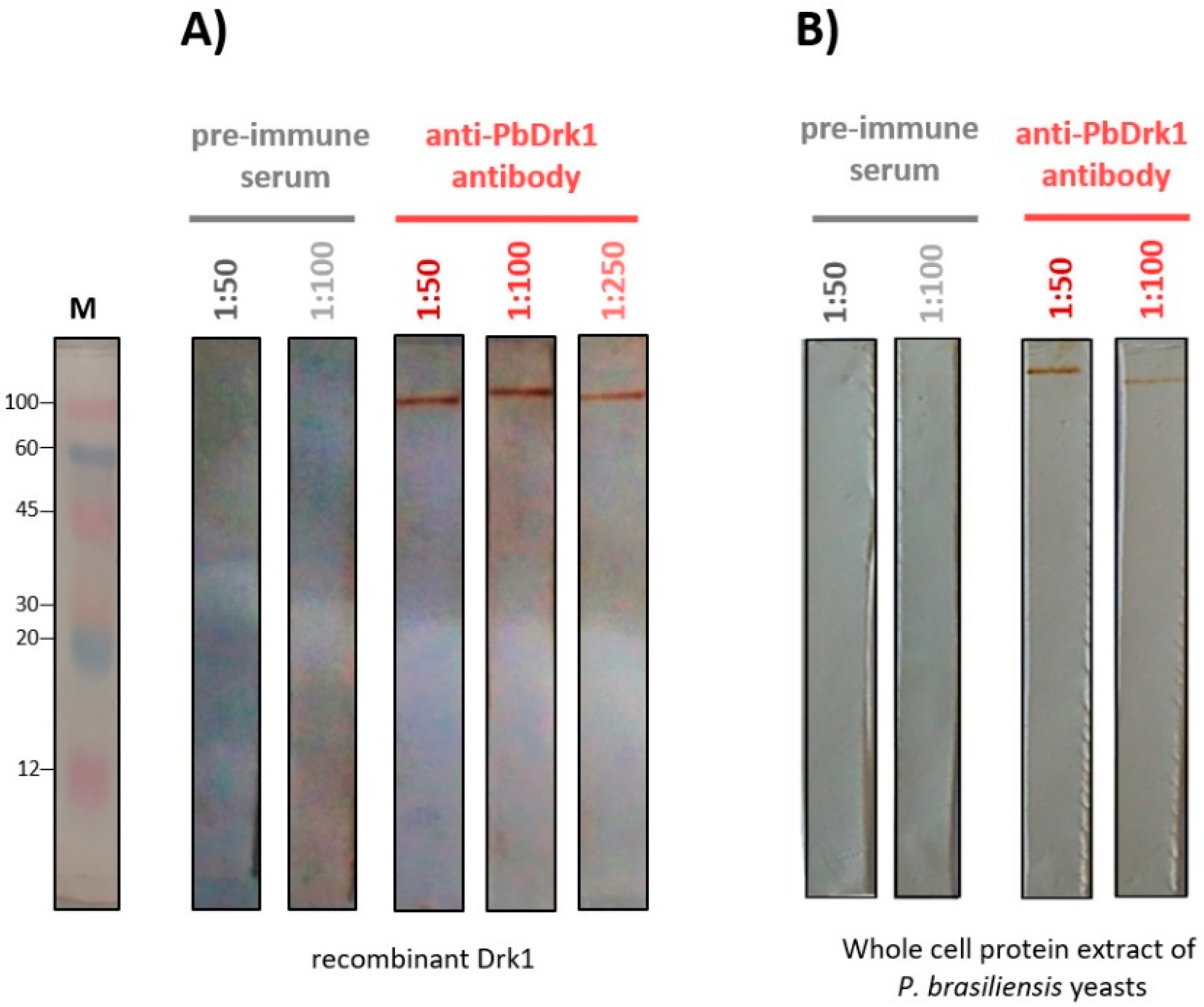
3.2. Anti-PbDrk1 Polyclonal Antibody Recognizes the rPbDRK1 and Native Protein in P. brasiliensis
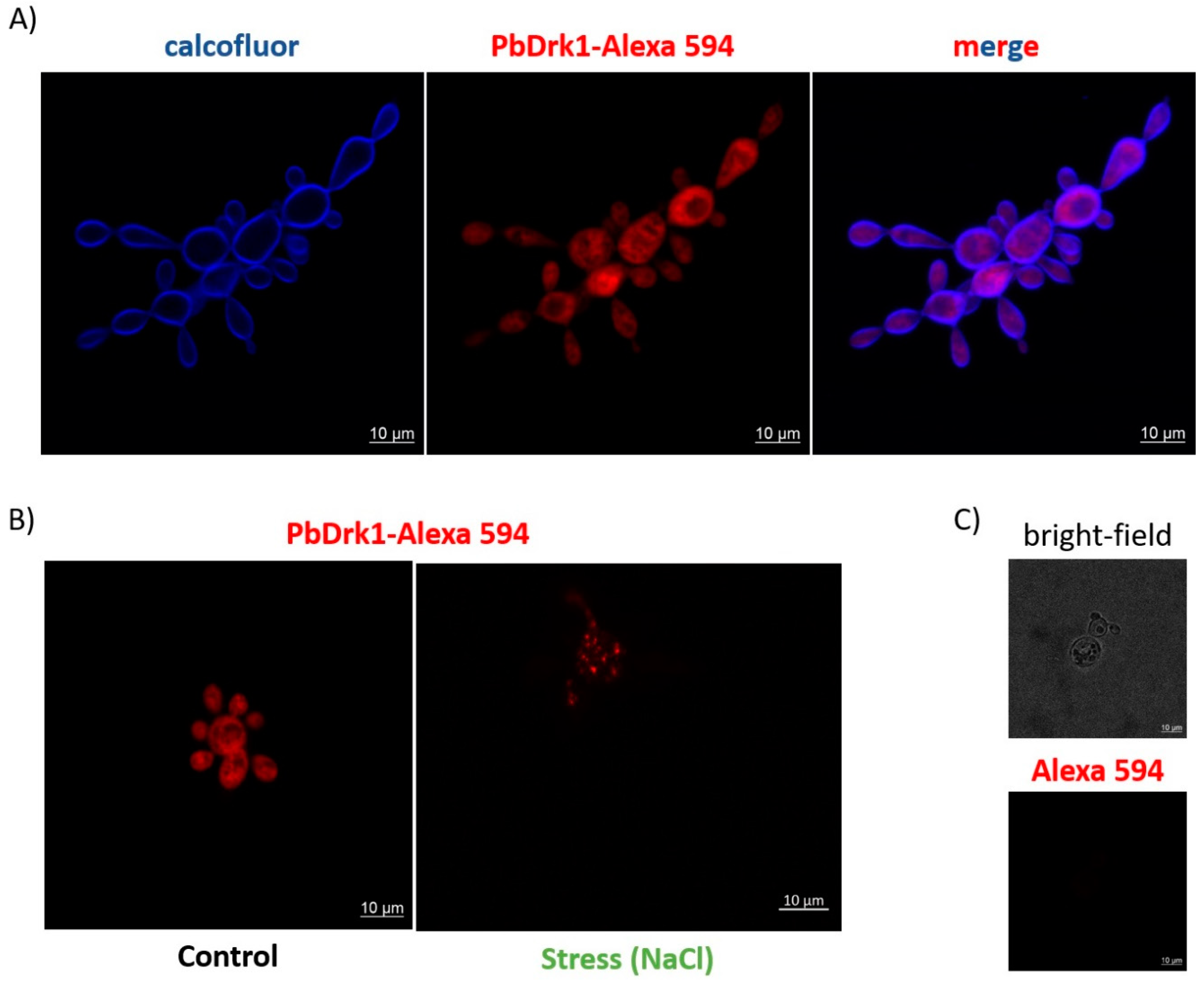
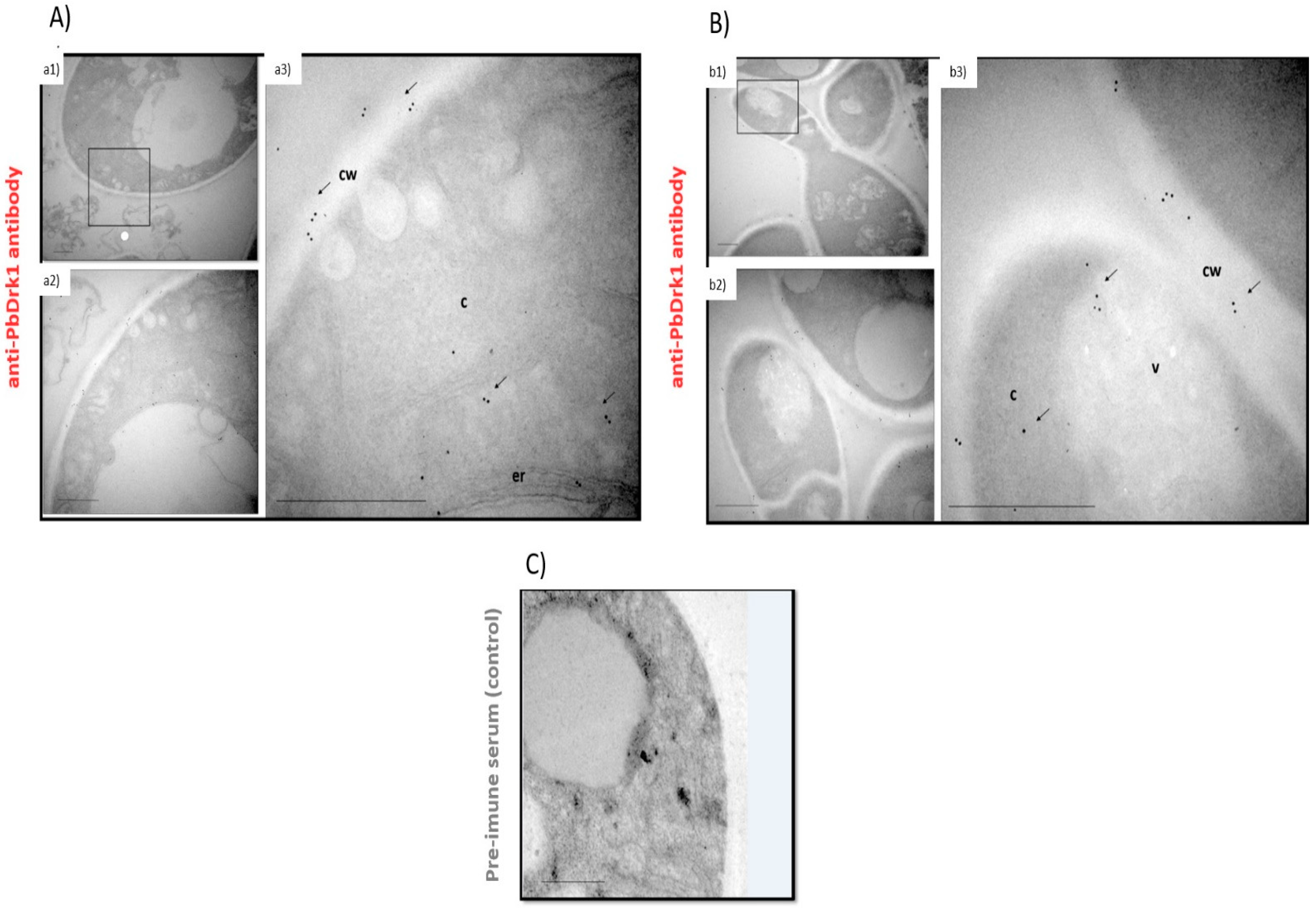
3.3. PbDrk1 Localization in the Yeast Cell Surface, Showing an Altered Pattern of Expression during Osmotic Stress Condition
3.4. Inhibition of Drk1 with Anti-PbDrk1 Antibody Alters the Interaction of P. brasiliensis with Pneumocytes and the Fungal Virulence in G. mellonella
3.5. PbDRK1 Knockdown Does Not Change the Growth Rate and Viability of P. brasiliensis
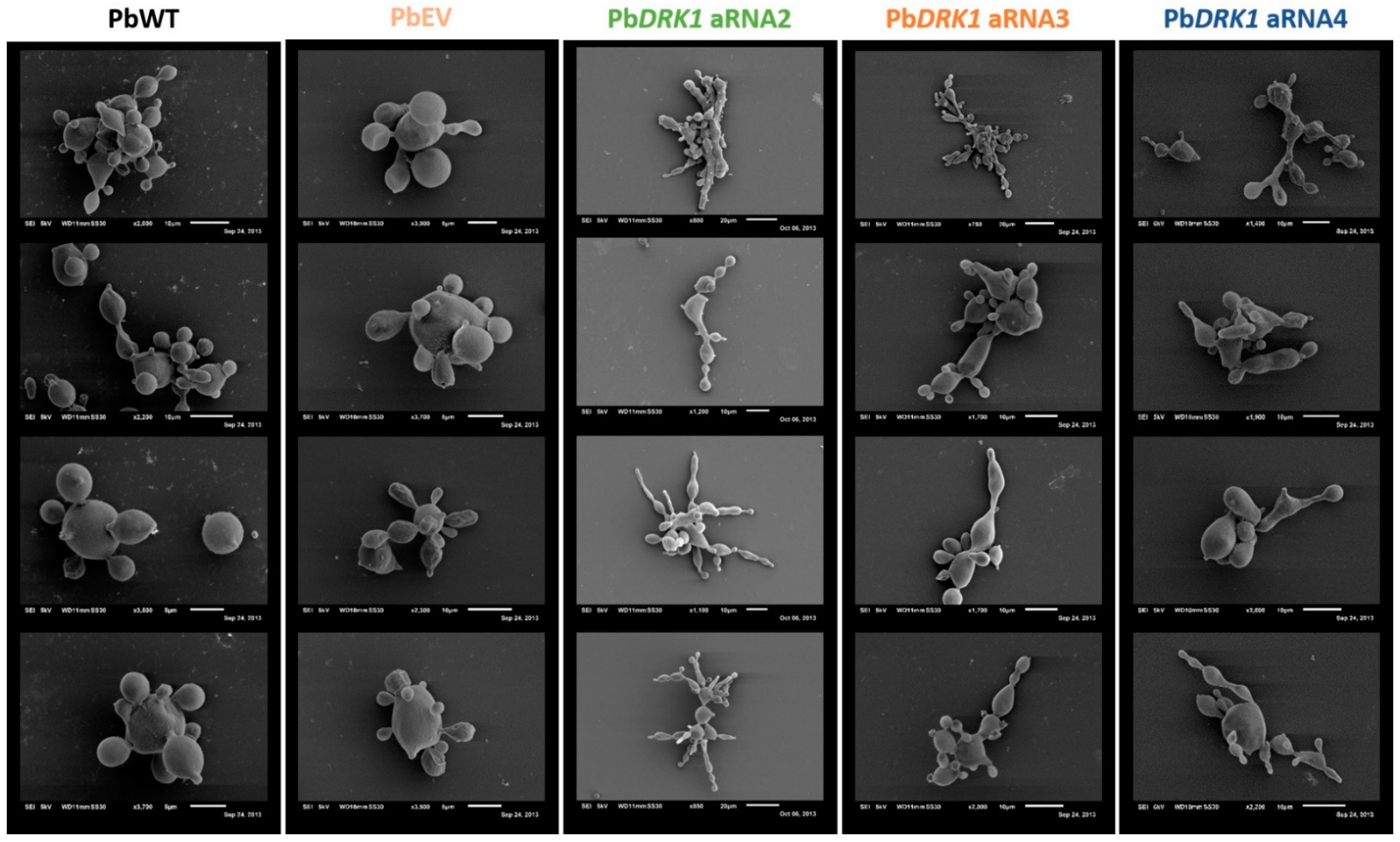
3.6. DRK1 Deficiency Alters the Morphology of P. brasiliensis, Resulting in Elongated Yeast Cells
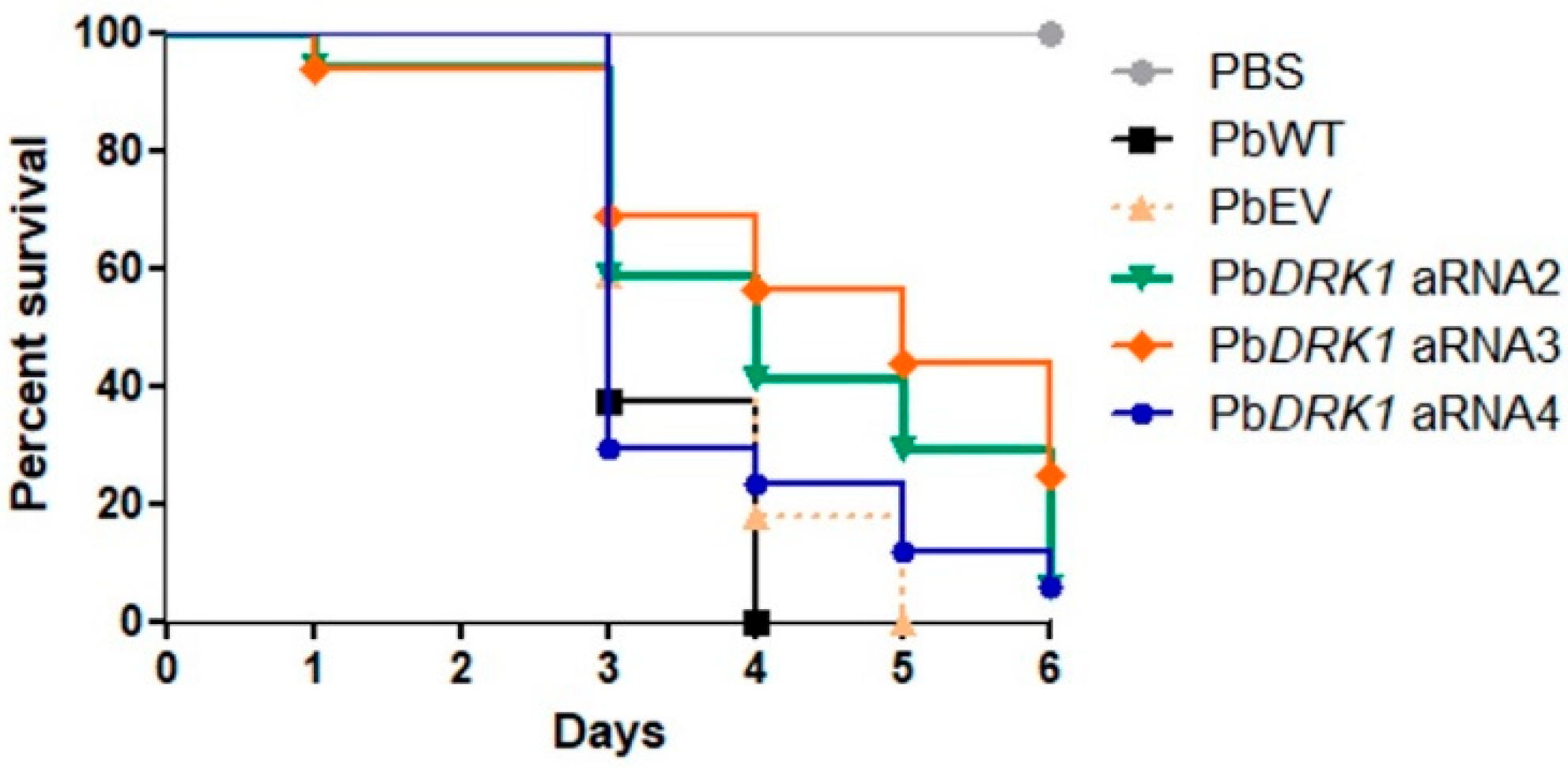
3.7. PbDRK1 Silence Affects the Virulence of P. brasiliensis in G. mellonella Model
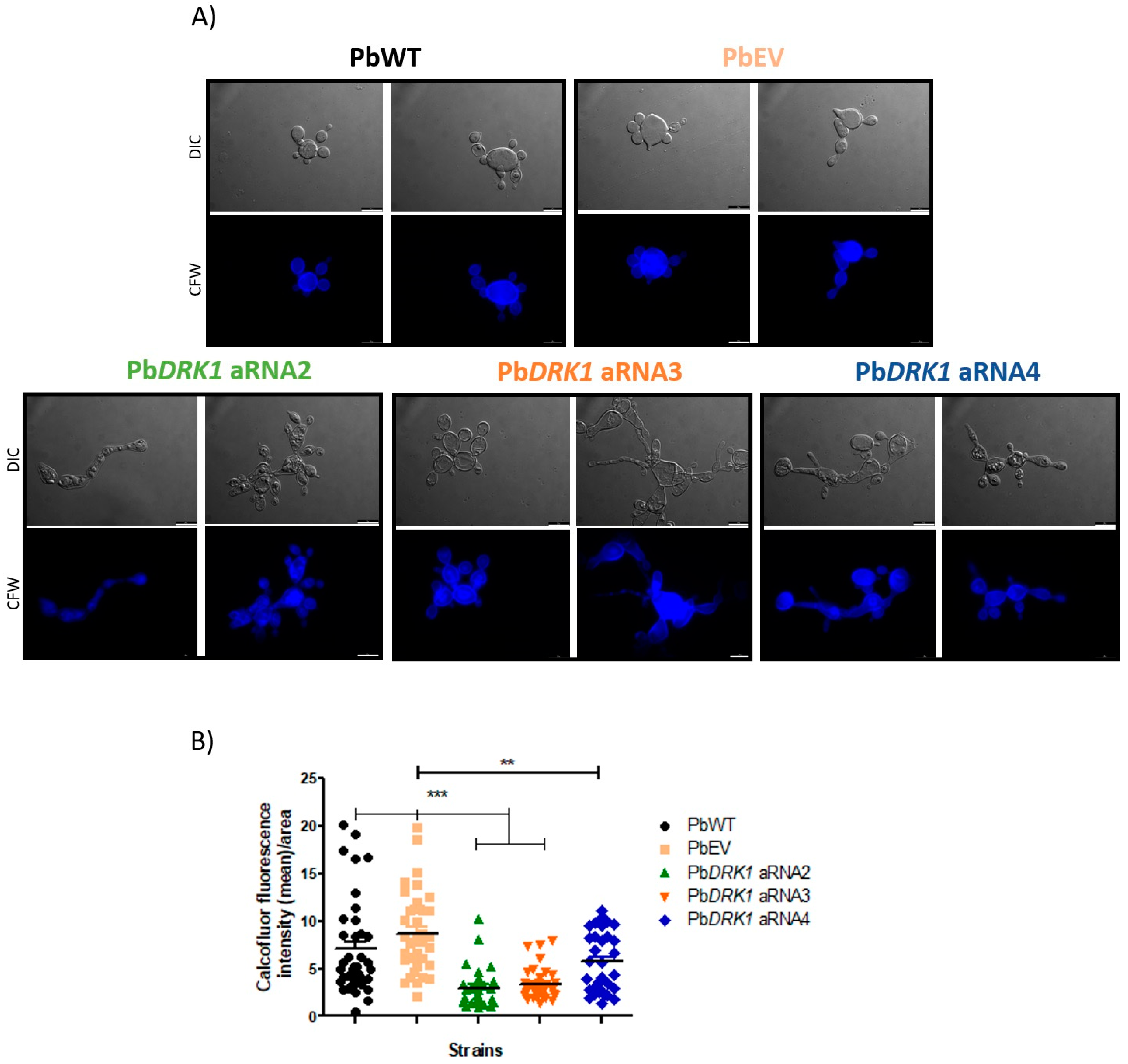
3.8. DRK1 Silencing Change the Chitin Level/Distribution in P. brasiliensis
3.9. Response to Osmotic Stress, Cell Wall Stressors and Antifungals Can Change due to Reduced Expression of Drk1 in P. brasiliensis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nemecek, J.C.; Wüthrich, M.; Klein, B.S. Global Control of Dimorphism and Virulence in Fungi. Science 2006, 312, 583–588. [Google Scholar] [CrossRef]
- Klein, B.S.; Tebbets, B. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 2007, 10, 314–319. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.F.; McEwen, J.G.; Clay, O.K.; Cuomo, C.A. Genome analysis reveals evolutionary mechanisms of adaptation in systemic dimorphic fungi. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques-Da-Silva, S.H.; Rodrigues, A.M.; Silveira-Gomes, F.; de Hoog, G.S.; de Camargo, Z.P. Occurrence of Paracoccidioides lutzii in the Amazon Region: Description of Two Cases. Am. J. Trop. Med. Hyg. 2012, 87, 710–714. [Google Scholar] [CrossRef] [Green Version]
