Harnessing Epigenetic Modifiers Reveals MAPK-Mediated Regulation Mechanisms in Hadal Fungi of Alternaria alternata Under High Hydrostatic Pressure
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungual Isolation and Identification
2.2. Quantification of Conidia
2.3. Culture of Chemical Epigenetic Modifier
2.4. RNA Extraction and Reverse Transcription
2.5. Quantitative Reverse Transcriptase PCR (qRT-PCR)
2.6. Extraction of Secondary Metabolites
2.7. UPLC-MS/MS Analysis of Secondary Metabolites
2.8. Antimicrobial Activity Assay
2.9. Transcriptome Analysis
2.10. Transcriptome Validation
2.11. Statistical Analysis
3. Results
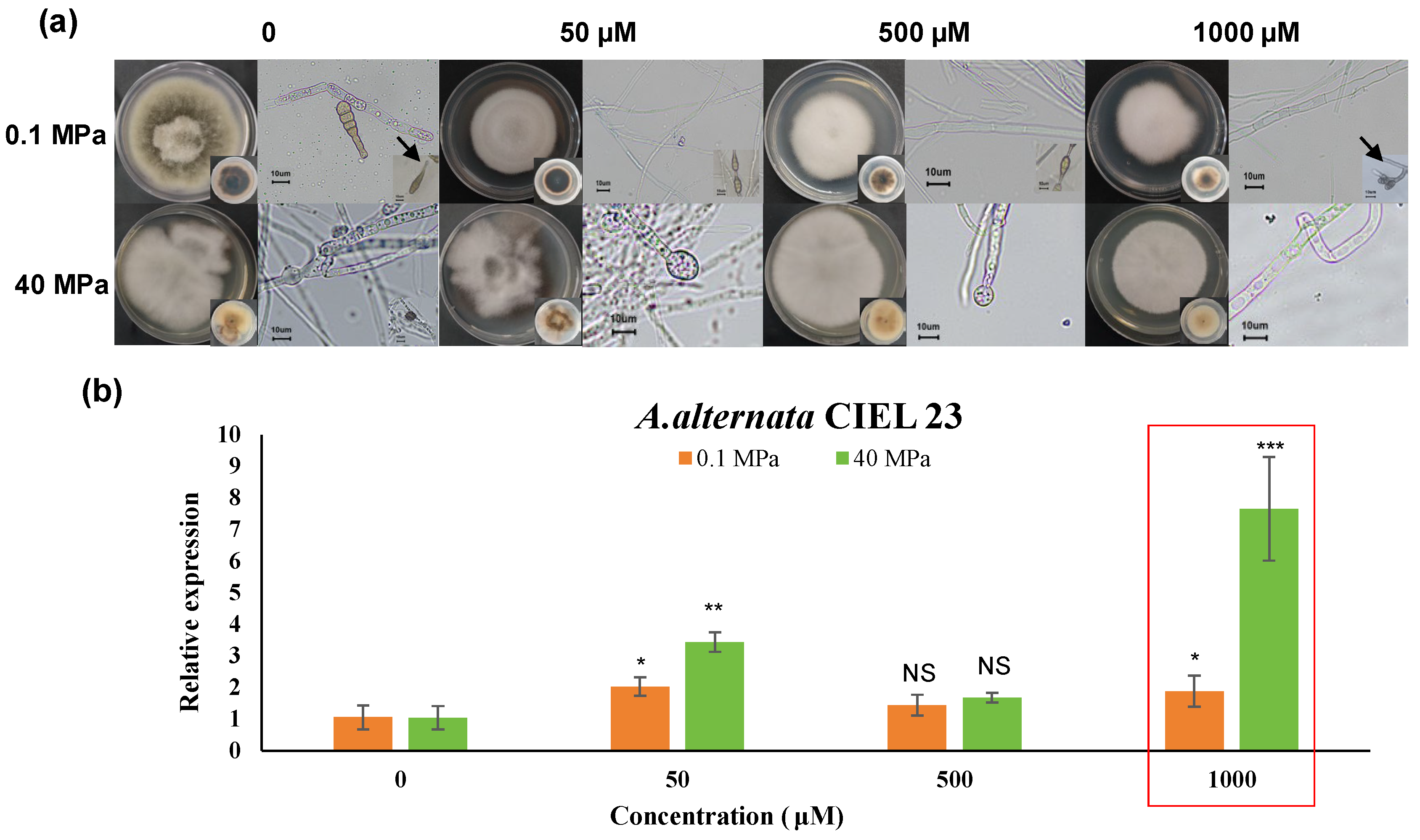
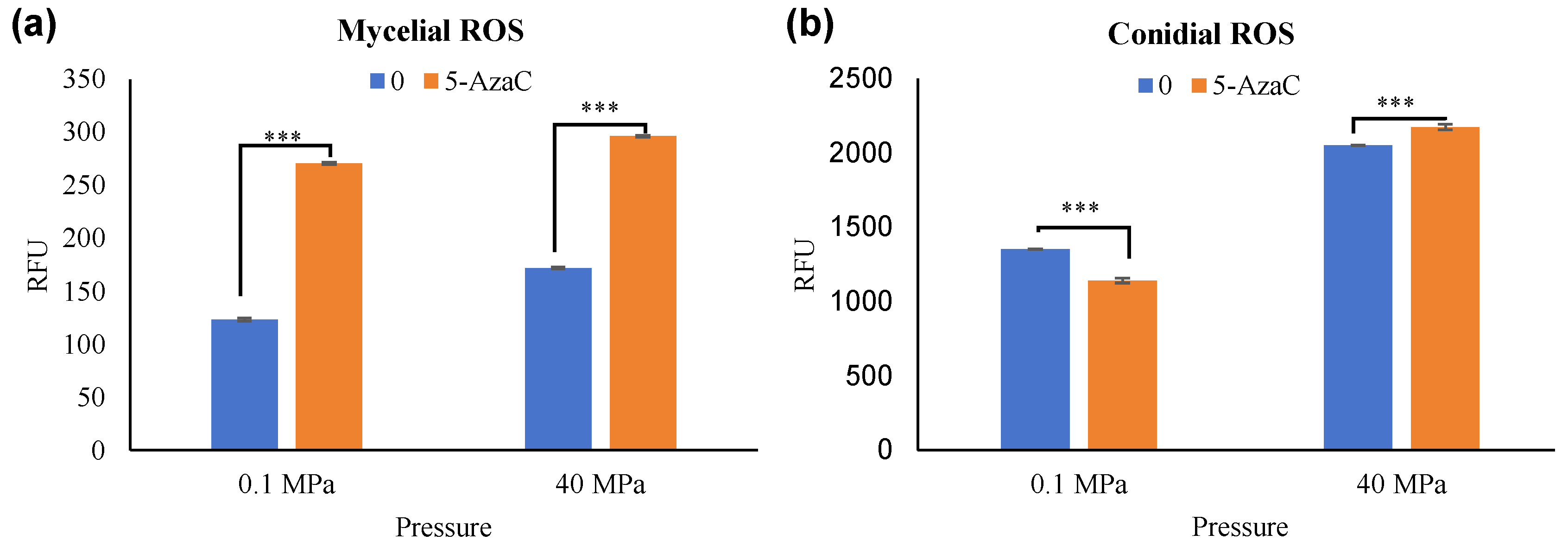
3.1. Epigenetic Responses of Hadal Fungi Under Different HHP Conditions
3.2. Impact of Epigenetic Modifiers and High Hydrostatic Pressure on Secondary Metabolite Profiles
3.3. Transcriptome Sequence Analysis and De Novo Assembly
3.4. Functional Annotation Analysis
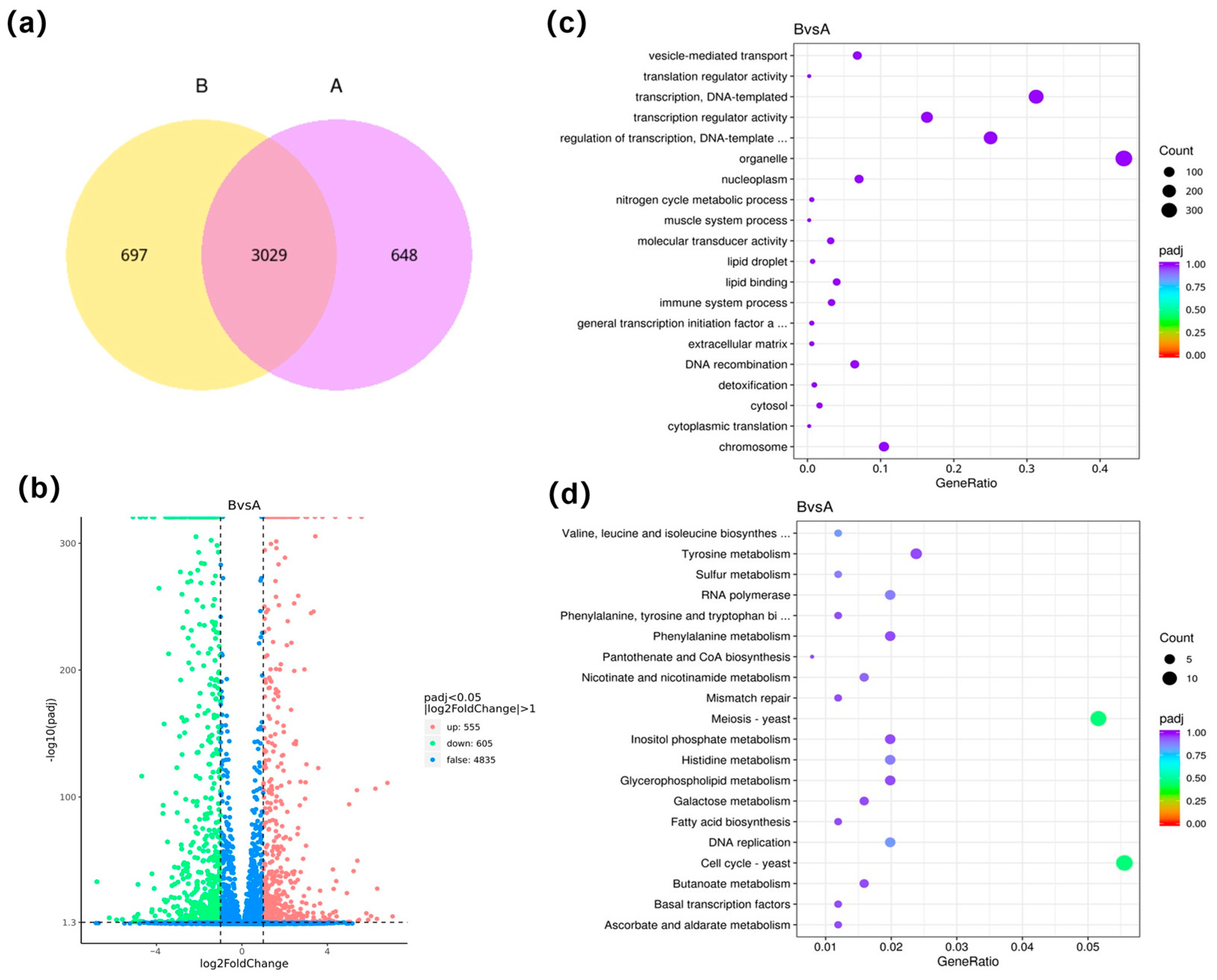
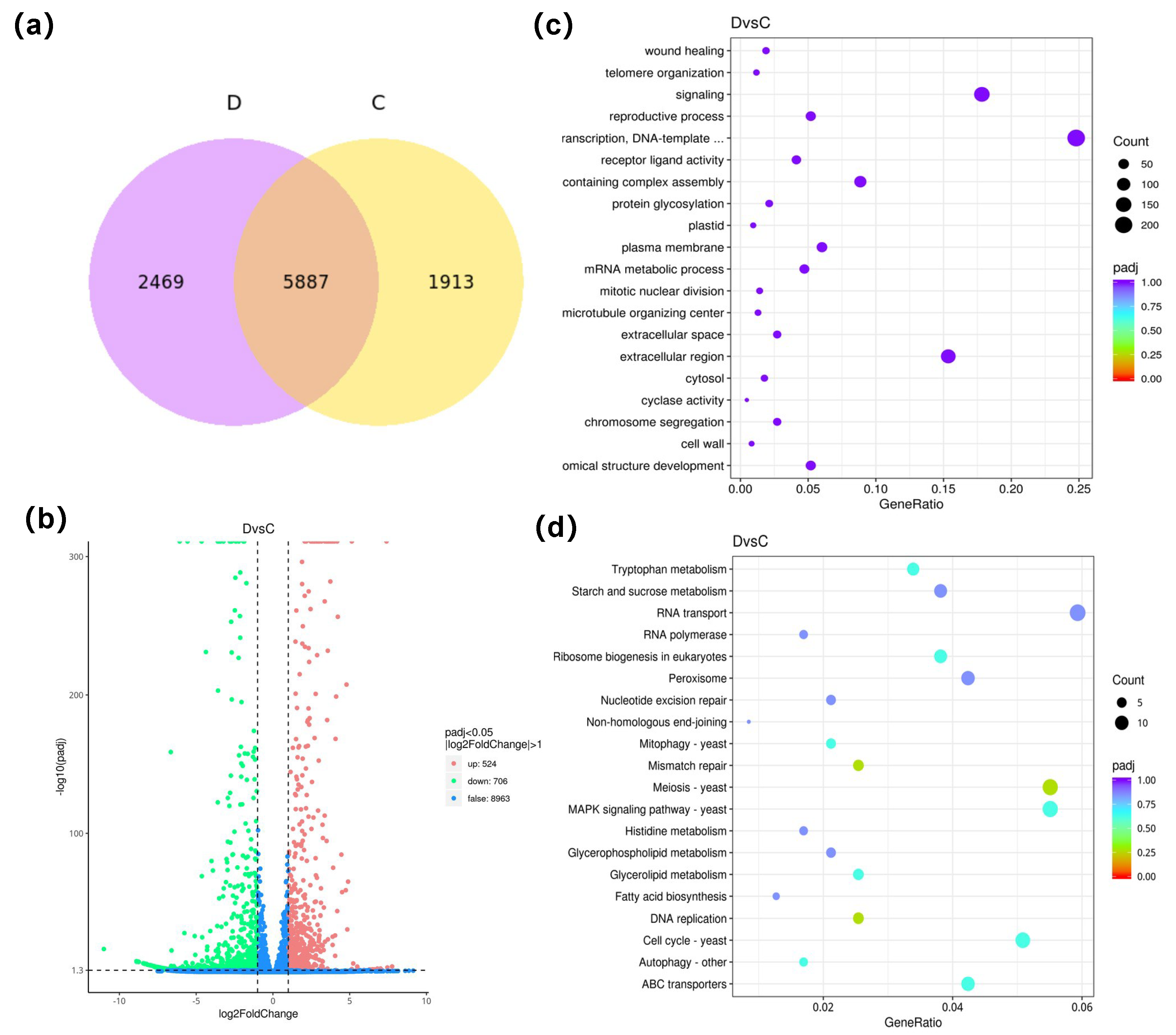
3.5. Analysis of Expression Differences and Enrichment
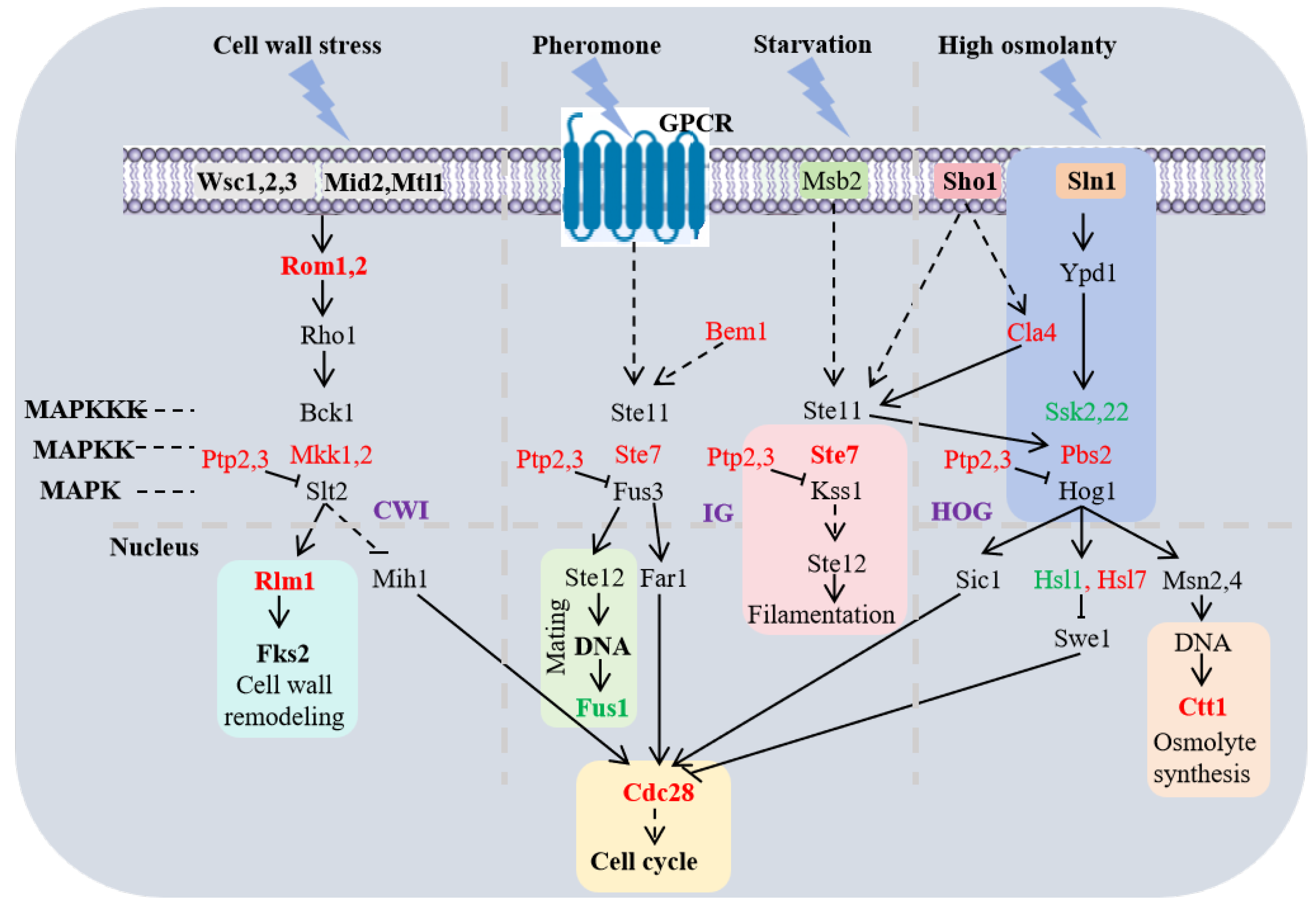
3.6. Epigenetic Modifier Regulated the MAPK Signaling Pathway in HHP
3.7. IG-MAPK Pathway Responsed to Epigenetic Modification Though Genes Involved in Filamentation, Mating and Cell Cycle
3.8. CWI-MAPK Pathway Responsed to Epigenetic Modification Though Genes Involved in Cell Wall Remdeling and Cell Cycle Under HHP Conditions
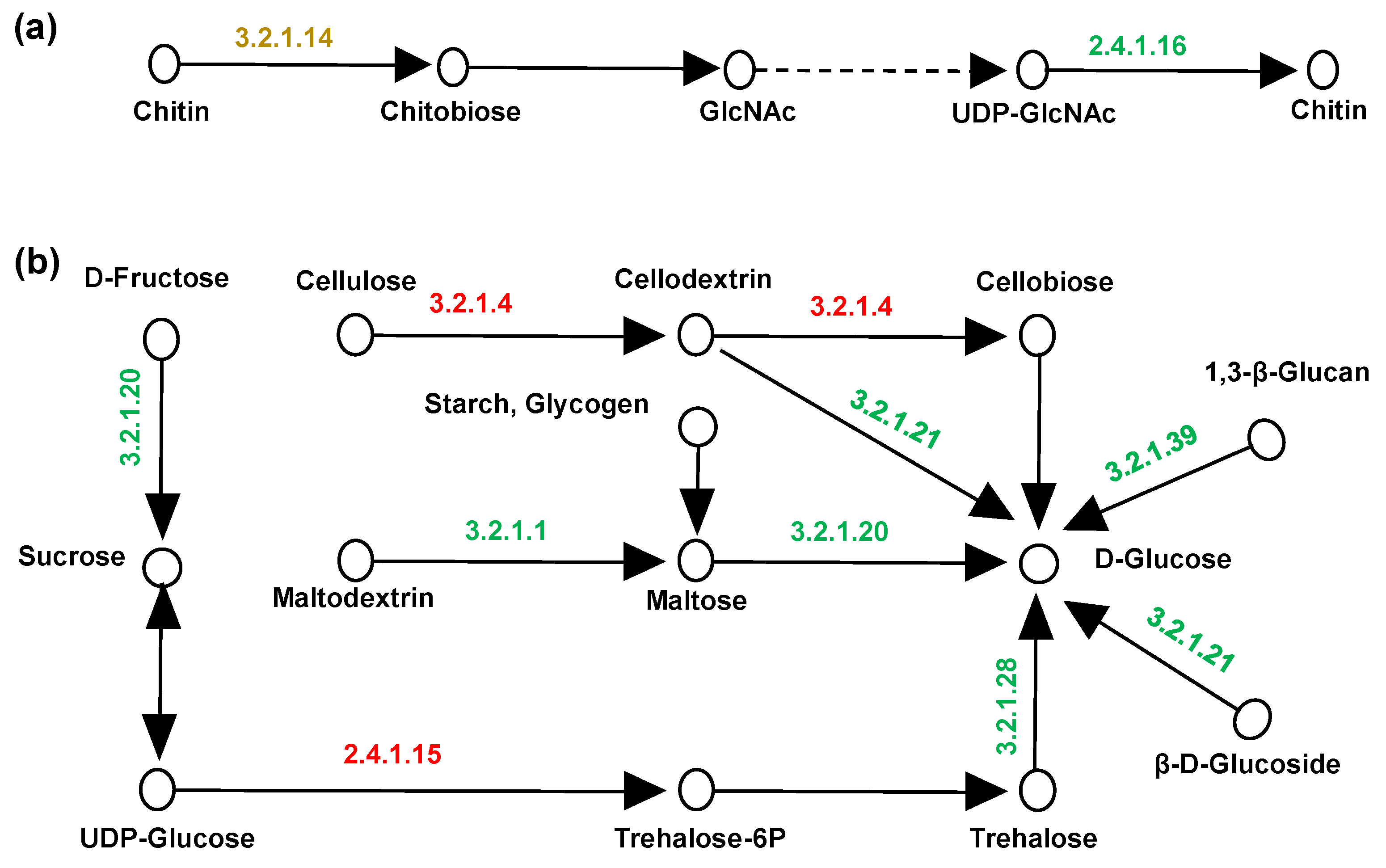
