Deletion of the SKO1 Gene in a hog1 Mutant Reverts Virulence in Candida albicans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
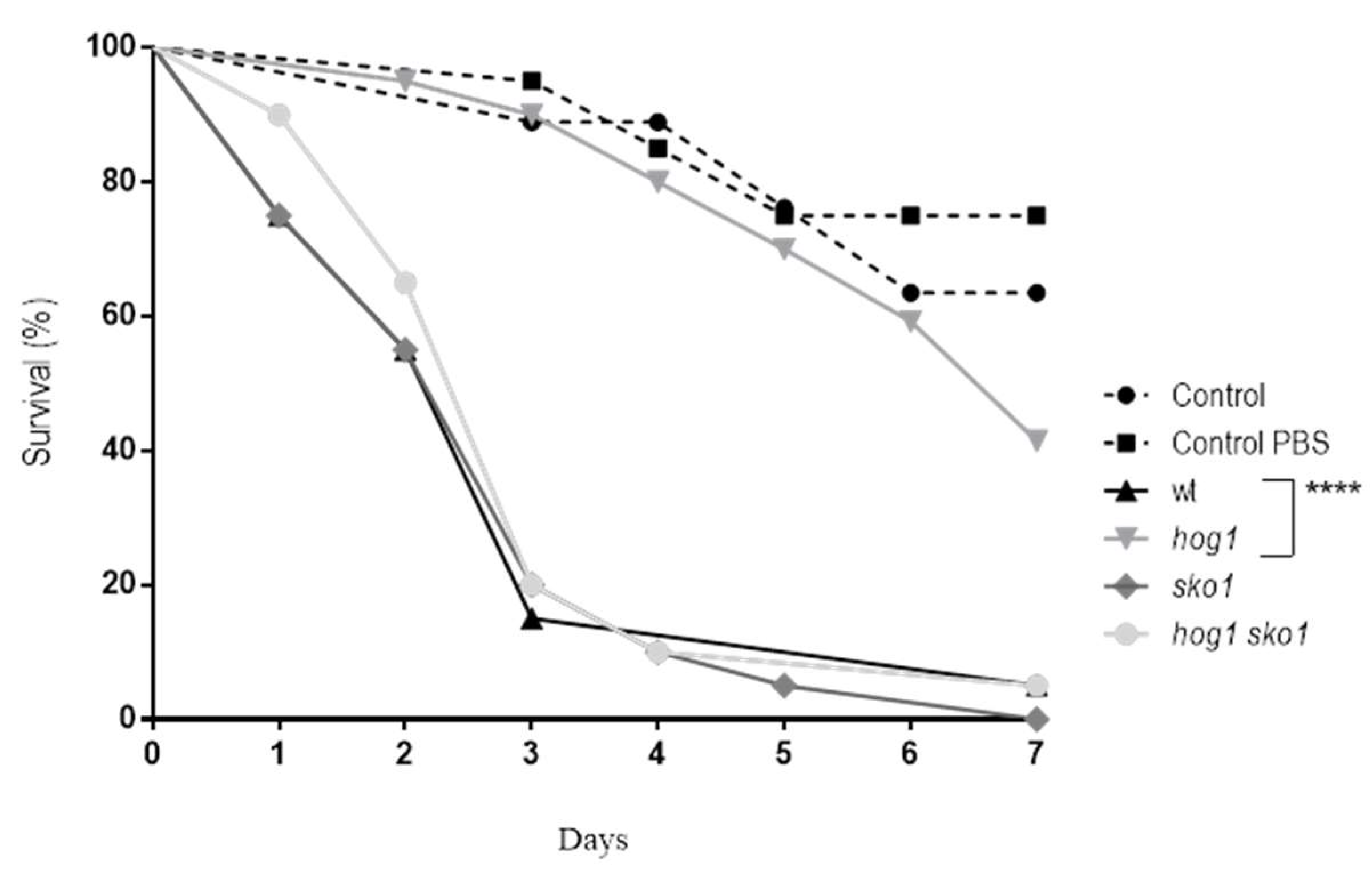
2.2. Virulence Assays in Galleria mellonella
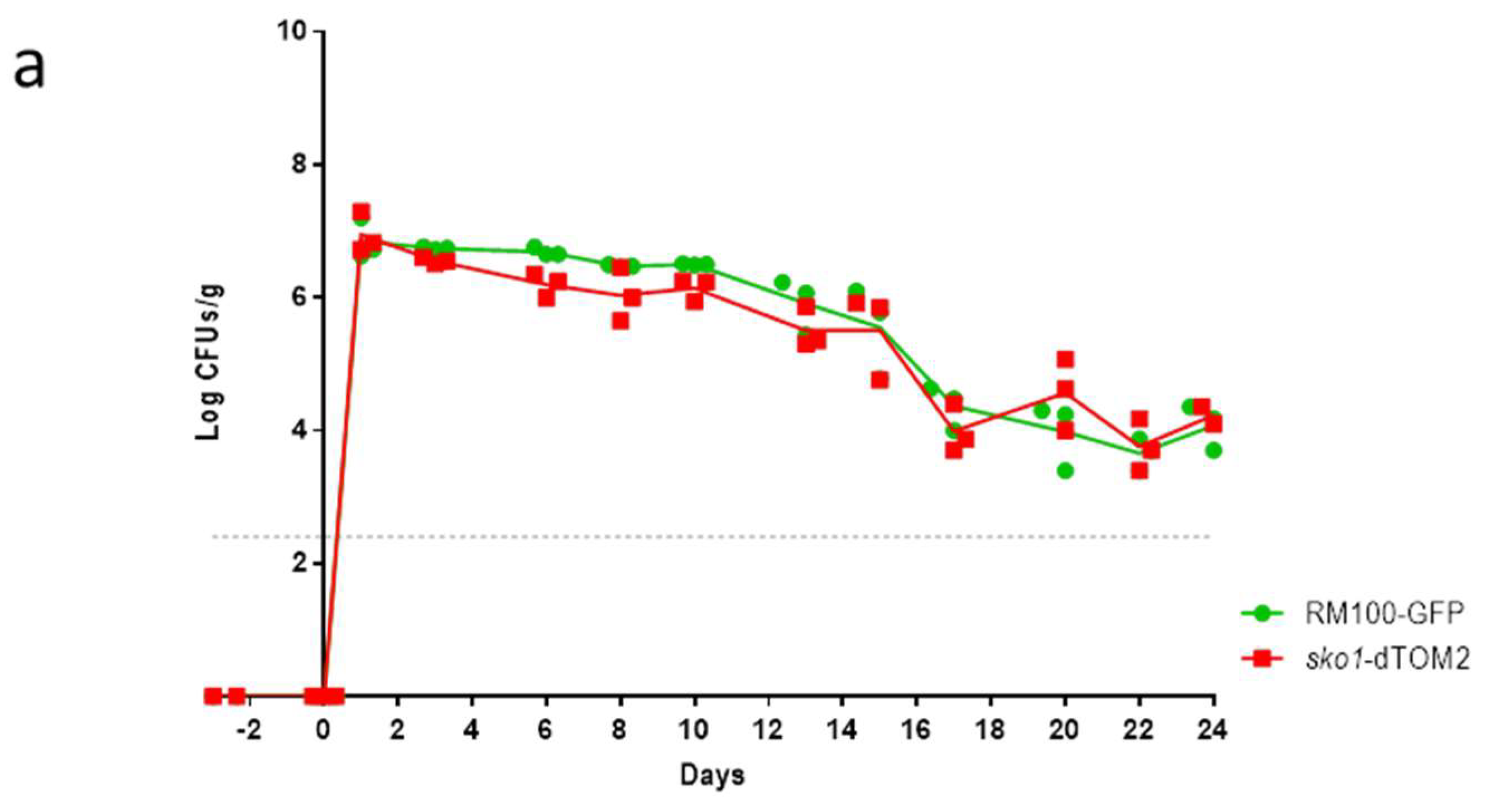
2.3. Murine Intestinal Commensalism Model and Adhesion Assay
3. Results
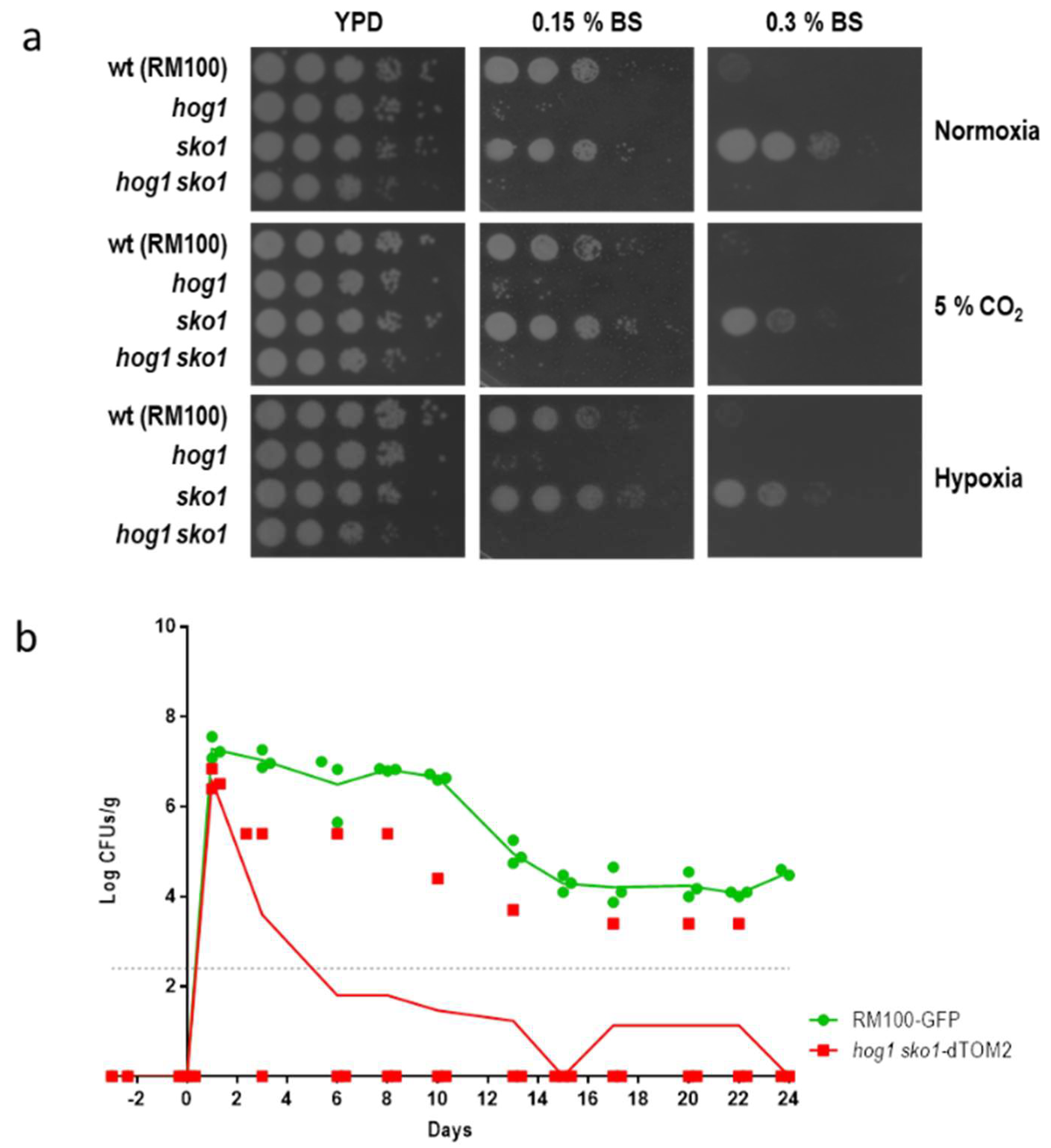
3.1. The Lack of Sko1 Does not Alter the Ability to Colonize the Murine Intestine
3.2. The sko1 Mutant is More Resistant to Bile Salts
3.3. Sko1 is not Required for Virulence, but Lack of Sko1 Enhances the Virulence of a hog1 Mutant
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Kultz, D. Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 1998, 46, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Day, A.M.; Quinn, J. Stress-Activated Protein Kinases in Human Fungal Pathogens. Front. Cell. Infect. Microbiol. 2019, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Candida species and virulence. ASM News 1994, 60, 313–318. [Google Scholar]
- Odds, F.C. Molecular phylogenetics and epidemiology of Candida albicans. Future Microbiol. 2010, 5, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Navarro-García, F.; Román, E.; Negredo, A.I.; Eisman, B.; Nombela, C.; Pla, J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2003, 2, 351–361. [Google Scholar] [CrossRef]
- Urrialde, V.; Prieto, D.; Pla, J.; Alonso-Monge, R. The Pho4 transcription factor mediates the response to arsenate and arsenite in Candida albicans. Front. Microbiol. 2015, 6, 118. [Google Scholar] [CrossRef]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.; Quinn, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 2006, 17, 1018–1032. [Google Scholar] [CrossRef]