- Martinez, R. Epidemiology of Paracoccidioidomycosis. Rev. Inst. Med. Trop. São Paulo 2015, 57, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 4, 11–24. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.-A. Host-Pathogen Interactions: Redefining the Basic Concepts of Virulence and Pathogenicity. Infect. Immun. 1999, 67, 3703–3713. [Google Scholar] [CrossRef] [Green Version]
- Cooney, N.M.; Klein, B.S. Fungal adaptation to the mammalian host: It is a new world, after all. Curr. Opin. Microbiol. 2008, 11, 511–516. [Google Scholar] [CrossRef] [Green Version]
- Ene, I.V.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.; Munro, C.A.; Brown, A.J.P. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986-15. [Google Scholar] [CrossRef] [Green Version]
- Jacob, S.; Thines, E. Multistep phosphorelay in fungi: The enigma of multiple signals and a limited number of signaling pathways. Mycol. Prog. 2017, 16, 1007–1013. [Google Scholar] [CrossRef]
- Smith, D.A.; Nicholls, S.; Morgan, B.A.; Brown, A.J.; Quinn, J. A Conserved Stress-activated Protein Kinase Regulates a Core Stress Response in the Human Pathogen Candida albicans. Mol. Biol. Cell 2004, 15, 4179–4190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, G.; Klein, B.S. Insights into Fungal Morphogenesis and Immune Evasion. Microbe Mag. 2008, 3, 416–423. [Google Scholar] [CrossRef]
- Saito, H. Regulation of the Osmoregulatory HOG MAPK Cascade in Yeast. J. Biochem. 2004, 136, 267–272. [Google Scholar] [CrossRef]
- Catlett, N.L.; Yoder, O.C.; Turgeon, B.G. Whole-Genome Analysis of Two-Component Signal Transduction Genes in Fungal Pathogens. Eukaryot. Cell 2003, 2, 1151–1161. [Google Scholar] [CrossRef] [Green Version]
- Defosse, T.A.; Sharma, A.; Mondal, A.K.; de Bernonville, T.D.; Latgé, J.-P.; Calderone, R.; Giglioli-Guivarc’H, N.; Courdavault, V.; Clastre, M.; Papon, N. Hybrid histidine kinases in pathogenic fungi. Mol. Microbiol. 2015, 95, 914–924. [Google Scholar] [CrossRef]
