Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa
Abstract
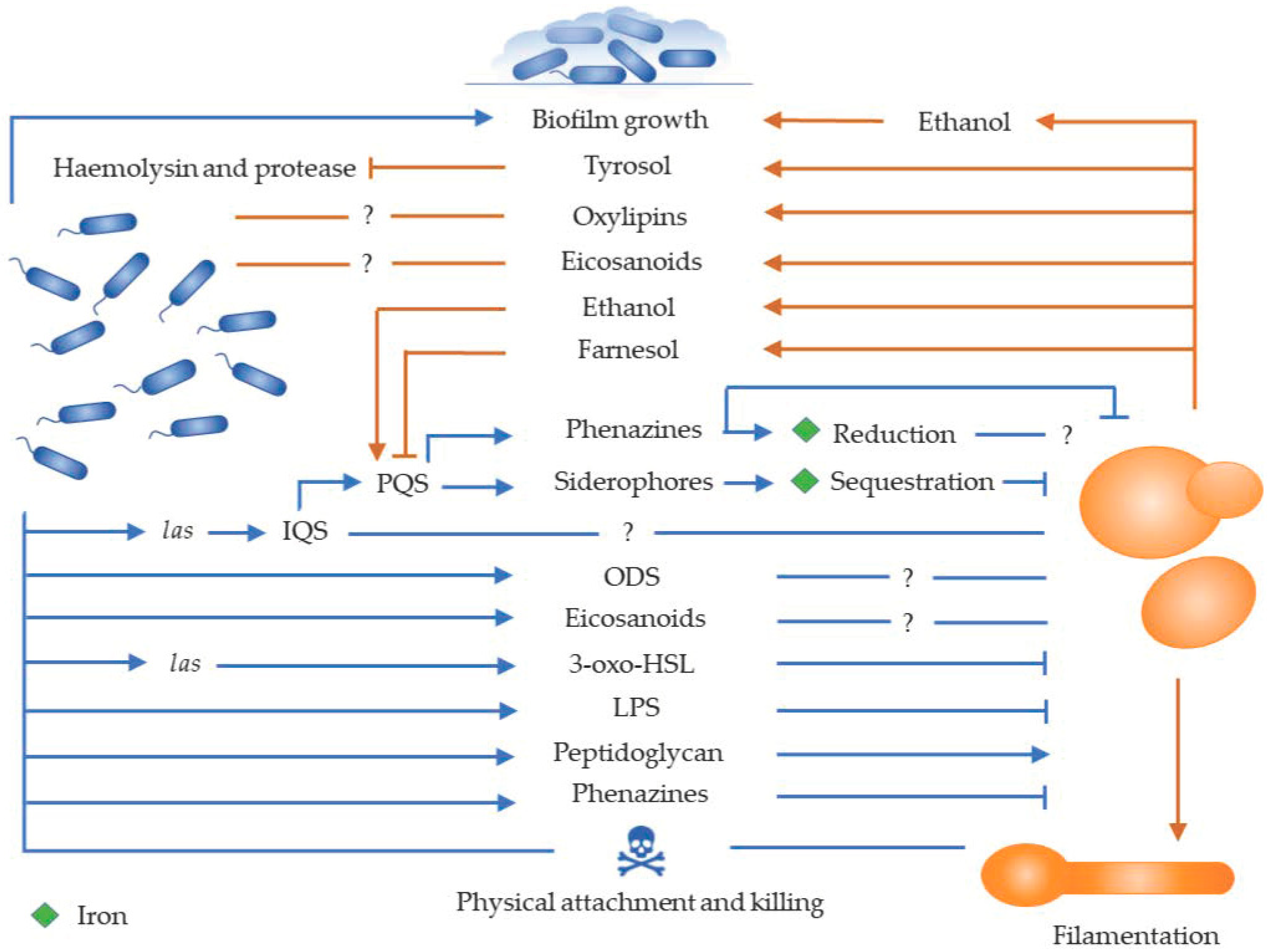
1. Introduction
2. In Vitro Interaction between Pseudomonas aeruginosa and Candida Species
2.1. Role of Cell Wall Components in the Interaction
2.2. Role of Quorum Sensing Molecules in the Interaction
2.3. Role of Phenazines in the Interaction
2.4. Role of Fatty Acid Metabolites in the Interaction
2.5. Role of Competition for Iron in the Interaction
3. In Vivo Interactions between Candida spp. and Pseudomonas aeruginosa
3.1. Interaction in the Lungs
3.2. Interaction in Wounds
3.3. Interaction in the Gastrointestinal Tract
4. Phenotypic Plasticity in Polymicrobial Interactions
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.H.; Kock, J.L.F. Oxidized fatty acids as inter-kingdom signaling molecules. Molecules 2014, 19, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Stöckli, M.; van Swaay, D.; Sabotič, J.; Kallio, P.T.; Künzler, M.; deMello, A.J.; Aebi, M. Probing bacterial-fungal interactions at the single cell level. Integr. Biol. (Camb.) 2014, 6, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.I.; Mujawar, L.H.; Shahzad, T.; Almeelbi, T.; Ismail, I.M.I.; Oves, M. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 2016, 183, 26–41. [Google Scholar] [CrossRef]
- Tay, W.H.; Chong, K.K.L.; Kline, K.A. Polymicrobial–host interactions during infection. J. Mol. Biol. 2016, 428, 3355–3371. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Kim, D.; Zhou, X.; Ahn, S.-J.; Burne, R.A.; Richards, V.P.; Koo, H. RNA-seq reveals enhanced sugar metabolism in Streptococcus mutans co-cultured with Candida albicans within mixed-species biofilms. Front. Microbiol. 2017, 8, 1036. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E. Synergistic effect of Candida albicans and Staphylococcus aureus on mouse mortality. Infect. Immun. 1982, 38, 921–924. [Google Scholar] [PubMed]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef]
- Auger, P.; Joly, J. Factors influencing germ tube production in Candida albicans. Mycopathologia 1977, 61, 183–186. [Google Scholar] [CrossRef]
- Hughes, W.T.; Kim, H.K. Mycoflora in cystic fibrosis: Some ecologic aspects of Pseudomonas aeruginosa and Candida albicans. Mycopathol. Mycol. Appl. 1973, 50, 261–269. [Google Scholar] [CrossRef]
- Kerr, J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J. Infect. 1994, 28, 305–310. [Google Scholar] [CrossRef]
- Kaleli, I.; Cevahir, N.; Demir, M.; Yildirim, U.; Sahin, R. Anticandidal activity of Pseudomonas aeruginosa strains isolated from clinical specimens. Mycoses 2006, 50, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.M.H.N.; Yau, J.Y.Y.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Pseudomonas aeruginosa inhibits in-vitro Candida biofilm development. BMC Microbiol. 2010, 10, 125. [Google Scholar] [CrossRef]
- Bandara, H.M.H.N.; Cheung, B.P.K.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol. Oral Microbiol. 2013, 28, 54–69. [Google Scholar] [CrossRef]
- Xu, X.L.; Lee, R.T.H.; Fang, H.M.; Wang, Y.M.; Li, R.; Zou, H.; Zhu, Y.; Wang, Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008, 4, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.; Barnes, J.D.; Mackenzie, K.S.; Odds, F.C.; Gow, N.A.R. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2008, 287, 48–55. [Google Scholar] [CrossRef]
- Holcombe, L.J.; McAlester, G.; Munro, C.A.; Enjalbert, B.; Brown, A.J.P.; Gow, N.A.R.; Ding, C.; Butler, G.; O’Gara, F.; Morrissey, J.P. Pseudomonas aeruginosa secreted factors impair biofilm development in Candida albicans. Microbiology 2010, 156, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Cugini, C.; Calfee, M.W.; Farrow, J.M.; Morales, D.K.; Pesci, E.C.; Hogan, D.A. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 896–906. [Google Scholar] [CrossRef]
