Abstract
Candida auris is an emerging fungal pathogen with cases reported in countries around the world and in 19 states within the United States as of August 2020. The CDC has recommended that hospitals perform active surveillance upon admission for patients with the appropriate risk factors. Currently, active surveillance requires that local hospitals send surveillance swabs to a public health laboratory for analysis. In this work, a real-time PCR assay was developed for the specific detection of C. auris from surveillance swabs, blood, and urine to enable rapid detection of this pathogen. The assay uses commercially available primers and reporter probes and it was verified on the LightCycler 480 PCR platform. Contrived specimens and prospectively collected composite groin/axilla surveillance swabs were used to validate the assay. The performance of the PCR assay on surveillance swabs was also compared to a second PCR assay targeting C. auris that was performed at the Minnesota Department of Health–Public Health Laboratory (MDH-PHL). Our PCR assay is able to detect and differentiate C. auris from closely related Candida species such as C. duobushaemulonii, C. haemulonii, and C. pseudohaemulonii on the basis of melting curve temperature differences.
1. Introduction
Candida auris is a globally emerging, multidrug-resistant fungal pathogen with the potential to cause serious invasive infections [1,2,3,4,5]. Outbreaks of C. auris have occurred in healthcare settings across the globe, including in the U.S. regions of California, Florida, New Jersey, New York City, and Chicago, Illinois. These outbreaks have been difficult to control due to the ability of C. auris to contaminate the patient care environment and survive on surfaces for several weeks. Rapid, accurate identification and immediate implementation of infection control measures for patients infected or colonized with C. auris are crucial to controlling the spread.
Several C. auris cases have been linked to receipt of healthcare in countries outside the United States. The CDC recognizes foreign healthcare exposure as a risk factor for C. auris, and currently recommends that U.S. healthcare facilities identify the species of all Candida isolates from patients who had an overnight stay in a healthcare facility outside the U.S. in the previous year, especially for patients with stays at those facilities with documented C. auris transmission [6]. As of 8 May 2020, 1122 clinical cases of C. auris have been identified in 19 U.S. states, and an additional 2253 patients have been found with C. auris colonization in 16 jurisdictions [7]. Testing in these states has shown axilla and groin composite swabs to be the most common and consistent sites for finding C. auris colonization [6].
While the CDC recommends admission screening for carbapenemase-producing carbapenem-resistant enterobacteriaceae (CP-CRE) and C. auris among patients who received healthcare abroad in the past year, this recommendation has been challenging to implement for C. auris largely due to a lack of laboratory capacity across the U.S. for C. auris colonization testing. The CDC and Antibiotic Resistance (AR) Lab Network regional public health laboratories in the U.S. have PCR methods available to use for rapid screening of surveillance swabs [1,8,9]. The literature also contains a few reports of PCR assays at independent laboratories that were verified using culture isolates of C. auris or contrived specimens [10,11,12,13]. Currently, many hospital laboratories must send surveillance swabs to the CDC or one of the AR Lab Network Regional laboratories performing the PCR assay or they must rely on routine fungal culture for screening of surveillance swabs. Fungal culture requires two or more days to complete and supplementation of the culture medium with dulcitol may be needed to enhance recovery in colonized individuals. Following growth in culture, identification of C. auris to the genus and species level requires use of either matrix-assisted, laser desorption-time of flight (MALDI-TOF) mass spectrometry (MS) or gene sequencing [14,15]. Laboratories must use caution in selecting the MALDI-TOF MS library to use for identification since some may not contain C. auris [16].
The goal of this work was to design and verify a real-time PCR method for the specific detection of C. auris directly from surveillance swabs, blood, and urine. Blood and urine were selected because these specimens are the sources most frequently positive in clinical cases when isolates were submitted to our laboratory for identification by clinical laboratories across the country. A PCR method for the direct detection and identification of C. auris is desirable because of its potential to identify colonized or infected patients more rapidly than fungal culture. The PCR can be performed on the day of patient admission while fungal culture results for Candida species typically require two or more days. Rapid information about C. auris colonization is used at our institution to inform infection prevention and control practices. Colonized patients are placed in contact precautions to reduce the likelihood of spread to other patients or staff. In addition, the PCR method was designed to differentiate C. auris from other closely related Candida species. Phenotypic and semi-automated identification methods have been reported to misidentify C. auris as other fungi such as C. duobushaemulonii, C. haemulonii, and C. psuedohaemulonii, whereas the PCR method developed is specific for C. auris. In this report, the PCR assay is described and its performance from blood and urine was characterized using contrived specimens because of a lack of clinical specimens. The PCR assay performance on fresh, prospectively collected surveillance swabs was also compared with the C. auris real-time PCR assay performed at the Minnesota Department of Health–Public Health Laboratory (MDH-PHL).
2. Materials and Methods
2.1. Specimens
Negative blood and urine specimens were waste specimens remaining after routine clinical care processes were completed. Composite groin/axilla surveillance swabs were collected prospectively following consent of the patient. The use of waste specimens and the prospective collection of surveillance swabs was approved by an Institutional Review Board of the Mayo Clinic on 5/22/2019 (19-002552).
Patient inclusion criteria for the collection of C. auris surveillance swabs required the patient to be admitted to an inpatient unit. In addition, the patient had to reside in or have received healthcare in a part of the U.S. or in a country with documented C. auris cases within the past year or the patient was positive for CP-CRE (KPC, NDM, OXA-48, VIM) on clinical testing or surveillance testing [17]. Australia, Canada, China, Colombia, France, Germany, India, Israel, Japan, Kenya, Kuwait, Oman, Pakistan, Panama, Russia, Saudi Arabia, Singapore, South Africa, South Korea, Spain, United Kingdom, United States “hot spots,” and Venezuela were included at the time of study. United States “hot spots” included the states of CA, CT, TX, OH, RI, NY, NJ, VA, PA, NC, as well as Washington DC, Puerto Rico, and Chicago IL patients with a zip code of 60601, 60007, 60018, 60068, 60106, 60131, 60176, and 60686. Patients who had tested negative for C. auris in the past six months or who were <18 years old were excluded.