- Cui, S.; Hassan, R.Y.; Heintz-Buschart, A.; Bilitewski, U. Regulation of Candida albicans Interaction with Macrophages through the Activation of HOG Pathway by Genistein. Molecules 2016, 21, 162. [Google Scholar] [CrossRef]
- Herrero-de-Dios, C.; Day, A.M.; Tillmann, A.T.; Kastora, S.L.; Stead, D.; Salgado, P.S.; Quinn, J.; Brown, A.J.P. Redox Regulation, Rather than Stress-Induced Phosphorylation, of a Hog1 Mitogen-Activated Protein Kinase Modulates Its Nitrosative-Stress-Specific Outputs. MBio 2018, 9, e02229-17. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Navarro-García, F.; Molero, G.; Díez-Orejas, R.; Gustin, M.; Pla, J.; Sánchez, M.; Nombela, C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999, 181, 3058–3068. [Google Scholar]
- Arana, D.M.; Alonso-Monge, R.; Du, C.; Calderone, R.; Pla, J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell. Microbiol. 2007, 9, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.D.; Román, E.; Correia, I.; Pla, J. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 2014, 9, e87128. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Jang, W.S.; Li, W.; Nayyar, N.; Edgerton, M. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot. Cell 2007, 6, 1876–1888. [Google Scholar] [CrossRef]
- Rauceo, J.M.; Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Deneault, J.S.; Smith, F.J.; Nantel, A.; Mitchell, A.P. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 2008, 19, 2741–2751. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Román, E.; Arana, D.M.; Prieto, A.D.; Urrialde, V.; Nombela, C.; Pla, J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 2010, 47, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Marotta, D.H.; Nantel, A.; Sukala, L.; Teubl, J.R.; Rauceo, J.M. Genome-wide transcriptional profiling and enrichment mapping reveal divergent and conserved roles of Sko1 in the Candida albicans osmotic stress response. Genomics 2013, 102, 363–371. [Google Scholar] [CrossRef]
- Sellam, A.; Hoog, M.V.; Tebbji, F.; Beaurepaire, C.; Whiteway, M.; Nantel, A. Modeling the transcriptional regulatory network that controls the early hypoxic response in Candida albicans. Eukaryot. Cell 2014, 13, 675–690. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Carvaihlo, S.; Nombela, C.; Rial, E.; Pla, J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology 2009, 155, 413–423. [Google Scholar] [CrossRef]
- Proft, M.; Pascual-Ahuir, A.; de Nadal, E.; Arino, J.; Serrano, R.; Posas, F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001, 20, 1123–1133. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol.Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Zapater, M.; Sohrmann, M.; Peter, M.; Posas, F.; de Nadal, E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 2007, 27, 3900–3910. [Google Scholar] [CrossRef]
- Rep, M.; Proft, M.; Remize, F.; Tamas, M.; Serrano, R.; Thevelein, J.M.; Hohmann, S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 2001, 40, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Negredo, A.; Monteoliva, L.; Gil, C.; Pla, J.; Nombela, C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 1997, 143, 297–302. [Google Scholar] [CrossRef] [PubMed]
- José, C.S.; Alonso-Monge, R.; Pérez-Díaz, R.M.; Pla, J.; Nombela, C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 1996, 178, 5850–5852. [Google Scholar] [CrossRef] [PubMed]