- de Kievit, T.R.; Iglewski, B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [CrossRef]
- Passador, L.; Cook, J.M.; Gambello, M.J.; Rust, L.; Iglewski, B.H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 1993, 260, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 1995, 92, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Wagner, V.E.; Bushnell, D.; Passador, L.; Brooks, A.I.; Iglewski, B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef]
- Ovchinnikova, E.S.; Krom, B.P.; van der Mei, H.C.; Busscher, H.J. Force microscopic and thermodynamic analysis of the adhesion between Pseudomonas aeruginosa and Candida albicans. Soft Matter 2012, 8, 6454. [Google Scholar] [CrossRef]
- Hogan, D.A.; Vik, Å.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef] [PubMed]
- McAlester, G.; O’Gara, F.; Morrissey, J.P. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 2008, 57, 563–569. [Google Scholar] [CrossRef]
- Trejo-Hernández, A.; Andrade-Domínguez, A.; Hernández, M.; Encarnación, S. Interspecies competition triggers virulence and mutability in Candida albicans-Pseudomonas aeruginosa mixed biofilms. ISME J. 2014, 8, 1974–1988. [Google Scholar] [CrossRef]
- Pesci, E.C.; Milbank, J.B.J.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 1999, 96, 11229–11234. [Google Scholar] [CrossRef]
- Lépine, F.; Déziel, E.; Milot, S.; Rahme, L.G. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta - Gen. Subj. 2003, 1622, 36–41. [Google Scholar] [CrossRef]
- De Sordi, L.; Mühlschlegel, F.A. Quorum sensing and fungal-bacterial interactions in Candida albicans: A communicative network regulating microbial coexistence and virulence. FEMS Yeast Res. 2009, 9, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Déziel, E.; Lépine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2004, 101, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.G.; Merritt, J.H.; Hampton, T.H.; Hodgkinson, J.T.; Janecek, M.; Spring, D.R.; Welch, M.; O’Toole, G.A. 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 6770–6780. [Google Scholar] [CrossRef] [PubMed]
- Phelan, V.V.; Moree, W.J.; Aguilar, J.; Cornett, D.S.; Koumoutsi, A.; Noble, S.M.; Pogliano, K.; Guerrero, C.A.; Dorrestein, P.C. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 1683–1693. [Google Scholar] [CrossRef]
- Reen, F.J.; Mooij, M.J.; Holcombe, L.J.; Mcsweeney, C.M.; Mcglacken, G.P.; Morrissey, J.P.; O’Gara, F. The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 2011, 77, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Hogan, D.A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Martínez, E.; Cosnahan, R.K.; Wu, M.; Gadila, S.K.; Quick, E.B.; Mobley, J.A.; Campos-Gómez, J. Oxylipins mediate cell-to-cell communication in Pseudomonas aeruginosa. Commun. Biol. 2019, 2, 66. [Google Scholar] [CrossRef]
- Nigam, S.; Ciccoli, R.; Ivanov, I.; Sczepanski, M.; Deva, R. On mechanism of quorum sensing in Candida albicans by 3(R)-hydroxy-tetradecaenoic acid. Curr. Microbiol. 2011, 62, 55–63. [Google Scholar] [CrossRef]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Jones-Dozier, S.L. Proteomic analysis of the response of Pseudomonas aeruginosa PAO1 to the cell to cell signaling molecule trans,trans-farnesol of Candida albicans. Ph.D. Thesis, Georgia State University, Atlanta, GA, USA, 2008. [Google Scholar]
- Abdel-Rhman, S.H.; El-Mahdy, A.M.; El-Mowafy, M. Effect of tyrosol and farnesol on virulence and antibiotic resistance of clinical isolates of Pseudomonas aeruginosa. Biomed Res. Int. 2015, 2015, 456463. [Google Scholar] [CrossRef]
- Fink, G.R.; Clardy, J.; Feng, Q.; Fujita, M.; Chen, H. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. 2004, 101, 5048–5052. [Google Scholar] [CrossRef]
- Alem, M.A.S.; Oteef, M.D.Y.; Flowers, T.H.; Douglas, L.J. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot. Cell 2006, 5, 1770–1779. [Google Scholar] [CrossRef]