2.2. Culture Isolates
In order to test the PCR assay’s ability to detect various strains of C. auris, type strains of Candida species were obtained from the Deutsche Sammlung von Mikroorganismen (DSMZ; Braunschweig, Germany) and the American Type Culture Collection (ATCC; Masassas, VA, USA). In addition, a panel of C. auris, C. haemulonii, C. duobushaemulonii, and other yeast isolates was obtained from the CDC and FDA Antibiotic Resistance Isolate Bank (AR Bank Panel #1099 C. auris, Atlanta, GA, USA). Clinical isolates sent to our reference laboratory for identification were also tested by the PCR assay and the source of each isolate is indicated in Appendix A Table A1. C. auris was identified by MALDI-TOF MS using a Bruker BioTyper system and the BDAL library with 8468 MSPs and supplemented with a custom library containing an additional 2631 MSPs. Isolates were freshly sub-cultured onto Inhibitory Mold Agar (IMA) (BBL, Sparks, MD, USA). Following growth, culture isolates were lysed by placing a 1 µL loopful of organism into a 1 mL tube containing 500 µL of sterilized water, 0.1 mm silica glass beads, and 2.4 mm Zirconia beads (BioSpec Products Inc, Bartlesville, OK, USA). The tubes were heated at 95 °C for 10 min, and then placed on a Disruptor Genie (Scientific Industries Inc., Bohemia, NY, USA) for 2 min to mechanically lyse the organisms and release the nucleic acid. 5 µL of nucleic acid was placed into 15 µL of PCR master mix (described below) and was tested without further processing using the PCR assay.
2.3. Specimen Processing and Nucleic Acid Extraction
Whole blood containing EDTA preservative, urine, and composite axilla/groin swabs were used to validate the PCR assay. Owing to a lack of patient specimens containing C. auris, each specimen type was validated using contrived samples spiked with C. auris near the limit of detection of the PCR assay.
Whole blood (200 µL) containing EDTA as a preservative was extracted on the MagNA Pure LC 2.0 Instrument (Roche Diagnostics, Indianapolis, IN, USA) using the MagNA Pure LC Total Nucleic Acid Isolation Kit, with an elution volume of 100 µL.
Urine was concentrated to 5 mL by centrifugation if the volume received was >10 mL. 250 µL of urine was heated at 95 °C for 5 min and 200 µL was extracted on the MagNA Pure LC 2.0 Instrument using the MagNA Pure LC Total Nucleic Acid Isolation Kit, and a final elution volume of 100 µL.
Surveillance swab types tested were (1) soft aluminum-wire swabs with a rayon head in liquid Stuart medium (Catalog #220133, BD Diagnostics, Franklin Lakes, NJ, USA), (2) plastic shafted culture swabs with a rayon head in liquid Stuart medium (catalog #220099, BD Diagnostics), and (3) nylon flocked Eswab in liquid Amies medium (Catalog #220245, BD Diagnostics). The aluminum wire swab and the plastic shafted swab were processed by cutting the swab above the swab head and placing the swab head into a tube containing 600 µL of Tris-EDTA neutralization buffer (NB). The NB tube was then placed on a thermomixer and shaken at 14,000 rpm for 6 min at 100 °C. Eswabs were processed by placing 60 µL of the liquid in the Eswab transport container into an NB tube and shaking at 14,000 rpm for 6 min at 100 °C on a thermomixer.
2.4. PCR Assay Conditions
The real-time PCR assay was developed for use on the LightCycler 480 instrument (Roche Life Science, Madison, WI, USA). C. auris primers and probe sequences were designed to detect a 269 bp region of the internal transcribed spacer 2 (ITS2) of the ribosomal gene (Figure 1a). The donor probe is labeled with fluorescein and the acceptor probe with a LightCycler® Red 610 nm fluorophore dye. The ITS target was chosen because of its highly conserved nature and the availability of sequence within public nucleotide databases [18].
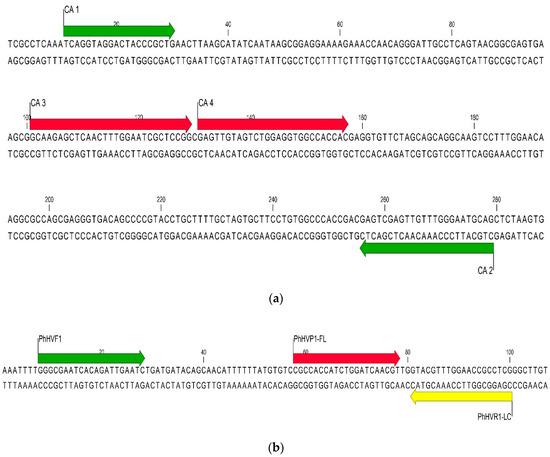
Figure 1.
(a) Sequence alignment of the C. auris internal transcribed spacer (ITS) target region with the PCR assay primers (green arrows) and probes (red arrows). (b) Sequence alignment of the Phocine herpesvirus type 1 internal control target region with the PCR assay primers (green and yellow arrows) and probes (red and yellow arrows).
Primers and probes were synthesized by TIB MOLBIOL (Adelphia, NJ, USA), and their sequences and product number are provided in Table 1. The PCR assay was performed using the LC FastStart DNA Master hybridization probe kit (Roche Diagnostics). Each reaction contained 0.8 µL of 25 mM MgCl2, 2 µL of 1× Roche LC FastStart mix, 0.03 µL of Recovery Template (PhHvµ DNA 106), 0.5 µM of each forward primer, 1 µM of each reverse primer, 0.2 µM of fluorescein-labeled probe, and 0.4 µM of Red 610-labeled probe. The total volume per reaction was 20 µL (15 µL master mix plus 5 µL of nucleic acid). PCR amplification with real-time detection was performed using the following cycling parameters: 1 template denaturing cycle at 95 °C for 10 min, followed by 45 amplification cycles at 95 °C for 10 s, 55 °C for 15 s, and 72 °C for 20 s. Following amplification, melting curve analysis was performed by measuring the fluorescent signal during the following cycling parameters: 95 °C for 30 s, 59 °C for 10 s, 45 °C for 15 s with a 0.1 °C/s transition, and 85 °C for 0 s with a 0.1 °C/s transition.