3.9. HOG-MAPK Pathway Responsed to Epigenetic Modification Though Osmolyte Synthesis and Cell Cycle Under HHP Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HHP | high hydrostatic pressure |
| PDA | potato dextrose agar |
| NCBI | National Centre for Biotechnology Information |
| 5-AzaC | 5-Azacytidine |
| PDB | potato dextrose broth |
| qRT-PCR | quantitative reverse transcriptase PCR |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| Cq | cyclic quantitative |
| MNE | mean normalized expression |
| SE | standard error |
| ANOVA | one-way analysis of variance |
| LSD | least significant difference |
| UPLC-MS/MS | ultra-performance liquid chromatography/tandem mass spectrometry |
| ESI | electrospray ionization |
| LB | Luria–Bertani |
| BNOZ | Beijing NOwo Zhiyuan |
| RNA-Seq | RNA sequencing |
| Nr | NCBI non-redundant protein sequences |
| Nt | NCBI nucleotide sequences |
| Pfam | Protein famil |
| KOG | EuKaryotic Ortholog Groups |
| Swiss-prot | Protein sequence database |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| DEGs | differentially expressed genes |
| PKS | polyketide synthase |
| IG | invasive growth |
| ROS | reactive oxidative stress |
| CWI | cell wall integrity |
| T6P | trehalose-6-phosphate |
| HOG | high-osmolarity glycerol |
| OSMAC | One Strain Many Compounds |
| DNMT | DNA methyltransferase |
References
- Etter, R.J.; Grassle, J.F. Patterns of species diversity in the deep sea as a function of sediment particle size diversity. Nature 1992, 360, 576–578. [Google Scholar] [CrossRef]