- Maeda, T.; Wurgler-Murphy, S.M.; Saito, H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 1994, 369, 242–245. [Google Scholar] [CrossRef]
- Posas, F.; Wurgler-Murphy, S.M.; Maeda, T.; Witten, E.A.; Thai, T.C.; Saito, H. Yeast HOG1 MAP Kinase Cascade Is Regulated by a Multistep Phosphorelay Mechanism in the SLN1–YPD1–SSK1 “Two-Component” Osmosensor. Cell 1996, 86, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Boyce, K.; Schreider, L.; Kirszenblat, L.; Andrianopoulos, A. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol. Microbiol. 2011, 82, 1164–1184. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Zhang, Z.; Zheng, F.; Liu, X. Molecular cloning, characterization and differential expression of DRK1 in Sporothrix schenckii. Int. J. Mol. Med. 2012, 31, 99–104. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, B.; Wu, Y.Z.; Wang, Y.; Liu, X.; Han, S. Two-component histidine kinase DRK1 is required for pathogenesis in Sporothrix schenckii. Mol. Med. Rep. 2017, 17, 721–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, A.F.A.; Navarro, M.V.; Castilho, D.G.; Calado, J.C.P.; Conceição, P.M.; Batista, W.L. A conserved dimorphism-regulating histidine kinase controls the dimorphic switching in Paracoccidioides brasiliensis. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni, G.; Lucciarini, R.; Amantini, C.; Jacobelli, J.; Spreghini, E.; Ballarini, P.; Piccoli, M.; Gismondi, A. Candida albicans Expresses a Focal Adhesion Kinase-Like Protein That Undergoes Increased Tyrosine Phosphorylation upon Yeast Cell Adhesion to Vitronectin and the EA.hy 926 Human Endothelial Cell Line. Infect. Immun. 2002, 70, 3804–3815. [Google Scholar] [CrossRef] [Green Version]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Torres, I.; Hernandez, O.; Tamayo, D.; Muñoz, J.F.; Jr, N.P.L.; García, A.M.; Restrepo, A.; Puccia, R.; McEwen, J.G. Inhibition of PbGP43 Expression May Suggest that gp43 is a Virulence Factor in Paracoccidioides brasiliensis. PLoS ONE 2013, 