- Hernandez, M.E.; Kappler, A.; Newman, D.K. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 2004, 70, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 6372–6381. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef]
- O’Malley, Y.Q.; Abdalla, M.Y.; McCormick, M.L.; Reszka, K.J.; Denning, G.M.; Britigan, B.E. Subcellular localization of Pseudomonas pyocyanin cytotoxicity in human lung epithelial cells. Am. J. Physiol. Cell. Mol. Physiol. 2003, 284, L420–L430. [Google Scholar] [CrossRef]
- Gloyne, L.S.; Grant, G.D.; Perkins, A.V.; Powell, K.L.; McDermott, C.M.; Johnson, P.V.; Anderson, G.J.; Kiefel, M.; Anoopkumar-Dukie, S. Pyocyanin-induced toxicity in A549 respiratory cells is causally linked to oxidative stress. Toxicol. Vitr. 2011, 25, 1353–1358. [Google Scholar] [CrossRef]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; Høiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Gibson, J.; Sood, A.; Hogan, D.A. Pseudomonas aeruginosa-Candida albicans interactions: Localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 2009, 75, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Tupe, S.G.; Kulkarni, R.R.; Shirazi, F.; Sant, D.G.; Joshi, S.P.; Deshpande, M.V. Possible mechanism of antifungal phenazine-1-carboxamide from Pseudomonas sp. against dimorphic fungi Benjaminiella poitrasii and human pathogen Candida albicans. J. Appl. Microbiol. 2015, 118, 39–48. [Google Scholar] [CrossRef]
- Morales, D.K.; Grahl, N.; Okegbe, C.; Dietrich, L.E.P.; Jacobs, N.J.; Hogan, D.A. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Briard, B.; Bomme, P.; Lechner, B.E.; Mislin, G.L.A.; Lair, V.; Prévost, M.-C.; Latgé, J.-P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 5, 8220. [Google Scholar] [CrossRef]
- Chen, A.I.; Dolben, E.F.; Okegbe, C.; Harty, C.E.; Golub, Y.; Thao, S.; Ha, D.G.; Willger, S.D.; O’Toole, G.A.; Harwood, C.S.; et al. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014, 10, e1004480. [Google Scholar] [CrossRef]
- Nishanth Kumar, S.; Nisha, G.V.; Sudaresan, A.; Venugopal, V.V.; Sree Kumar, M.M.; Lankalapalli, R.S.; Dileep Kumar, B.S. Synergistic activity of phenazines isolated from Pseudomonas aeruginosa in combination with azoles against Candida species. Med. Mycol. 2014, 52, 482–490. [Google Scholar] [CrossRef]
- Deva, R.; Ciccoli, R.; Kock, L.; Nigam, S. Involvement of aspirin-sensitive oxylipins in vulvovaginal candidiasis. FEMS Microbiol. Lett. 2001, 198, 37–43. [Google Scholar] [CrossRef]
- Grózer, Z.; Tóth, A.; Tóth, R.; Kecskeméti, A.; Vágvölgyi, C.; Nosanchuk, J.D.; Szekeres, A.; Gácser, A. Candida parapsilosis produces prostaglandins from exogenous arachidonic acid and OLE2 is not required for their synthesis. Virulence 2015, 6, 85–92. [Google Scholar] [CrossRef]
- Noverr, M.C.; Phare, S.M.; Toews, G.B.; Coffey, M.J.; Huffnagle, G.B. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 2001, 69, 2957–2963. [Google Scholar] [CrossRef]
- Noverr, M.C.; Toews, G.B.; Huffnagle, G.B. Production of prostaglandins and leukotrienes by pathogenic fungi. Infect. Immun. 2002, 70, 400–402. [Google Scholar] [CrossRef]
- Alem, M.A.; Douglas, L.J. Prostaglandin production during growth of Candida albicans biofilms. J. Med. Microbiol. 2005, 54, 1001–1005. [Google Scholar] [CrossRef]
- Erb-Downward, J.R.; Noverr, M.C. Characterization of prostaglandin E2 production by Candida albicans. Infect. Immun. 2007, 75, 3498–3505. [Google Scholar] [CrossRef] [PubMed]