- Arístegui, J.; Gasol, J.M.; Duarte, C.M.; Herndld, G.J. Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 2009, 54, 1501–1529. [Google Scholar] [CrossRef]
- Debbab, A.; Aly, A.H.; Lin, W.H.; Proksch, P. Bioactive compounds from marine bacteria and fungi. Microb. Biotechnol. 2010, 3, 544. [Google Scholar] [CrossRef]
- Guo, C.; Yang, Z.; Chen, J.; Zhang, D. Ran, L. Wang, C. Lu, B. Cheng, Q. Study on food sources and trophic levels of benthic organisms in Yap Trench based on stable carbon and nitrogen isotopes. Acta Oceanol. Sin. 2018, 40, 51–60. [Google Scholar]
- Peng, Q.; Li, Y.; Deng, L.; Fang, J.; Yu, X. High hydrostatic pressure shapes the development and production of secondary metabolites of Mariana Trench sediment fungi. Sci. Rep. 2021, 11, 11436. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, H.; Zhu, W. New natural products from the marine-derived Aspergillus fungi-A review. Wei Sheng Wu Xue Bao Acta Microbiol. Sin. 2016, 56, 331–362. [Google Scholar]
- Holler, U.; Konig, G.M.; Wright, A.D. Three new metabolites from marine-derived fungi of the genera coniothyrium and microsphaeropsis. J. Nat. Prod. 1999, 62, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Wang, G.; Jin, S.; Zheng, T. Advances in Marine Microbial Diversity Research. Adv. Mar. Sci. 2004, 377–384. [Google Scholar] [CrossRef]
- Rosario, N.; Antonio, T. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin. Mar. Drugs 2016, 14, 37–41. [Google Scholar] [CrossRef]