- Prieto, D.; Roman, E.; Alonso-Monge, R.; Pla, J. Overexpression of the Transcriptional Regulator WOR1 Increases Susceptibility to Bile Salts and Adhesion to the Mouse Gut Mucosa in Candida albicans. Front. Cell. Infect. Microbiol. 2017, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Mayer, F.L.; Miramon, P.; Citiulo, F.; Slesiona, S.; Jacobsen, I.D.; Hube, B. Distinct roles of Candida albicans-specific genes in host-pathogen interactions. Eukaryot. Cell 2014, 13, 977–989. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Koffel, R.; Tiwari, R.; Falquet, L.; Schneiter, R. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell. Biol. 2005, 25, 1655–1668. [Google Scholar] [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Hofs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Richardson, J.P.; Mogavero, S.; Moyes, D.L.; Blagojevic, M.; Kruger, T.; Verma, A.H.; Coleman, B.M.; Diaz, J.D.; Schulz, D.; Ponde, N.O.; et al. Processing of Candida albicans Ece1p Is Critical for Candidalysin Maturation and Fungal Virulence. MBio 2018, 9, e02178-17. [Google Scholar] [CrossRef]
- Swidergall, M.; Khalaji, M.; Solis, N.V.; Moyes, D.L.; Drummond, R.A.; Hube, B.; Lionakis, M.S.; Murdoch, C.; Filler, S.G.; Naglik, J.R. Candidalysin Is Required for Neutrophil Recruitment and Virulence During Systemic Candida albicans Infection. J. Infect. Dis. 2019, 220, 1477–1488. [Google Scholar] [CrossRef] [PubMed]

| Microorganism | Strain | Genotype | Nomenclature in the Manuscript and Figures | Source |
|---|---|---|---|---|
| C. albicans | RM100 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG-URA3-hisG | wt | [23] |