- Haas-Stapleton, E.J.; Lu, Y.; Hong, S.; Arita, M.; Favoreto, S.; Nigam, S.; Serhan, C.N.; Agabian, N. Candida albicans modulates host defense by biosynthesizing the pro-resolving mediator resolvin E1. PLoS ONE 2007, 2, e1316. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, Y.; Ishibashi, Y.; Hiruma, M.; Nishikawa, A.; Ikeda, S. Candida albicans abrogates the expression of interferon-γ-inducible protein-10 in human keratinocytes. FEMS Immunol. Med. Microbiol. 2008, 54, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Ells, R.; Kock, J.L.F.; Albertyn, J.; Kemp, G.; Pohl, C.H. Effect of inhibitors of arachidonic acid metabolism on prostaglandin E2 production by Candida albicans and Candida dubliniensis biofilms. Med. Microbiol. Immunol. 2011, 200, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Ali, S.; Shukla, P.K. Arachidonic acid affects biofilm formation and PGE2 level in Candida albicans and non-albicans species in presence of subinhibitory concentration of fluconazole and terbinafine. Braz. J. Infect. Dis. 2014, 18, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Geginat, G.; Tammer, I. Prostaglandin E2 from Candida albicans stimulates the growth of Staphylococcus aureus in mixed biofilms. PLoS ONE 2015, 10, e0135404. [Google Scholar] [CrossRef]
- Romani, L. Innate and adaptive immunity in Candida albicans infections and saprophytism. J. Leukoc. Biol. 2000, 68, 175–179. [Google Scholar]
- Smeekens, S.P.; van de Veerdonk, F.L.; van der Meer, J.W.M.; Kullberg, B.J.; Joosten, L.A.B.; Netea, M.G. The Candida Th17 response is dependent on mannanand B-glucan-induced prostaglandin E2. Int. Immunol. 2010, 22, 889–895. [Google Scholar] [CrossRef]
- Martínez, E.; Campos-Gómez, J. Oxylipins produced by Pseudomonas aeruginosa promote biofilm formation and virulence. Nat. Commun. 2016, 7, 13823. [Google Scholar] [CrossRef]
- Serhan, C.N. Lipoxins and aspirin-triggered 15-epi-lipoxin biosynthesis: An update and role in anti-inflammation and pro-resolution. Prostaglandins Other Lipid Mediat. 2002, 68–69, 433–455. [Google Scholar] [CrossRef]
- Vance, R.E.; Hong, S.; Gronert, K.; Serhan, C.N.; Mekalanos, J.J. The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proc. Natl. Acad. Sci. USA 2004, 101, 2135–2139. [Google Scholar] [CrossRef]
- Fourie, R.; Ells, R.; Kemp, G.; Sebolai, O.M.; Albertyn, J.; Pohl, C.H. Pseudomonas aeruginosa produces aspirin insensitive eicosanoids and contributes to the eicosanoid profile of polymicrobial biofilms with Candida albicans. Prostaglandins Leukot. Essent. Fat. Acids 2017, 117, 36–46. [Google Scholar] [CrossRef]
- Hou, C.T. New bioactive fatty acids. Asia Pac. J. Clin. Nutr. 2008, 17 (Suppl. 1), 192–195. [Google Scholar]
- Noverr, M.C.; Huffnagle, G.B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 2004, 72, 6206–6210. [Google Scholar] [CrossRef]
- Purschke, F.G.; Hiller, E.; Trick, I.; Rupp, S. Flexible survival strategies of Pseudomonas aeruginosa in biofilms result in increased fitness compared with Candida albicans. Mol. Cell. Proteomics 2012, 11, 1652–1669. [Google Scholar] [CrossRef]
- Nazik, H.; Joubert, L.-M.; Secor, P.R.; Sweere, J.M.; Bollyky, P.L.; Sass, G.; Cegelski, L.; Stevens, D.A. Pseudomonas phage inhibition of Candida albicans. Microbiology 2017, 163, 1568–1577. [Google Scholar] [CrossRef]
- Jabra-Rizk, M.A.; Kong, E.F.; Tsui, C.; Nguyen, M.H.; Clancy, C.J.; Fidel, P.L.; Noverr, M. Candida albicans pathogenesis: Fitting within the host-microbe damage response framework. Infect. Immun. 2016, 84, 2724–2739. [Google Scholar] [CrossRef]
- Kerr, J.R. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J. Clin. Microbiol. 1994, 32, 525–527. [Google Scholar]
- Azoulay, E.; Timsit, J.-F.; Tafflet, M.; de Lassence, A.; Darmon, M.; Zahar, J.-R.; Adrie, C.; Garrouste-Orgeas, M.; Cohen, Y.; Mourvillier, B.; et al. Candida colonization of the respiratory tract and subsequent Pseudomonas ventilator-associated pneumonia. Chest 2006, 129, 110–117. [Google Scholar] [CrossRef]