- Ibrahim, S.R.; Abdallah, H.M.; Mohamed, G.A.; Deshmukh, S.K. Exploring potential of Aspergillus sclerotio rum: Secondary metabolites and biotechnological rel evance. Mycol. Prog. 2023, 22, 8. [Google Scholar] [CrossRef]
- Kamat, S.; Kumar, S.; Philip, S.; Kumari, M. Secondary metabolites from marine fungi: Current status and application. In Microbial Biomolecules; Academic Press: Cambridge, UK, 2023; pp. 181–209. [Google Scholar]
- Tiwari, P.; Bae, H. Endophytic fungi: Key insights, emerging prospects, and challenges in natural product drug discovery. Microorganisms 2022, 10, 360. [Google Scholar] [CrossRef]
- Li, X.; Xia, Z.; Tang, J.; Wu, J.; Tong, J.; Li, M.; Ju, J.; Chen, H.; Wang, L. Identification and Biological Evaluation of Secondary Metabolites from Marine Derived Fungi-Aspergillus sp. SCSIOW3, Cultivated in the Presence of Epigenetic Modifying Agents. Molecules 2017, 22, 1302. [Google Scholar] [CrossRef]
- VanderMolen, K.M.; Darveaux, B.A.; Chen, W.-L.; Swanson, S.M.; Pearce, C.J.; Oberlies, N.H. Epigenetic Manipulation of a Filamentous Fungus by the Proteasome-Inhibitor Bortezomib Induces the Production of an Additional Secondary Metabolite. RSC Adv. 2014, 4, 18329–18335. [Google Scholar] [CrossRef]
- Williams, R.B.; Henrikson, J.C.; Hoover, A.R.; Lee, A.E.; Cichewicz, R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 6, 1895–1897. [Google Scholar] [CrossRef]
- Peng, Q.; Li, Y.; Fang, J.; Yu, X. Effects of Epigenetic Modification and High Hydrostatic Pressure on Polyketide Synthase Genes and Secondary Metabolites of Alternaria alternata Derived from the Mariana Trench Sediments. Mar. Drugs 2023, 21, 585. [Google Scholar] [CrossRef]