| C. albicans | CNC13 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG hog1::hisG-URA3-hisG/hog1::hisG | hog1 | [24] |
| C. albicans | VIC100 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG | sko1 | [15] |
| C. albicans | VIC200 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG | hog1 sko1 | [15] |
| C. albicans | RM-GFP | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG-URA3-hisG ADH1/adh1::tTA pTET-MoGFP-SAT1 | RM100-GFP | This work |
| C. albicans | VIC101 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG ADH1/adh1::tTA pTET-dTOM2-SAT1 | sko1-dTOM2 | This work |
| C. albicans | VIC201 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG ADH1/adh1::tTA pTET-dTOM2-SAT1 | hog1 sko1-dTOM2 | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urrialde, V.; Prieto, D.; Hidalgo-Vico, S.; Román, E.; Pla, J.; Alonso-Monge, R. Deletion of the SKO1 Gene in a hog1 Mutant Reverts Virulence in Candida albicans. J. Fungi 2019, 5, 107. https://doi.org/10.3390/jof5040107
Urrialde V, Prieto D, Hidalgo-Vico S, Román E, Pla J, Alonso-Monge R. Deletion of the SKO1 Gene in a hog1 Mutant Reverts Virulence in Candida albicans. Journal of Fungi. 2019; 5(4):107. https://doi.org/10.3390/jof5040107
Chicago/Turabian StyleUrrialde, Verónica, Daniel Prieto, Susana Hidalgo-Vico, Elvira Román, Jesús Pla, and Rebeca Alonso-Monge. 2019. "Deletion of the SKO1 Gene in a hog1 Mutant Reverts Virulence in Candida albicans" Journal of Fungi 5, no. 4: 107. https://doi.org/10.3390/jof5040107
APA StyleUrrialde, V., Prieto, D., Hidalgo-Vico, S., Román, E., Pla, J., & Alonso-Monge, R. (2019). Deletion of the SKO1 Gene in a hog1 Mutant Reverts Virulence in Candida albicans. Journal of Fungi, 5(4), 107. https://doi.org/10.3390/jof5040107