- Roux, D.; Gaudry, S.; Dreyfuss, D.; El-Benna, J.; De Prost, N.; Denamur, E.; Saumon, G.; Ricard, J.D. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit. Care Med. 2009, 37, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Hamet, M.; Pavon, A.; Dalle, F.; Pechinot, A.; Prin, S.; Quenot, J.P.; Charles, P.E. Candida spp. airway colonization could promote antibiotic-resistant bacteria selection in patients with suspected ventilator-associated pneumonia. Intensive Care Med. 2012, 38, 1272–1279. [Google Scholar] [CrossRef]
- Xu, L.; Wang, F.; Shen, Y.; Hou, H.; Liu, W.; Liu, C.; Jian, C.; Wang, Y.; Sun, M.; Sun, Z. Pseudomonas aeruginosa inhibits the growth of pathogenic fungi: In vitro and in vivo studies. Exp. Ther. Med. 2014, 7, 1516–1520. [Google Scholar] [CrossRef][Green Version]
- Nseir, S.; Jozefowicz, E.; Cavestri, B.; Sendid, B.; Di Pompeo, C.; Dewavrin, F.; Favory, R.; Roussel-Delvallez, M.; Durocher, A. Impact of antifungal treatment on Candida-Pseudomonas interaction: A preliminary retrospective case-control study. Intensive Care Med. 2007, 33, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Gaudry, S.; Khoy-Ear, L.; Aloulou, M.; Phillips-Houlbracq, M.; Bex, J.; Skurnik, D.; Denamur, E.; Monteiro, R.C.; Dreyfuss, D.; et al. Airway fungal colonization compromises the immune system allowing bacterial pneumonia to prevail. Crit. Care Med. 2013, 41, e191–e199. [Google Scholar] [CrossRef]
- Sun, K.; Metzger, D.W. Inhibition of pulmonary antibacterial defense by interferon-γ during recovery from influenza infection. Nat. Med. 2008, 14, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, S.S.; Zhao, X.; Hua, L.; Wang, J.F.; Nelson, S.; Ouyang, J. Ethanol inhibits lung clearance of Pseudomonas aeruginosa by a neutrophil and nitric oxide-dependent mechanism, in vivo. Alcohol. Clin. Exp. Res. 1999, 23, 735–744. [Google Scholar] [CrossRef]
- Ader, F.; Jawhara, S.; Nseir, S.; Kipnis, E.; Faure, K.; Vuotto, F.; Chemani, C.; Sendid, B.; Poulain, D.; Guery, B. Short term Candida albicans colonization reduces Pseudomonas aeruginosa-related lung injury and bacterial burden in a murine model. Crit. Care 2011, 15, R150. [Google Scholar] [CrossRef] [PubMed]
- Mear, J.B.; Gosset, P.; Kipnis, E.; Faure, E.; Dessein, R.; Jawhara, S.; Fradin, C.; Faure, K.; Poulain, D.; Sendid, B.; et al. Candida albicans airway exposure primes the lung innate immune response against Pseudomonas aeruginosa infection through innate lymphoid cell recruitment and interleukin-22-associated mucosal response. Infect. Immun. 2014, 82, 306–315. [Google Scholar] [CrossRef]
- Bergeron, A.C.; Seman, B.G.; Hammond, J.H.; Archambault, L.S.; Hogan, D.A.; Wheeler, R.T. Candida albicans and Pseudomonas aeruginosa interact to enhance virulence of mucosal infection in transparent zebrafish. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Conrad, D.J.; Bailey, B.A. Multidimensional clinical phenotyping of an adult cystic fibrosis patient population. PLoS ONE 2015, 10, e0122705. [Google Scholar] [CrossRef] [PubMed]
- Grahl, N.; Dolben, E.L.; Filkins, L.M.; Crocker, A.W.; Willger, S.D.; Morrison, H.G.; Sogin, M.L.; Ashare, A.; Gifford, A.H.; Jacobs, N.J.; et al. Profiling of bacterial and fungal microbial communities in cystic fibrosis sputum using RNA. mSphere 2018, 3, e00292-18. [Google Scholar] [CrossRef] [PubMed]
- Güngör, Ö.; Tamay, Z.; Güler, N.; Erturan, Z. Frequency of fungi in respiratory samples from Turkish cystic fibrosis patients. Mycoses 2013, 56, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Valenza, G.; Tappe, D.; Turnwald, D.; Frosch, M.; König, C.; Hebestreit, H.; Abele-Horn, M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst. Fibros. 2008, 7, 123–127. [Google Scholar] [CrossRef]
- Williamson, D.R.; Albert, M.; Perreault, M.M.; Delisle, M.-S.; Muscedere, J.; Rotstein, C.; Jiang, X.; Heyland, D.K. The relationship between Candida species cultured from the respiratory tract and systemic inflammation in critically ill patients with ventilator-associated pneumonia. Can. J. Anesth. Can. d’anesthésie 2011, 58, 275–284. [Google Scholar] [CrossRef]