- Rispail, N.; Soanes, D.M.; Ant, C.; Czajkowski, R.; Grünler, A.; Huguet, R.; Perez-Nadales, E.; Poli, A.; Sartorel, E.; Valiante, V.; et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 2009, 46, 287–298. [Google Scholar] [CrossRef]
- Khan, A.; Shah, S.T.; Basit, A.; Mohamed, H.I.; Li, Y. Mitogen-Activated Protein Kinase: A Potent Signaling Protein that Combats Biotic and Abiotic Stress in Plants. J. Plant Growth Regul. 2024, 43, 1762–1786. [Google Scholar] [CrossRef]
- Lin, L.; Wu, J.; Jiang, M.; Wang, Y. Plant mitogen-activated protein kinase cascades in environmental stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef]
- Raghukumar, C.; Muraleedharan, U.; Gaud, V.R.; Mishra, R. Xylanases of marine fungi of potential use for biobleaching of paper pulp. J. Ind. Microbiol. Biotechnol. 2004, 31, 433–441. [Google Scholar] [CrossRef]
- Li, Y.F.; Sun, T.T.; Guo, D.G.; Gao, J.; Zhang, J.A.; Cai, F.; Fischer, R.; Shen, Q.R.; Yu, Z.Z. Comprehensive analysis of the regulatory network of blue-light-regulated conidiation and hydrophobin production in Trichoderma guizhouense. Environ. Microbiol. 2021, 23, 6241–6256. [Google Scholar] [CrossRef]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A. Major Challenges and Perspectives in the Diagnostics and Treatment of Dermatophyte nfections. J. Appl. Microbiol. 2020, 129, 212–232. [Google Scholar] [CrossRef] [PubMed]