8, e68434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goes, T.; Bailão, E.F.L.C.; Correa, C.R.; Bozzi, A.; Dos Santos, L.I.; Gomes, D.; Soares, C.M.A.; Goes, A.M. New Developments of RNAi in Paracoccidioides brasiliensis: Prospects for High-Throughput, Genome-Wide, Functional Genomics. PLOS Negl. Trop. Dis. 2014, 8, e3173. [Google Scholar] [CrossRef] [Green Version]
- Marcos, C.M.; De Oliveira, H.C.; Silva, J.; Assato, P.; Yamazaki, D.S.; Da Silva, R.A.M.; Santos, C.T.; Santos-Filho, N.; Portuondo, D.L.; Giannini, M.J.M.; et al. Identification and characterisation of elongation factor Tu, a novel protein involved in Paracoccidioides brasiliensis–host interaction. FEMS Yeast Res. 2016, 16, fow079. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meshulam, T.; Levitz, S.M.; Christin, L.; Diamond, R.D. A Simplified New Assay for Assessment of Fungal Cell Damage with the Tetrazolium Dye, (2,3)-bis-(2-Methoxy-4-Nitro-5-Sulphenyl)-(2H)-Tetrazolium-5-Carboxanilide (XTT). J. Infect. Dis. 1995, 172, 1153–1156. [Google Scholar] [CrossRef] [PubMed]
- Marcos, C.M.; Tamer, G.; De Oliveira, H.C.; Assato, P.A.; Scorzoni, L.; Santos, C.T.; Singulani, J.D.L.; De Silva, J.D.F.; De Almeida, R.; E Silva, A.C.A.D.P.; et al. Down-regulation of TUFM impairs host cell interaction and virulence by Paracoccidioides brasiliensis. Sci. Rep. 2019, 9, 17206–17215. [Google Scholar] [CrossRef] [PubMed]
- Scorzoni, L.; E Silva, A.C.A.D.P.; Singulani, J.D.L.; Leite, F.S.; De Oliveira, H.C.; Da Silva, R.A.M.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Comparison of virulence between Paracoccidioides brasiliensis and Paracoccidioides lutzii using Galleria mellonella as a host model. Virulence 2015, 6, 766–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, A.; Carmona, J.; Cunha, C.; Carvalho, A.; Rappleye, C.; Goldman, W.; Hooykaas, P.; Leão, C.; Ludovico, P.; Rodrigues, F. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 2007, 44, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Menino, J.; Almeida, A.J.; Rodrigues, F. Gene Knockdown in Paracoccidioides brasiliensis Using Antisense RNA. Methods Mol. Biol. 2012, 845, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Cleary, I.A.; MacGregor, N.B.; Saville, S.P.; Thomas, D.P. Investigating the Function of Ddr48p in Candida albicans. Eukaryot. Cell 2012, 11, 718–724. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Marcos, C.M.; Silva, J.; De Oliveira, H.C.; Assato, P.; Singulani, J.D.L.; Lopez, A.M.; Tamayo, D.P.; Hernandez-Ruiz, O.; McEwen, J.G.; Giannini, M.J.M.; et al. Decreased expression of 14-3-3 in Paracoccidioides brasiliensis confirms its involvement in fungal pathogenesis. Virulence 2016, 7, 72–84. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, H.C.; Joffe, L.S.; Simon, K.S.; Castelli, R.F.; Reis, F.C.G.; Bryan, A.M.; Borges, B.S.; Medeiros, L.C.S.; Bocca, A.L.; Del Poeta, M.; et al. Fenbendazole Controls In Vitro Growth, Virulence Potential, and Animal Infection in the Cryptococcus Model. Antimicrob. Agents Chemother. 2020, 64, e00286-20. [Google Scholar] [CrossRef]
- De Paula e Silva, A.C.A.; Oliveira, H.C.; Silva, J.; Sangalli-Leite, F.; Scorzoni, L.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Microplate alamar Blue Assay for Paracoccidioides Susceptibility Testing. J. Clin. Microbiol. 2013, 51, 1250–1252. [Google Scholar] [CrossRef] [Green Version]
- Ram, A.F.J.; Klis, F.M. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 2006, 1, 2253–2256. [Google Scholar] [CrossRef] [PubMed]
- Fassler, J.S.; West, A.H. Histidine Phosphotransfer Proteins in Fungal Two-Component Signal Transduction Pathways. Eukaryot. Cell 2013, 12, 1052–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hérivaux, A.; So, Y.-S.; Gastebois, A.; Latgé, J.-P.; Bouchara, J.-P.; Bahn, Y.-S.; Papon, N. Major Sensing Proteins in Pathogenic Fungi: The Hybrid Histidine Kinase Family. PLoS Pathog. 2016, 12, e1005683. [Google Scholar] [CrossRef] [Green Version]