- Leclair, L.W.; Hogan, D.A. Mixed bacterial-fungal infections in the CF respiratory tract. Med. Mycol. 2010, 48, S125–S132. [Google Scholar] [CrossRef][Green Version]
- Haiko, J.; Saeedi, B.; Bagger, G.; Karpati, F.; Özenci, V. Coexistence of Candida species and bacteria in patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Pendleton, K.M.; Erb-Downward, J.R.; Bao, Y.; Branton, W.R.; Falkowski, N.R.; Newton, D.W.; Huffnagle, G.B.; Dickson, R.P. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am. J. Respir. Crit. Care Med. 2017, 196, 1610–1612. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; O’Donoghue, E.; Bennett, K.; Gunaratnam, C.; O’Neill, S.J.; McElvaney, N.G. Sputum Candida albicans presages FEV 1 decline and hospital-treated exacerbations in cystic fibrosis. Chest 2010, 138, 1186–1195. [Google Scholar] [CrossRef]
- Kim, S.H.; Clark, S.T.; Surendra, A.; Copeland, J.K.; Wang, P.W.; Ammar, R.; Collins, C.; Tullis, D.E.; Nislow, C.; Hwang, D.M.; et al. Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor Nrg1 as a mechanism of pathogen adaptation. PLOS Pathog. 2015, 11, e1005308. [Google Scholar] [CrossRef]
- Murad, A.M.A.; Leng, P.; Straffon, M.; Wishart, J.; Macaskill, S.; MacCallum, D.; Schnell, N.; Talibi, D.; Marechal, D.; Tekaia, F.; et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001, 20, 4742–4752. [Google Scholar] [CrossRef] [PubMed]
- Delisle, M.-S.; Williamson, D.R.; Albert, M.; Perreault, M.M.; Jiang, X.; Day, A.G.; Heyland, D.K. Impact of Candida species on clinical outcomes in patients with suspected ventilator-associated pneumonia. Can. Respir. J. 2011, 18, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.M.; Huffnagle, G.B.; Dickson, R.P. The significance of Candida in the human respiratory tract: Our evolving understanding. Pathogens and Disease 2017, 75, fxt029. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, M.P.; Pham, D.; Fein, M.; Kong, L.; Weisman, L.E. Neonatal coinfection model of coagulase-negative Staphylococcus (Staphylococcus epidermidis) and Candida albicans: Fluconazole prophylaxis enhances survival and growth. Antimicrob. Agents. Chemother. 2007, 51, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Lindau, S.; Nadermann, M.; Ackermann, H.; Bingold, T.M.; Stephan, C.; Kempf, V.A.J.; Herzberger, P.; Beiras-Fernandez, A.; Zacharowski, K.; Meybohm, P. Antifungal therapy in patients with pulmonary Candida spp. colonization may have no beneficial effects. J. Intensive Care 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Terraneo, S.; Ferrer, M.; Martin-Loeches, I.; Esperatti, M.; Di Pasquale, M.; Giunta, V.; Rinaudo, M.; de Rosa, F.; Li Bassi, G.; Centanni, S.; Torres, A. Impact of Candida spp. isolation in the respiratory tract in patients with intensive care unit-acquired pneumonia. Clin. Microbiol. Infect. 2016, 22, 94.e1–94.e8. [Google Scholar] [CrossRef]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Townsend, E.M.; Sherry, L.; Kean, R.; Hansom, D.; Mackay, W.G.; Williams, C.; Butcher, J.; Ramage, G. Implications of antimicrobial combinations in complex wound biofilms containing fungi. Antimicrob. Agents Chemother. 2017, 61, e00672-17. [Google Scholar] [CrossRef]
- Chellan, G.; Shivaprakash, S.; Karimassery Ramaiyar, S.; Varma, A.K.; Varma, N.; Thekkeparambil Sukumaran, M.; Rohinivilasam Vasukutty, J.; Bal, A.; Kumar, H. Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes. J. Clin. Microbiol. 2010, 48, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Church, D.; Elsayed, S.; Reid, O.; Winston, B.; Lindsay, R. Burn wound infections. Clin. Microbiol. Rev. 2006, 19, 403–434. [Google Scholar] [CrossRef]
- Neely, A.N.; Law, E.J.; Holder, I.A. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect. Immun. 1986, 52, 200–204. [Google Scholar] [PubMed]