- Scott Chialvo, C.H.; Griffin, L.H.; Reed, L.K.; Ciesla, L. Exhaustive extraction of cyclopeptides from Amanita phalloides: Guidelines for working with complex mixtures of secondary metabolites. Nat. Ecol. Evol. 2020, 10, 4233–4240. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Osh lack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Peidro-Guzmán, H.; Pérez-Llano, Y.; González-Abradelo, D.; Fernández-López, M.G.; Dávila-Ramos, S.; Aranda, E.; Olicón-Hernández, D.; García, A.; Lira-Ruan, V.; Pliego, O.; et al. Transcriptomic analysis of polyaromatic hydrocarbon degradation by the halophilic fungus Aspergillus sydowii at hypersaline conditions. Environ. Microbiol. 2020, 23, 3435–3459. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Bartlett, D.H.; Xiao, X. Transcriptomic analysis reveals common adaptation mechanisms under different stresses for moderately piezophilic bacteria. Microb. Ecol. 2021, 81, 617–629. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef]
- He, P.; Wang, Y.; Wang, X.; Zhang, X.; Tian, C. The mitogen-activated protein kinase CgMK1 governs appressorium formation, melanin synthesis, and plant infection of Colletotrichum gloeosporioides. Front. Microbiol. 2017, 8, 2216. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, J. Stress and plant DNA methylation: Potential applications and challenges in breeding. Adv. Nat. Sci. 2009, 19, 248–256. [Google Scholar]
- Dichtl, K.; Samantaray, S.; Wagener, J. Cell wall integrity signalling in human pathogenic fungi. Cell. Microbiol. 2016, 18, 1228–1238. [Google Scholar] [CrossRef]
- Hohmann, S. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett. 2009, 583, 4025–4029. [Google Scholar] [CrossRef] [PubMed]