- Brandhorst, T.T.; Wüthrich, M.; Warner, T.; Klein, B. Targeted Gene Disruption Reveals an Adhesin Indispensable for Pathogenicity of Blastomyces dermatitidis. J. Exp. Med. 1999, 189, 1207–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isaac, D.T.; Berkes, C.A.; English, B.C.; Murray, D.H.; Lee, Y.N.; Coady, A.; Sil, A. Macrophage cell death and transcriptional response are actively triggered by the fungal virulence factor C bp1 during H. capsulatum infection. Mol. Microbiol. 2015, 98, 910–929. [Google Scholar] [CrossRef] [PubMed]
- Sebghati, T.S.; Engle, J.T.; Goldman, W.E. Intracellular Parasitism by Histoplasma capsulatum: Fungal Virulence and Calcium Dependence. Science 2000, 290, 1368–1372. [Google Scholar] [CrossRef]
- Rappleye, C.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum -(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [Green Version]
- Foureau, E.; Clastre, M.; Montoya, E.J.O.; Besseau, S.; Oudin, A.; Glévarec, G.; Simkin, A.J.; Crèche, J.; Atehortùa, L.; Giglioli-Guivarc’H, N.; et al. Subcellular localization of the histidine kinase receptors Sln1p, Nik1p and Chk1p in the yeast CTG clade species Candida guilliermondii. Fungal Genet. Biol. 2014, 65, 25–36. [Google Scholar] [CrossRef]
- Hagiwara, D.; Takahashi-Nakaguchi, A.; Toyotome, T.; Yoshimi, A.; Abe, K.; Kamei, K.; Gonoi, T.; Kawamoto, S. NikA/TcsC Histidine Kinase is Involved in Conidiation, Hyphal Morphology, and Responses to Osmotic Stress and Antifungal Chemicals in Aspergillus fumigatus. PLoS ONE 2013, 8, e80881. [Google Scholar] [CrossRef]
- Lawry, S.; Tebbets, B.; Kean, I.; Stewart, D.; Hetelle, J.; Klein, B.S. Fludioxonil Induces Drk1, a Fungal Group III Hybrid Histidine Kinase, To Dephosphorylate Its Downstream Target, Ypd1. Antimicrob. Agents Chemother. 2017, 61, e01414-16. [Google Scholar] [CrossRef] [Green Version]