Table 1.
Candida auris PCR assay primer and probe sequences.
2.5. PCR Assay Controls
A positive control plasmid containing the ITS2 target of C. auris was purchased from TIB MOLBIOL (TIB #4605). An internal control plasmid was also purchased from TIB MOLBIOL (TIB #30-8393-02). The plasmid contains the PhHV1 glycoprotein B gene from Phoca vitulina, a herpes virus that infects harbor seals. Primers and probes were designed to detect a 93 base pair region within the PhHV1 gene (Table 1 and Figure 1b). The donor probe is labeled with fluorescein while the acceptor probe is labeled with LightCycler® Red 670 fluorophore.
The negative control for extracted samples (blood, urine) consists of Escherichia coli ATCC #25922 in 50% Stool Transport and Recovery (S.T.A.R.) buffer (Product #03335208001, Roche Diagnostics). A sterile culture swab clipped into an NB tube is used for the surveillance swab negative control.
2.6. PCR Assay Limit of Detection
The limit of detection (LOD) for the PCR assay was determined for the blood, urine, and surveillance swabs. A 0.5 McFarland solution of C. auris was spiked into negative specimens at concentrations ranging from 1 colony forming unit (CFU) to 733 CFU. The CFU of each dilution was confirmed by plating the inoculum onto SAB plates and enumerating the colonies following growth at 30 °C. 200 µL of each dilution was extracted three times using the MagNA Pure 2.0 extraction system as described previously. Each DNA extract was tested in duplicate for a total of six PCR replicates per dilution. The limit of detection was defined as the concentration that was positive by the PCR assay in 6 out of 6 replicates. Positive and negative extraction controls were also included for quality assurance.
2.7. PCR Assay Precision
Intra-day assay precision was tested by spiking negative whole blood specimens with three concentrations (low ~50 CFU/reaction, intermediate ~200 CFU/reaction, high ~550 CFU/reaction) of C. auris suspended in sterile water. 200 µL of each spiked specimen was extracted on the MagNA Pure 2.0 instrument and was tested in triplicate with the PCR assay on the LC 480 instrument. All 9 replicates were tested on the same day.
Inter-day assay precision was tested by spiking negative whole blood with the same three concentrations (low, intermediate, high) of C. auris suspended in sterile water. On 3 separate days and using 3 different laboratory technologists, 200 µL of each of the spiked specimens were extracted on the MagNA Pure 2.0 instrument and tested in triplicate with the PCR assay on the LC 480 instrument (n = 9 replicates per day, n = 27 replicates total).
2.8. PCR Assay Accuracy
Thirty negative specimens for each specimen type (surveillance swabs, blood, urine) were spiked with C. auris at a concentration within 1 log of the LOD determined for that specimen type. 6 different clinical isolates of C. auris were used for spiking each type of specimen (i.e., 5 specimens per isolate) to examine the effect, if any, of different C. auris isolate strains on detection by the PCR assay. Specimens were processed as described previously and tested by the PCR assay.
2.9. PCR Assay Specificity
The analytical specificity of the C. auris PCR assay was examined in silico by performing a BLAST search of each primer, each probe, and the entire amplicon sequence using the National Center for Biotechnology Information (NCBI) GenBank BLAST search website [18]. 50 organisms from the NCBI database that were closest in sequence homology to the target region of the C. auris PCR assay were identified to predict potential cross-reactivity with the PCR assay. An additional 9 unrelated but common organisms were analyzed in silico (Debaryomyces hansenii, Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, Candida krusei, Aspergillus fumigatus, Penicillium sp., and Fusarium sp.).
A panel of genomic DNA from 72 bacterial, fungal, and viral organisms found on skin, in blood, in urine, and in the environment were tested using the PCR assay for potential cross-reactivity with the primers and probes. Amplification and Sanger dideoxy sequencing of either 16S (bacteria), D2 LSU (fungi) rDNA, or viral-specific PCR assays [19,20] were utilized to confirm the presence of amplifiable nucleic acids in the specificity panel.
2.10. PCR Assay Comparison with MDH-PHL PCR Assay
The performance of the laboratory-developed PCR assay on surveillance swabs was compared with a real-time PCR assay performed by the MDH-PHL, the AR Lab Network Central Region laboratory. The assay was developed at the MDH-PHL (modifications to a previously described assay) [1]. Surveillance swabs were collected in duplicate from each consenting patient who met the CDC surveillance criteria. One swab was tested by the laboratory-developed PCR assay while the second swab was tested using the real-time PCR assay at the MDH-PHL. The swabs were stored and shipped at 4–25 °C and were tested within 4 days of collection.
2.11. Stability Studies
PCR master mix stability was tested up to 35 days of storage at 2 to 8 °C and −15 to −25 °C using the positive control plasmid. Specimen stability in each matrix type (blood, urine, surveillance swabs) was tested by spiking each matrix with 250 CFU/µl of C. auris and storing in individual aliquots at 2 to 8 °C and −15 to −25 °C. Individual aliquots were tested in triplicate after 0, 1, 8, and 14 days of storage. Extracted nucleic acid stability was tested by spiking negative extracts with 250 CFU/µL of C. auris and storing in individual aliquots at 2 to 8 °C and −15 to −25 °C. Individual aliquots were tested in triplicate after 0, 1, 8, and 14 days of storage.
3. Results
3.1. Detection of C. auris Culture Isolates
The PCR assay detected 32 of 32 (100%) culture isolates of C. auris (Appendix A Table A1) with an average Cp of 19.12 ± 1.94 cycles and an average melting temperature of 70.59 ± 0.13 °C. A representative melting curve is shown in Figure 2.
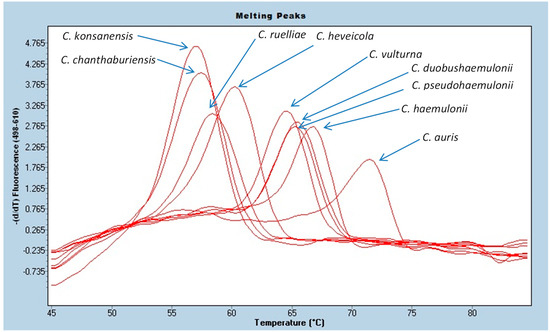
Figure 2.