- de Macedo, J.L.S.; Santos, J.B. Bacterial and fungal colonization of burn wounds. Mem. Inst. Oswaldo Cruz 2005, 100, 535–539. [Google Scholar] [CrossRef]
- Gupta, N.; Haque, A.; Mukhopadhyay, G.; Narayan, R.P.; Prasad, R. Interactions between bacteria and Candida in the burn wound. Burns 2005, 31, 375–378. [Google Scholar] [CrossRef]
- Neville, B.A.; d’Enfert, C.; Bougnoux, M.-E. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015, 15, fov081. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Medina, E.; Koh, A.Y. The complexities of bacterial-fungal interactions in the mammalian gastrointestinal tract. Microb. Cell 2016, 3, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Tancrède, C.H.; Andremont, A.O. Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J. Infect. Dis. 1985, 152, 99–103. [Google Scholar] [CrossRef]
- Lopez-Medina, E.; Fan, D.; Coughlin, L.A.; Ho, E.X.; Lamont, I.L.; Reimmann, C.; Hooper, L.V.; Koh, A.Y. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog. 2015, 11, e1005129. [Google Scholar] [CrossRef]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Lan, C.-Y.; Newport, G.; Murillo, L.A.; Jones, T.; Scherer, S.; Davis, R.W.; Agabian, N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 2002, 99, 14907–14912. [Google Scholar] [CrossRef]
- Pande, K.; Chen, C.; Noble, S.M. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 2013, 45, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Cao, C.; Xie, J.; Ni, J.; Guan, G.; Tao, L.; Zhang, L.; Huang, G. N-acetylglucosamine-induced white-to-opaque switching in Candida albicans is independent of the Wor2 transcription factor. Fungal Genet. Biol. 2014, 62, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Zavala, B.; Reuß, O.; Park, Y.-N.; Ohlsen, K.; Morschhäuser, J. Environmental induction of white–opaque switching in Candida albicans. PLoS Pathog. 2008, 4, e1000089. [Google Scholar] [CrossRef] [PubMed]
- Malavia, D.; Lehtovirta-Morley, L.E.; Alamir, O.; Weiß, E.; Gow, N.A.R.; Hube, B.; Wilson, D. Zinc limitation induces a hyper-adherent goliath phenotype in Candida albicans. Front. Microbiol. 2017, 8, 2238. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.T.; Mall, M.A.; Kicic, A.; Stick, S.M. AREST CF Hypoxia and sterile inflammation in cystic fibrosis airways: Mechanisms and potential therapies. Eur. Respir. J. 2017, 49, 1600903. [Google Scholar] [CrossRef] [PubMed]
- Faure, E.; Kwong, K.; Nguyen, D. Pseudomonas aeruginosa in chronic lung infections: How to adapt within the host? Front. Immunol. 2018, 9, 2416. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Cantón, R.; Campo, P.; Baquero, F.; Blázquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000, 288, 1251–1254. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.E.; Buckley, D.G.; Wu, Z.; Saenphimmachak, C.; Hoffman, L.R.; D’Argenio, D.A.; Miller, S.I.; Ramsey, B.W.; Speert, D.P.; Moskowitz, S.M.; et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. 2006, 103, 8487–8492. [Google Scholar] [CrossRef]
- Workentine, M.L.; Sibley, C.D.; Glezerson, B.; Purighalla, S.; Norgaard-Gron, J.C.; Parkins, M.D.; Rabin, H.R.; Surette, M.G. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS ONE 2013, 8, e60225. [Google Scholar] [CrossRef]
- Clark, S.T.; Diaz Caballero, J.; Cheang, M.; Coburn, B.; Wang, P.W.; Donaldson, S.L.; Zhang, Y.; Liu, M.; Keshavjee, S.; Yau, Y.C.W.; et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci. Rep. 2015, 5, 10932. [Google Scholar] [CrossRef]
- DeVault, J.D.; Kimbara, K.; Chakrabarty, A.M. Pulmonary dehydration and infection in cystic fibrosis: Evidence that ethanol activates alginate gene expression and induction of mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 1990, 4, 737–745. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect. Immun. 2015. [Google Scholar] [CrossRef]