- Stojanovski, K.; Ferrar, T.; Benisty, H.; Uschner, F.; Delgado, J.; Jimenez, J.; Solé, C.; de Nadal, E.; Klipp, E.; Posas, F.; et al. Interaction dynamics determine signaling and output pathway responses. Cell Rep. 2017, 19, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Yamamoto, K.; Oyama, M.; Kozuka-Hata, H.; Saito, H.; Tatebayashi, K. Scaffold protein Ahk1, which associates with Hkr1, Sho1, Ste11, and Pbs2, inhibits cross talk signaling from the Hkr1 osmosensor to the Kss1 mitogen-activated protein kinase. Mol. Cell Biol. 2016, 36, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Jamalzadeh, S.; Pujari, A.N.; Cullen, P.J. A Rab escort protein regulates the MAPK pathway that controls filamentous growth in yeast. Sci. Rep. 2020, 10, 22184. [Google Scholar] [CrossRef]
- Zhong, M.; Li, Y.; Deng, L.; Fang, J.; Yu, X. Insight into the adaptation mechanisms of high hydrostatic pressure in physiology and metabolism of hadal fungi from the deepest ocean sediment. mSystems 2023, 9, e0108523. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]
- Begani, J.; Lakhani, J.; Harwani, D. Current strategies to induce secondary metabolites from microbial biosynthetic cryptic gene clusters. Ann. Microbiol. 2018, 68, 419–432. [Google Scholar] [CrossRef]
- Nielsen, K.F.; Larsen, T.O. The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front. Microbiol. 2015, 6, 71. [Google Scholar] [CrossRef]
- Toghueo, R.; Sahal, D.; Boyom, F.F. Recent advances in inducing endophytic fungal specialized metabolites using small molecule elicitors including epigenetic modififiers. Phytochemistry 2020, 174, 112338. [Google Scholar] [CrossRef]