PCR melting curves for C. auris and closely related Candida species.
All of the C. auris isolates in the CDC and FDA Antibiotic Resistance Isolate Bank panel (#1099) as of 5/22/2020 were detected by the PCR assay (n = 12). C. haemulonii (n = 1) and C. duobushaemulonii (n = 2) were also detected by the PCR but were differentiated from C. auris on the basis of melting temperature differences. Other yeast isolates in the panel were not detected by PCR assay.
3.2. PCR Assay Limit of Detection
The LOD of the PCR assay using C. auris spiked into EDTA whole blood samples was determined to be 54 CFU/reaction. The LOD in urine was 37 CFU/reaction. The LOD in aluminum-shafted rayon, plastic-shafted rayon, and Eswabs was 4, 11, and 37 CFU/reaction, respectively.
3.3. PCR Assay Precision
The precision results are presented in Appendix A Table A2 and Table A3. For intra-day precision, all 9 replicates were detected (100%). The average crossing point (Cp) was 27.79 ± 0.18 cycles at the high concentration (550 CFU/reaction), 28.54 ± 0.35 cycles at the intermediate concentration (200 CFU/reaction), and 35.22 ± 0.99 cycles at the low concentration (50 CFU/reaction). All Cps for the 9 replicates were within ± 2 cycles of the average. The average Tm was 69.99 ± 0.06 °C for the high concentration, 70.24 ± 0.26 °C for the intermediate concentration, and 70.47 ± 0.12 °C for the low concentration.
For inter-day precision, all 27 replicates were detected (100%). The average Cp was 29.15 ± 0.86 cycles at the high concentration, 30.47 ± 0.78 cycles at the intermediate concentration, and 33.08 ± 0.82 cycles at the low concentration. The average Tm was 70.68 ± 0.20 °C for the high concentration, 70.69 ± 0.20 °C for the intermediate concentration, and 70.96 ± 0.32 °C for the low concentration. No differences in results between days or between technologists were noted.
3.4. PCR Assay Accuracy
The PCR assay detected C. auris in each specimen matrix type with an accuracy of ≥93.3% (Table 2). The assay detected 29/30 whole blood specimens (96.7%), 29/30 urine specimens (96.7%), 30/30 aluminum-shafted rayon swab specimens (100%), 28/30 plastic-shafted rayon swab specimens (93.3%), and 28/30 Eswab specimens (93.3%). One of the six specimens that was not detected, a plastic-shafted rayon swab, was inhibited, as demonstrated by a negative internal control. Each specimen type had an amplification curve standard deviation of ≤2.56 cycles and a melting temperature standard deviation of ≤0.39 °C for the 30 contrived specimens.

Table 2.
Summary Cp and Tm (°C) results for contrived specimens spiked with C. auris.
The agreement of the laboratory-developed PCR assay with the MDH-PHL PCR assay was 100% (Table 3). Sixty-five surveillance swab specimens were negative by both PCR assays and one swab was inhibited in both PCR assays.

Table 3.
Comparison of the laboratory-developed PCR assay with the real-time PCR assay performed by the MDH-PHL.
3.5. PCR Assay Specificity
The laboratory-developed PCR was found to be highly specific for C. auris. As expected, none of the bacteria, fungi, or viruses tested were positive in the PCR assay with the exception of the closely related Candida species such as C. duobushaemulonii, C. haemulonii, and C. pseudohaemulonii (Appendix A Table A4). These 3 species can be misidentified as C. auris using some identification systems. While they are detected by the laboratory-developed PCR assay, their melting peak temperatures (Tm = 65.09–66.50 °C) are sufficiently different from C. auris (Tm = 70.59 °C) to readily allow differentiation from C. auris based on melting temperature differences. The melting temperatures of C. duobushaemulonii, C. haemulonii, and C. pseudohaemulonii are within 1.5 °C of each other so they cannot be differentiated from each other by melting curve analysis, but they are distinct from C. auris (Figure 2). Other closely related but infrequently isolated Candida species (i.e., Candida chanthaburiensis, Candida heveicola, Candida konsanensis, Candida ruelliae, Candida vulturna) also produced melting curves that overlapped each other but that were distinct from C. auris by several degrees (Figure 2). Using an in silico analysis of 60 organisms in GeneBank with the closest ITS2 sequence homology did not identify any homology in the probe binding region that would cause concern for cross-reactivity (Appendix A Table A4 and Figure 3).
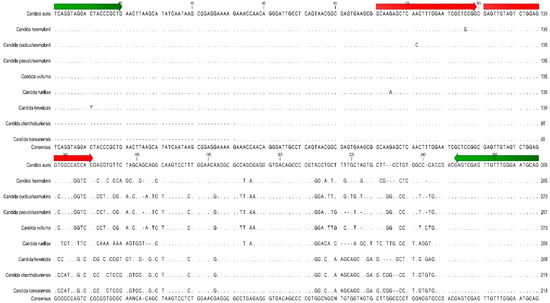
Figure 3.
Sequence alignment of C. auris ITS target region with 8 closely related Candida species. Green arrows indicate the position of the PCR assay primers. Red arrows indicate the position of the PCR assay reporter probes.
3.6. Stability Studies
The PCR master mix was stable at 2 to 8 °C and −15 to −25 °C up to 35 days. Specimen stability and extract stability varied by specimen type. EDTA whole blood, urine, aluminum-shafted rayon swabs, and Eswab specimens and nucleic acid extracts were stable after storage at 2 to 8 °C and −15 to −25 °C for 14 days. Plastic-shafted rayon swabs specimen stability was only good for 1 day at both 2 to 8 °C and −15 to −25 °C, so that swab type must be tested on the day collected to avoid loss of sensitivity of the assay and the other swabs types are preferred due to better specimen stability over time.