- Dettman, J.R.; Rodrigue, N.; Aaron, S.D.; Kassen, R. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 2013, 110, 21065–21070. [Google Scholar] [CrossRef]
- Jain, M.; Ramirez, D.; Seshadri, R.; Cullina, J.F.; Powers, C.A.; Schulert, G.S.; Bar-Meir, M.; Sullivan, C.L.; McColley, S.A.; Hauser, A.R. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 2004, 42, 5229–5237. [Google Scholar] [CrossRef]
- Tart, A.H.; Blanks, M.J.; Wozniak, D.J. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 2006, 188, 6483–6489. [Google Scholar] [CrossRef]
| Infection Site | Type of Study/Model | Observations | Effect of P. aeruginosa on Candida | Effect of Candida on P. aeruginosa | Number(s) in Reference List |
|---|---|---|---|---|---|
| Lungs | Postoperative monitoring of surgery patients | Inhibition of C. albicans after subsequent colonization with P. aeruginosa. Reversed with antibiotic treatment | Inhibited | - | [80] |
| Lungs | Monitoring of patients with mechanical ventilation | Colonization with Candida spp. associated with increased risk of P. aeruginosa VAP1 | - | Promoted | [81] |
| Lungs | Monitoring of patients with VAP 1 | Increase in isolation of multidrug resistant bacteria such as P. aeruginosa when Candida spp. are present. Reduced risk for P. aeruginosa VAP 1 with antifungal treatment | - | Promoted | [83,85] |
| Lungs | Analysis of sputum samples from CF 2 patients | Higher incidence of co-existence between P. aeruginosa and Candida spp. in CF2 patients compared to other respiratory disorders | - | - | [98,100] |
| Lungs | Analysis of sputum samples from CF 2 patients | Presence of hyperfilamentous C. albicans with loss of function mutation in NGR1 in presence of P. aeruginosa | No inhibition of filamentation | _ | [101] |
| Lungs | Wistar rat model | Fungal colonization promoted pneumonia by P. aeruginosa Reversed with antifungal treatment | - | Promoted | [82,86] |
| Lungs | Mouse model | Prior C. albicans colonization promoted P. aeruginosa clearance | - | Clearance enhanced | [89,90] |
| Mucosa | Zebrafish swimbladder model | Synergistic increase in virulence in co-infection compared to single species infection | Increased virulence | Increased virulence | [91] |
| Wounds | Microbial populations of deep tissue wounds of patients with type 2 diabetes cultured | C. albicans not found in combination with P. aeruginosa | Inhibited | - | [106] |
| Wounds | Mouse model | Pre-infection with P. aeruginosa predisposed mice to lethal C. albicans infection | P. aeruginosa proteolytic enzymes promote lethal C. albicans infection | - | [108] |
| Wounds | Microbial analysis of wounds of burn patients | Inhibition of Candida spp. when Pseudomonas spp. were present | Inhibited | - | [110] |
| Wound model | In vitro biofilm model to mimic wounds | Biofilms of C. albicans, Staphylococcus aureus and P. aeruginosa created. Monotreatment with antifungal/antibiotic only shifted population dynamics without affecting overall biofilm bioburden | Provides physical support and protection to bacteria | [105] | |
| Gastro-intestinal tract | Neutropenic mouse model | Levels of both P. aeruginosa and C. albicans unaffected by co-incubation | Decrease in siderophore production and virulence | [114] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fourie, R.; Pohl, C.H. Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa. J. Fungi 2019, 5, 34. https://doi.org/10.3390/jof5020034
Fourie R, Pohl CH. Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa. Journal of Fungi. 2019; 5(2):34. https://doi.org/10.3390/jof5020034
Chicago/Turabian StyleFourie, Ruan, and Carolina H. Pohl. 2019. "Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa" Journal of Fungi 5, no. 2: 34. https://doi.org/10.3390/jof5020034
APA StyleFourie, R., & Pohl, C. H. (2019). Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa. Journal of Fungi, 5(2), 34. https://doi.org/10.3390/jof5020034





