Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification
Abstract
1. Introduction
2. Methods
2.1. Ethics’ Statement
2.2. Characteristics of Strains Used to Make the in-House Library
2.3. Construction of In-House Library “C. auris Colombia”
2.4. Validation of Colombia Candida auris Library
3. Results
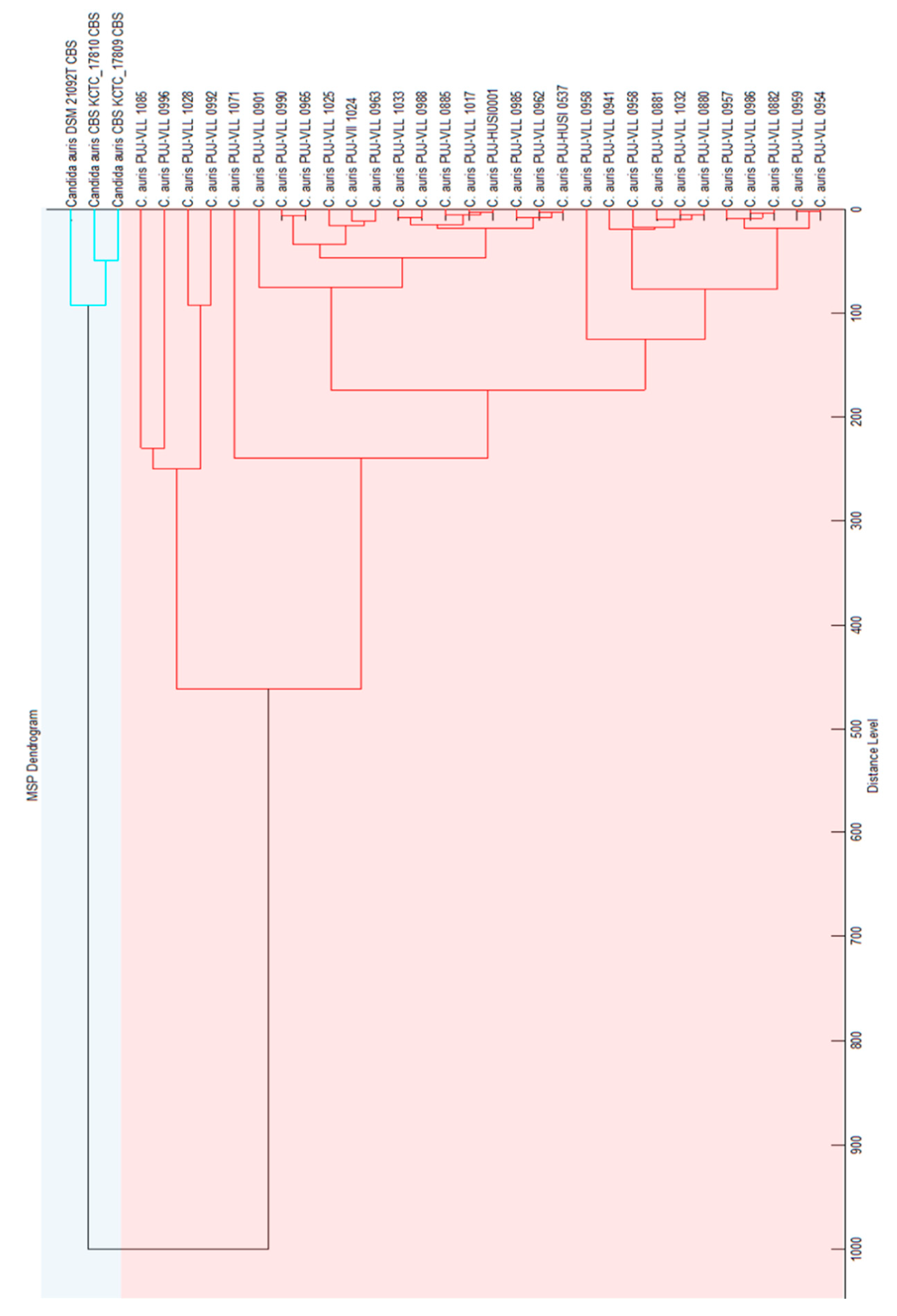
3.1. Construction of the in-House Library “C. auris Colombia”
3.2. Candida auris Colombia Library Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of data and material
Abbreviations
| MDR | Multidrug resistant |
| MALDI-TOF MS | Matrix Assisted Laser Desorption and Ionization-Time of Flight Mass Spectrometry |
| MSP | Main spectra library |
| HCCA | α-Cyano-4-hydroxycinnamic acid |
Footnote
References
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef]
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73, i4–i13. [Google Scholar] [CrossRef]
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014, 10, 95–105. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Parra-Giraldo, C.M.; Ceballos-Garzón, A.; Martínez, H.P.; Rodríguez, G.J.; Álvarez-Moreno, C.A.; Rodríguez, J.Y. Invasive Infections with Multidrug-Resistant Yeast Candida auris, Colombia. Emerg. Infect. Dis. 2017, 23, 162–164. [Google Scholar] [CrossRef]
- Parra-Giraldo, C.M.; Valderrama, S.L.; Cortes-Fraile, G.; Garzón, J.R.; Ariza, B.E.; Morio, F.; Linares-Linares, M.Y.; Ceballos-Garzón, A.; de la Hoz, A.; Hernandez, C.; et al. First report of sporadic cases of Candida auris in Colombia. Int. J. Infect. Dis. 2018, 69, 63–67. [Google Scholar] [CrossRef]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabhaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019, 57, 1–12. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention Identification of Candida auris-Fungal Diseases. Available online: https://www.cdc.gov/fungal/candida-auris/recommendations.html (accessed on 3 December 2019).