4. Discussion
The laboratory-developed PCR assay was found to have good sensitivity from blood, urine, and a variety of surveillance swab types. The Eswab is the preferred type for surveillance due to its good sensitivity, extended specimen stability, and its simple processing requirements. The flocked Eswab releases the specimen into the liquid of the swab container, allowing laboratory staff to sample the liquid without the requirement to cut the swab head off first into the liquid. This is ergonomically preferred for laboratory staff who can avoid potential repetitive motion injuries associated with clipping of swabs, and it also reduces the potential for contamination of the laboratory work area with target nucleic acid during the cutting process.
The sensitivity of the assay from blood and urine using contrived specimens was ≤100 CFU/reaction, so the PCR assay can be useful for the direct detection of C. auris from these specimen types. The PCR test should be ordered on blood and urine specimens only from those patients who are strongly suspected to have C. auris disseminated infection based on a review of symptoms and risk factors, such as recent foreign hospitalization [6]. Clinicians must not order the PCR assay on blood or urine for surveillance of colonization or for suspected infections at other local sites (e.g., respiratory, wound) because the sensitivity of these specimen types to detect colonization is unproven at this time. Although these specimens are easily obtained, especially urine, these are not appropriate or optimal specimen types for the detection of colonization or localized infections. Collection of composite axilla/groin surveillance swabs is encouraged for surveillance purposes.
The PCR assay detects C. auris isolates that belong to each of the currently recognized clades. In silico analysis of the target sequence and the primers and probes utilized in the PCR assay predicted that the assay would detect all clades. Testing of the CDC and FDA Antibiotic Resistance Isolate Bank panel, which contains C. auris strains from all clades (East Asia (n = 1), South Asia (n = 5), Africa (n = 2), South America (n = 3), and Iran (n = 1)), confirmed that the PCR assay detects all clades identified to date.
The ability to rapidly and directly detect C. auris from surveillance swabs and specimens such as blood and urine is important to provide another diagnostic tool to assist with containing the spread of this emerging fungal pathogen. The MDH-PHL is one of seven labs in CDC’s AR Lab Network to receive funding support for enhanced capacity to detect and respond to emerging antimicrobial resistance threats, including C. auris. One tool the AR Lab Network labs use to stop the spread of C. auris is colonization testing, which has been implemented as targeted screening in response to clinical cases, as well as for admission screening for specifically defined patients.
To date, C. auris has been identified in one patient in Minnesota who travelled and received healthcare outside of the U.S. MDH-PHL and epidemiologists continue to work with the CDC, as well as local and regional healthcare facilities to ensure that the state is prepared to identify and respond to cases of C. auris. As part of these efforts, the MDH-PHL AR Lab Network laboratory and MDH epidemiologists worked with the Mayo Clinic to implement C. auris colonization screening among patients admitted from foreign countries and U.S. regions with ongoing C. auris transmission because the Mayo Clinic is more likely than many hospitals in the state to admit patients with C. auris colonization. While the MDH-PHL/Mayo Clinic partnership works well, there is also a need for healthcare facilities to have the ability to perform C. auris screening on site. Testing of surveillance swabs at a centralized public health lab requires local, trained resources at healthcare facilities for packaging and shipping of specimens. This results in delayed testing of the surveillance swabs by at least one day and potentially longer depending on local hospital resources and the need to batch swabs prior to shipping. In addition, public health resources are not limitless, so having alternative avenues for testing and surveillance can be useful when public health resources are strained. For those healthcare facilities with the need and the capacity, the laboratory-developed PCR assay described in this work can provide a method that can be implemented locally following verification. The commercial availability of the primers and probes and description of assay conditions in this work can assist with local adoption.
Author Contributions
Conceptualization, K.A.R., P.S.V., P.S., and N.L.W.; Data curation, R.C.W., S.P.B., N.M.Z., K.M.H., K.A.R., and J.M.R.; Formal analysis, R.C.W., S.P.B., N.M.Z., K.M.H., K.M.J., J.M.K., S.A.H., J.E.B., J.L.F., and N.L.W.; Funding acquisition, L.K.S., P.S.V., P.S., and N.L.W.; Investigation, N.M.Z., L.K.S., K.A.R., J.E.B., J.M.R., P.S.V., P.S., and N.L.W.; Methodology, R.C.W., S.P.B., N.M.Z., K.M.H., K.M.J., J.M.K., S.A.H., J.M.R., J.L.F., P.S., and N.L.W.; Project administration, L.K.S., K.A.R., P.S.V., P.S., and N.L.W.; Resources, L.K.S., K.A.R., P.S.V., P.S., and N.L.W.; Supervision, L.K.S. and J.E.B.; Validation, R.C.W., S.P.B., N.M.Z., K.M.H., K.M.J., J.M.K., S.A.H., J.M.R., J.L.F., P.S.V., P.S., and N.L.W.; Writing—original draft, R.C.W., P.S.V., and N.L.W.; Writing—review and editing, S.P.B., N.M.Z., K.M.H., K.M.J., J.M.K., S.A.H., L.K.S., K.M.R., J.E.B., J.M.R., J.L.F., and P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Candida auris isolates used to evaluate the PCR test.
Table A1.
Candida auris isolates used to evaluate the PCR test.