- Omura, F.; Takagi, M.; Kodama, Y. Compromised chitin synthesis in lager yeast affects its Congo red resistance and release of mannoproteins from the cells. FEMS Microbiol. Lett. 2020. [Google Scholar] [CrossRef]
- Imai, K.; Noda, Y.; Adachi, H.; Yoda, K. A Novel Endoplasmic Reticulum Membrane Protein Rcr1 Regulates Chitin Deposition in the Cell Wall of Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 8275–8284. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zhang, L.; Wakabayashi, H.; Myers, J.; Jiang, Y.; Cao, Y.; Jimenez-Ortigosa, C.; Perlin, D.S.; Rustchenko, E. Tolerance to Caspofungin in Candida albicans is Associated with at Least Three Distinctive Mechanisms That Govern Expression of FKS Genes and Cell Wall Remodeling. Antimicrob. Agents Chemother. 2017, 61, e00071-17. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.K.; MacCallum, D.M.; Jacobsen, M.D.; Walker, L.A.; Odds, F.C.; Gow, N.A.R.; Munro, C.A. Elevated Cell Wall Chitin in Candida albicans Confers Echinocandin Resistance In Vivo. Antimicrob. Agents Chemother. 2012, 56, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Kelly, J.; Rowan, R.; McCann, M.; Kavanagh, K. Exposure to caspofungin activates Cap and Hog pathways in Candida albicans. Med. Mycol. 2009, 47, 697–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Chan, K.L.; Faria, N.C.G.; Martins, M.D.L.; Campbell, B.C. Targeting the Oxidative Stress Response System of Fungi with Redox-Potent Chemosensitizing Agents. Front. Microbiol. 2012, 3, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.; Lee, D.G. Reactive oxygen species modulate itraconazole-induced apoptosis via mitochondrial disruption in Candida albicans. Free Radic. Res. 2017, 52, 39–50. [Google Scholar] [CrossRef] [PubMed]



| Primers | Sequences | References |
|---|---|---|
| DRK1-F | 5′-CATATGACTCGTGGTGAT-3′ | Current work |
| DRK1-R | 5′-GCGGCCGCGCACAGAGACAG-3′ | Current work |
| DRK1AS2-F | 5′-CCGCTCGAGCGGCTACCAACGGCGATGTGAC-3′ | Current work |
| DRK1AS2-R | 5′-GGCGCGCCCACAAATGATTGCATGGTC-3′ | Current work |