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef]
- Ceballos-Garzón, A.; Cortes, G.; Morio, F.; Zamora-Cruz, E.L.; Linares, M.Y.; Ariza, B.E.; Valderrama, S.L.; Garzón, J.R.; Alvarez-Moreno, C.A.; Le Pape, P.; et al. Comparison between MALDI-TOF MS and MicroScan in the identification of emerging and multidrug resistant yeasts in a fourth-level hospital in Bogotá, Colombia. BMC Microbiol. 2019, 19, 106. [Google Scholar] [CrossRef]
- Panda, A.; Ghosh, A.K.; Mirdha, B.R.; Xess, I.; Paul, S.; Samantaray, J.C.; Srinivasan, A.; Khalil, S.; Rastogi, N.; Dabas, Y. MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J. Microbiol. Methods 2015, 109, 93–105. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- Vatanshenassan, M.; Boekhout, T.; Meis, J.F.; Berman, J.; Chowdhary, A.; Ben-Ami, R.; Sparbier, K.; Kostrzewa, M. Candida auris Identification and Rapid Antifungal Susceptibility Testing Against Echinocandins by MALDI-TOF MS. Front. Cell. Infect. Microbiol. 2019, 9, 20. [Google Scholar] [CrossRef]
- Escandón, P.; Chow, N.A.; Caceres, D.H.; Gade, L.; Berkow, E.L.; Armstrong, P.; Rivera, S.; Misas, E.; Duarte, C.; Moulton-Meissner, H.; et al. Molecular Epidemiology of Candida auris in Colombia Reveals a Highly Related, Countrywide Colonization With Regional Patterns in Amphotericin B Resistance. Clin. Infect. Dis. 2018, 68, 15–21. [Google Scholar] [CrossRef]
- Rhodes, J.; Fisher, M.C. Global epidemiology of emerging Candida auris. Curr. Opin. Microbiol. 2019, 52, 84–89. [Google Scholar] [CrossRef]
- Marklein, G.; Josten, M.; Klanke, U.; Müller, E.; Horré, R.; Maier, T.; Wenzel, T.; Kostrzewa, M.; Bierbaum, G.; Hoerauf, A.; et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 2009, 47, 2912–2917. [Google Scholar] [CrossRef]
- Pinto, A.; Halliday, C.; Zahra, M.; van Hal, S.; Olma, T.; Maszewska, K.; Iredell, J.R.; Meyer, W.; Chen, S.C.A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS ONE 2011, 6, 1–7. [Google Scholar] [CrossRef]
- Kostrzewa, M.; Schubert, S. MALDI-TOF Mass Spectrometry in Microbiology. In British Library Cataloguing in Publication Data; Caister Academic Press: Norfolk, UK, 2016; Volume 1, pp. 1–10. [Google Scholar]
- Yaman, G.; Akyar, I.; Can, S. Evaluation of the MALDI TOF-MS method for identification of Candida strains isolated from blood cultures. Diagn. Microbiol. Infect. Dis. 2012, 73, 65–67. [Google Scholar] [CrossRef]
- Rodríguez-Leguizamón, G.; Fiori, A.; López, L.F.; Gómez, B.L.; Parra-Giraldo, C.M.; Gómez-López, A.; Suárez, C.F.; Ceballos, A.; Van Dijck, P.; Patarroyo, M.A. Characterising atypical Candida albicans clinical isolates from six third-level hospitals in Bogotá, Colombia. BMC Microbiol. 2015, 15, 199. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Armstrong, P.A.; Rivera, S.M.; Escandon, P.; Caceres, D.H.; Chow, N.; Stuckey, M.J.; Díaz, J.; Gomez, A.; Vélez, N.; Espinosa-Bode, A.; et al. Hospital-Associated Multicenter Outbreak of Emerging Fungus Candida auris, Colombia, 2016. Emerg. Infect. Dis. 2019, 25, 1339–1346. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceballos-Garzon, A.; Amado, D.; Vélez, N.; Jiménez-A, M.J.; Rodríguez, C.; Parra-Giraldo, C.M. Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification. J. Fungi 2020, 6, 72. https://doi.org/10.3390/jof6020072
Ceballos-Garzon A, Amado D, Vélez N, Jiménez-A MJ, Rodríguez C, Parra-Giraldo CM. Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification. Journal of Fungi. 2020; 6(2):72. https://doi.org/10.3390/jof6020072
Chicago/Turabian StyleCeballos-Garzon, Andrés, Daniela Amado, Norida Vélez, María José Jiménez-A, Crescencio Rodríguez, and Claudia Marcela Parra-Giraldo. 2020. "Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification" Journal of Fungi 6, no. 2: 72. https://doi.org/10.3390/jof6020072
APA StyleCeballos-Garzon, A., Amado, D., Vélez, N., Jiménez-A, M. J., Rodríguez, C., & Parra-Giraldo, C. M. (2020). Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification. Journal of Fungi, 6(2), 72. https://doi.org/10.3390/jof6020072