| Isolate ID | Organism | Source of Isolate | Geographic Origin | PCR Amplification Cp (Cycles) | PCR Melting Temperature Tm (°C) |
|---|---|---|---|---|---|
| CA1 | Candida auris | Blood | Illinois | 21.69 | 70.51 |
| CA2 | Candida auris | Urine | Illinois | 20.14 | 70.46 |
| CA3 | Candida auris | Blood | Illinois | 21.57 | 70.41 |
| CA4 | Candida auris | Blood | New Jersey | 19.75 | 70.44 |
| CA5 | Candida auris | Axilla Wound | New Jersey | 18.88 | 70.47 |
| CA6 | Candida auris | Groin Wound | New Jersey | 19.30 | 70.48 |
| CA7 | Candida auris | Blood | New Jersey | 19.72 | 70.44 |
| CA8 | Candida auris | Blood | New Jersey | 19.90 | 70.34 |
| CA9 | Candida auris | Paranasal Sinus | New Jersey | 19.78 | 70.50 |
| CA10 | Candida auris | Peritoneal Fluid | Massachusetts | 17.04 | 70.58 |
| CA11 | Candida auris | Blood | Massachusetts | 17.78 | 70.49 |
| CA12 | Candida auris | Catheter Tip PICC line | New Jersey | 20.68 | 70.50 |
| CA13 | Candida auris | Urine | New Jersey | 19.17 | 70.75 |
| CA14 | Candida auris | Perirectal | Massachusetts | 18.41 | 70.76 |
| CA15 | Candida auris | Pleural Fluid | Saudi Arabia | 20.88 | 70.66 |
| CA16 | Candida auris | Blood | New Jersey | 21.68 | 70.52 |
| CA17 | Candida auris | Blood | Saudi Arabia | 19.80 | 70.58 |
| CA18 | Candida auris | Blood | Saudi Arabia | 18.28 | 70.48 |
| CA19 | Candida auris | Urine | Saudi Arabia | 18.74 | 70.67 |
| CA20 | Candida auris | Blood | New Jersey | 22.23 | 70.55 |
| CA21 | Candida auris | Axilla/Groin Swab | New Jersey | 22.88 | 70.56 |
| CA22 | Candida auris | Urine | New Jersey | 21.40 | 70.69 |
| CA23 | Candida auris | Axilla/Groin Swab | New Jersey | 20.74 | 70.53 |
| CA24 | Candida auris | Axilla/Groin Swab | New Jersey | 15.91 | 70.91 |
| CA25 | Candida auris | Urine | New Jersey | 16.22 | 70.67 |
| CA26 | Candida auris | Axilla/Groin Swab | New Jersey | 16.81 | 70.70 |
| CA27 | Candida auris | Sacrum Ulcer Swab | Saudi Arabia | 17.33 | 70.75 |
| CA28 | Candida auris | Axilla/Groin Swab | New Jersey | 16.51 | 70.72 |
| CA29 | Candida auris | Abdominal Wound | New Jersey | 16.94 | 70.72 |
| CA30 | Candida auris | Blood | New Jersey | 16.79 | 70.74 |
| CA31 | Candida auris | Urine | Saudi Arabia | 17.66 | 70.71 |
| CA32 | Candida auris | Urine | New Jersey | 17.35 | 70.72 |
| Average Range—Min Range—Max SD | 19.12 | 70.59 | |||
| 15.91 | 70.34 | ||||
| 22.88 | 70.91 | ||||
| 1.94 | 0.13 | ||||

Table A2.
Intra-day precision: 3 replicates tested at 3 concentrations (high, medium, low) on the same day.
Table A2.
Intra-day precision: 3 replicates tested at 3 concentrations (high, medium, low) on the same day.
| Replicate # | High Concentration (550 CFU/Rxn) | Medium Concentration (200 CFU/Rxn) | Low Concentration (50 CFU/Rxn) | |||
|---|---|---|---|---|---|---|
| Cp (cycles) | Tm (°C) | Cp (cycles) | Tm (°C) | Cp (cycles) | Tm (°C) | |
| 1 | 27.83 | 70.02 | ||||
| 2 | 27.60 | 69.92 | ||||
| 3 | 27.95 | 70.02 | ||||
| 4 | 28.83 | 70.02 | ||||
| 5 | 28.15 | 70.52 | ||||
| 6 | 28.63 | 70.17 | ||||
| 7 | 35.35 | 70.59 | ||||
| 8 | 34.18 | 70.35 | ||||
| 9 | 36.14 | 70.46 | ||||
| Average | 27.79 | 69.99 | 28.54 | 70.24 | 35.22 | 70.47 |
| Range—Min | 27.60 | 69.92 | 28.15 | 70.02 | 34.18 | 70.35 |
| Range—Max | 27.95 | 70.02 | 28.63 | 70.52 | 36.14 | 70.59 |
| SD | 0.18 | 0.06 | 0.35 | 0.26 | 0.99 | 0.12 |
CFU = colony forming unit; Rxn = reaction.

Table A3.
Inter-day precision: 3 replicates tested at 3 concentrations (high, medium, low) over 3 days, with testing performed by 3 technologists.
Table A3.
Inter-day precision: 3 replicates tested at 3 concentrations (high, medium, low) over 3 days, with testing performed by 3 technologists.
| High Concentration (550 CFU/Rxn) | |||
| Date Assayed | Replicate # | Cp (cycles) | Tm (°C) |
| Day 1 Tech: 1 | 1 | 30.06 | 70.90 |
| 2 | 30.30 | 71.05 | |
| 3 | 29.37 | 70.59 | |
| Day 2 Tech: 2 | 4 | 29.08 | 70.40 |
| 5 | 29.34 | 70.47 | |
| 6 | 29.75 | 70.66 | |
| Day 3 Tech: 3 | 7 | 28.48 | 70.70 |
| 8 | 28.24 | 70.70 | |
| 9 | 27.70 | 70.65 | |
| Average | 29.15 | 70.68 | |
| Range—Min | 27.70 | 70.40 | |
| Range—Max | 30.30 | 71.05 | |
| SD | 0.86 | 0.20 | |
| Medium Concentration (200 CFU/Rxn) | |||
| Date Assayed | Replicate # | Cp (cycles) | Tm (°C) |
| Day 1 Tech: 1 | 1 | 31.11 | 70.90 |
| 2 | 31.52 | 71.05 | |
| 3 | 30.83 | 70.59 | |
| Day 2 Tech: 2 | 4 | 30.65 | 70.63 |
| 5 | 30.43 | 70.40 | |
| 6 | 30.96 | 70.48 | |
| Day 3 Tech: 3 | 7 | 30.11 | 70.75 |
| 8 | 29.10 | 70.75 | |
| 9 | 29.48 | 70.70 | |
| Average | 30.47 | 70.69 | |
| Range—Min | 29.10 | 70.40 | |
| Range—Max | 31.52 | 71.05 | |
| SD | 0.78 | 0.20 | |
| Low Concentration (50 CFU/Rxn) | |||
| Date Assayed | Replicate # | Cp (cycles) | Tm (°C) |
| Day 1 Tech: 1 | 1 | 33.82 | 71.04 |
| 2 | 34.24 | 71.10 | |
| 3 | 33.20 | 70.91 | |
| Day 2 Tech: 2 | 4 | 32.94 | 70.24 |
| 5 | 31.76 | 70.77 | |
| 6 | 33.34 | 70.90 | |
| Day 3 Tech: 3 | 7 | 32.54 | 71.25 |
| 8 | 33.76 | 71.24 | |
| 9 | 32.12 | 71.20 | |
| Average | 33.08 | 70.96 | |
| Range—Min | 31.76 | 70.24 | |
| Range—Max | 34.24 | 71.25 | |
| SD | 0.82 | 0.32 | |
CFU = colony forming unit; Rxn = reaction.