| DRK1AS3-F | 5′-CCGCTCGAGCGGCCTCCATCTCGACGATCT -3′ | Current work |
| DRK1AS3-R | 5′-GGCGCGCCTGTAGTTTGTGTCAGACT-3′ | Current work |
| DRK1-AS4-F | 5′-CCGCTCGAGCGGTGACACAAACTACAAAGA-3′ | Current work |
| DRK1-AS4-R | 5′-GGCGCGCCTCATCAATTTTCACCCTC--3′ | Current work |
| Hph-F | 5′-AACTCACCGCGACGTCTGTCGA-3′ | [27] |
| Hph-R | 5′-CTACACAGCCATCGGTCCAGA-3′ | [27] |
| DRK1-qPCR-F | 5′-CTTTTCCCTTTGGCGGCTACAA-3′ | Current work |
| DRK1-qPCR-R | 5′-ACTATCCTCCGGATTAGGTCTTCTAAC-3′ | Current work |
| L34-F | 5′-TCAATCTCTCCCGCGAATCC-3′ | [28] |
| L34-R | 5′-AGTTGGCGATTGTTGTGCGG-3′ | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcos, C.M.; de Oliveira, H.C.; Assato, P.A.; Castelli, R.F.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Drk1, a Dimorphism Histidine Kinase, Contributes to Morphology, Virulence, and Stress Adaptation in Paracoccidioides brasiliensis. J. Fungi 2021, 7, 852. https://doi.org/10.3390/jof7100852
Marcos CM, de Oliveira HC, Assato PA, Castelli RF, Fusco-Almeida AM, Mendes-Giannini MJS. Drk1, a Dimorphism Histidine Kinase, Contributes to Morphology, Virulence, and Stress Adaptation in Paracoccidioides brasiliensis. Journal of Fungi. 2021; 7(10):852. https://doi.org/10.3390/jof7100852
Chicago/Turabian StyleMarcos, Caroline Maria, Haroldo Cesar de Oliveira, Patrícia Akemi Assato, Rafael Fernando Castelli, Ana Marisa Fusco-Almeida, and Maria José Soares Mendes-Giannini. 2021. "Drk1, a Dimorphism Histidine Kinase, Contributes to Morphology, Virulence, and Stress Adaptation in Paracoccidioides brasiliensis" Journal of Fungi 7, no. 10: 852. https://doi.org/10.3390/jof7100852
APA StyleMarcos, C. M., de Oliveira, H. C., Assato, P. A., Castelli, R. F., Fusco-Almeida, A. M., & Mendes-Giannini, M. J. S. (2021). Drk1, a Dimorphism Histidine Kinase, Contributes to Morphology, Virulence, and Stress Adaptation in Paracoccidioides brasiliensis. Journal of Fungi, 7(10), 852. https://doi.org/10.3390/jof7100852