Table A4.
Specificity panel of organisms tested by the PCR assay.
Table A4.
Specificity panel of organisms tested by the PCR assay.
| Organism | Source | PCR Assay Result | Melting Peak Tm (°C) | Genomes/µL |
|---|---|---|---|---|
| Bacteria | ||||
| Acinetobacter baumannii | ATCC 19606 | Negative | ND | 2.01 × 1011 |
| Bacillus subtilis group | Patient isolate | Negative | ND | 6.83 × 107 |
| Borrelia burgdorferi | ATCC 35210 | Negative | ND | 2.06 × 1010 |
| Bartonella henselae | ATCC 25583 | Negative | ND | 1.10 × 108 |
| Clostridium difficile | ATCC 9689d-5 | Negative | ND | 4.69 × 109 |
| Corynebacterium amycolatum | Patient isolate | Negative | ND | 3.41 × 108 |
| Corynebacterium aurimucosum/minutissimum | Patient isolate | Negative | ND | 1.65 × 108 |
| Corynebacterium striatum | Patient isolate | Negative | ND | 5.23 × 107 |
| Cutibacterium acnes | ATCC 6919 | Negative | ND | 6.91 × 107 |
| Enterobacter cloacae | ATCC 13047 | Negative | ND | 5.51 × 108 |
| Enterococcus faecalis | ATCC 19433-U | Negative | ND | 3.41 × 109 |
| Enterococcus faecium | ATCC 19434 | Negative | ND | 4.67 × 109 |
| Escherichia coli | ATCC 25922 | Negative | ND | 1.33 × 1010 |
| Haemophilus influenzae | ATCC 10211 | Negative | ND | 4.36 × 109 |
| Klebsiella pneumoniae | ATCC 700603 | Negative | ND | 3.41 × 1010 |
| Legionella pneumophila | ATCC 33152 | Negative | ND | 6.31 × 109 |
| Listeria monocytogenes | ATCC 15313 | Negative | ND | 1.63 × 109 |
| Mycobacterium avium | ATCC 700398 | Negative | ND | 4.15 × 107 |
| Mycobacterium haemophilum | ATCC 29548 | Negative | ND | 5.47 × 107 |
| Mycoplasma pneumoniae | ATCC 15531 | Negative | ND | 4.00 × 108 |
| Pseudomonas aeruginosa | ATCC 27853 | Negative | ND | 1.55 × 1010 |
| Staphylococcus aureus | ATCC 25923 | Negative | ND | 4.24 × 109 |
| Staphylococcus epidermidis | ATCC 14990 | Negative | ND | 1.42 × 109 |
| Streptococcus mitis group | Patient isolate | Negative | ND | 1.29 × 108 |
| Streptococcus pyogenes | ATCC 19615 | Negative | ND | 1.21 × 109 |
| Tropheryma whipplei | Patient blood | Negative | ND | 6.41 × 109 |
| Filamentous Fungi | ||||
| Alternaria species | Patient isolate | Negative | ND | 2.72 × 107 |
| Aspergillus flavus | ATCC 204304 | Negative | ND | 1.58 × 107 |
| Aspergillus fumigatus | ATCC MYA-3626 | Negative | ND | 4.83 × 107 |
| Blastomyces dermatitidis | Patient isolate | Negative | ND | 2.27 × 106 |
| Curvularia pallescens | ATCC 60941 | Negative | ND | 2.77 × 107 |
| Emergomyces africanus | Patient isolate | Negative | ND | 1.15 × 107 |
| Epidermophyton floccosum | CBS 970.25 | Negative | ND | 4.04 × 107 |
| Fusarium species | NIH-54 | Negative | ND | 3.91 × 107 |
| Histoplasma capsulatum | Patient isolate | Negative | ND | 1.47 × 107 |
| Microsporum canis | NIH-25 | Negative | ND | 3.37 × 107 |
| Paecilomyces species | NIH-56 | Negative | ND | 1.99 × 107 |
| Penicillium species | NIH-34 | Negative | ND | 3.95 × 107 |
| Rhizopus species | DSMZ-1834 | Negative | ND | 7.02 × 106 |
| Scopulariopsis species | NIH-49 | Negative | ND | 2.80 × 107 |
| Trichophyton rubrum | ATCC MYA-4438 | Negative | ND | 3.65 × 107 |
| Yeast | ||||
| Candida albicans | ATCC 60193 | Negative | ND | 5.70 × 108 |
| Candida duobushaemulonii | CBS 7798 | Positive | 65.09 | 3.92 × 106 |
| Candida glabrata | ATCC 15126 | Negative | ND | 1.20 × 107 |
| Candida haemulonii | ATCC 22991 | Positive | 66.50 | 8.57 × 106 |
| Candida intermedia | ATCC 14439 | Negative | ND | 3.21 × 106 |
| Candida krusei | ATCC 6258 | Negative | ND | 3.20 × 106 |
| Candida parapsilosis | ATCC 22019 | Negative | ND | 2.58 × 106 |
| Candida pseudohaemulonii | CBS 12370 | Positive | 65.13 | 5.16 × 106 |
| Candida pseudointermedia | ATCC 60126 | Negative | ND | 3.19 × 106 |
| Candida tropicalis | ATCC 1369 | Negative | ND | 1.53×106 |
| Clavispora lusitaniae | Patient isolate | Negative | ND | 3.25 × 106 |
| Cryptococcus gattii | ATCC 32269 | Negative | ND | 1.66 × 108 |
| Cryptococcus neoformans | ATCC 28957 | Negative | ND | 4.85 × 107 |
| Debaryomyces hansenii | Patient isolate | Negative | ND | 9.64 × 106 |
| Malassezia furfur | ATCC 12078 | Negative | ND | 1.31 × 107 |
| Pneumocystis species | Patient isolate | Negative | ND | 4.54 × 106 |
| Rhodotorula mucilaginosa | Patient isolate | Negative | ND | 1.44 × 107 |
| Saccharomyces cerevisiae | ATCC 4126 | Negative | ND | 5.42 × 106 |
| Trichosporon cutaneum | ATCC 28592 | Negative | ND | 1.26 × 107 |
| Trichosporon dermatis | ATCC MYA-4294 | Negative | ND | 1.59 × 107 |
| Parasite/Virus/Human | ||||
| Acanthamoeba polyphaga | ATCC 50371 | Negative | ND | 1.00 × 105 |
| Balamuthia mandrillaris | ATCC PRA290 | Negative | ND | 1.00 × 103 |
| Cyclospora species | Patient stool | Negative | ND | 6.96 × 105 |
| Dientamoeba fragilis | ATCC 30948 | Negative | ND | 2.74 × 105 |
| Giardia lamblia | ATCC 30957 | Negative | ND | 3.21 × 108 |
| Naegleria fowleri | ATCC 30896 | Negative | ND | 1.00 × 105 |
| Plasmodium falciparum | WHO STD | Negative | ND | 1.00 × 106 |
| Cytomegalovirus | AcroMetrix | Negative | ND | 5.00 × 108 |
| Epstein-Barr virus | AcroMetrix | Negative | ND | 5.00 × 108 |
| Human DNA | MRC-5 cells | Negative | ND | 3.42 × 107 |
AcroMetrix = ThermoFisher AcroMetrix controls, Waltham, MA; ATCC = American Type Culture Collection, Manassas, VA; CBS = CBS-KNAW Collections, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; DSMZ = Leibniz Institute DSMZ—German Collection of Microorganisms and Cell Cultures GmbH, Germany; NIH = National Institutes of Health, Bethesda, MD; ND = not detected; WHO = World Health Organization.
References
- Leach, L.; Zhu, Y.; Chaturvedi, S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Spivak, E.S.; Hanson, K.E. Candida auris: An Emerging Fungal Pathogen. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Saris, K.; Meis, J.F.; Voss, A. Candida auris. Curr. Opin. Infect. Dis. 2018, 31, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Navalkele, B.D.; Revankar, S.; Chandrasekar, P. Candida auris: A worrisome, globally emerging pathogen. Expert Rev. Anti-Infect. Ther. 2017, 15, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.; Quinn, M.; Tsay, S.; Poirot, E.; Chaturvedi, S.; Southwick, K.; Greenko, J.; Fernandez, R.; Kallen, A.; Vallabhaneni, S.; et al. Candida auris in Healthcare Facilities, New York, USA, 2013–2017. Emerg. Infect. Dis. 2018, 24, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- CDC. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-screening.html (accessed on 5 July 2020).
- CDC. Available online: https://www.cdc.gov/fungal/candida-auris/index.html (accessed on 5 July 2020).
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef] [PubMed]
- CDC. Available online: https://www.cdc.gov/drugresistance/laboratories/AR-lab-network-testing-details.html (accessed on 5 July 2020).
- Ruiz-Gaitán, A.C.; Fernández-Pereira, J.; Valentin, E.; Tormo-Mas, M.A.; Eraso, E.; Pemán, J.; de Groot, P.W.J. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 2018, 308, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Theill, L.; Dudiuk, C.; Morales-Lopez, S.; Berrio, I.; Rodríguez, J.Y.; Marin, A.; Gamarra, S.; Garcia-Effron, G. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev. Iberoam. Micol. 2018, 35, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Fang, W.; Daneshnia, F.; Al-Hatmi, A.M.; Liao, W.; Pan, W.; Khan, Z.; Ahmad, S.; Rosam, K.; Lackner, M.; et al. Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol. 2019, 14, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Sexton, D.J.; Kordalewska, M.; Bentz, M.L.; Welsh, R.M.; Perlin, D.S.; Litvintseva, A.P. Direct Detection of Emergent Fungal Pathogen Candida auris in Clinical Skin Swabs by SYBR Green-Based Quantitative PCR Assay. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Vatanshenassan, M.; Boekhout, T.; Meis, J.F.; Berman, J.; Chowdhary, A.; Ben-Ami, R.; Sparbier, K.; Kostrzewa, M. Candida auris Identification and Rapid Antifungal Susceptibility Testing Against Echinocandins by MALDI-TOF MS. Front. Cell. Infect. Microbiol. 2019, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Situ, S.F.; Steven, A.; Razak, M.F.A. The Pitfall of Utilizing a Commercial Biochemical Yeast Identification Kit to Detect Candida auris. Ann. Clin. Lab. Sci. 2019, 49, 546–549. [Google Scholar] [PubMed]
- CDC. Available online: https://www.cdc.gov/fungal/candida-auris/identification.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Ffungal%2Fcandida-auris%2Frecommendations.html (accessed on 5 July 2020).
- CDC. Available online: https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html#world (accessed on 5 July 2020).
- NCBI GenBank. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 5 July 2020).
- Binnicker, M.J.; Espy, M.E. Comparison of six real-time PCR assays for qualitative detection of cytomegalovirus in clinical specimens. J. Clin. Microbiol. 2013, 51, 3749–3752. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Espy, M.J.; Mandrekar, J.; Jones, M.F.; Cockerill, F.R.; Patel, R. Quantitative real-time polymerase chain reaction for evaluating DNAemia due to cytomegalovirus, Epstein-Barr virus, and BK virus in solid-organ transplant recipients. Clin. Infect. Dis. 2007, 45, 1056–1061. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).