- Pocas-Fonseca, M.J.; Cabral, C.G.; Manfrao-Netto, J.H.C. Epigenetic manipulation of filamentous fungi for biotechnological applications: A systematic review. Biotechnol. Lett. 2020, 42, 885–904. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latgé, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef]
- Pontón, J. The fungal cell wall and the mechanism of action of anidulafungin. Rev. Iberoam. Micol. 2008, 25, 78–82. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, cryptococcus, and Aspergillus species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef]
- Mouriño-Pérez, R.R. Septum development in filamentous ascomycetes. Fungal. Biol. Rev. 2013, 27, 1–9. [Google Scholar] [CrossRef]
- Wang, F.; Xiao, X.; Ou, H.-Y.; Gai, Y.; Wang, F. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J. Bacteriol. 2009, 191, 2574–2584. [Google Scholar] [CrossRef]
- Peter, J. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nature reviews. Genetics 2012, 13, 484–492. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Q.; Wei, Q.; Yu, X. Harnessing Epigenetic Modifiers Reveals MAPK-Mediated Regulation Mechanisms in Hadal Fungi of Alternaria alternata Under High Hydrostatic Pressure. J. Fungi 2025, 11, 650. https://doi.org/10.3390/jof11090650
Peng Q, Wei Q, Yu X. Harnessing Epigenetic Modifiers Reveals MAPK-Mediated Regulation Mechanisms in Hadal Fungi of Alternaria alternata Under High Hydrostatic Pressure. Journal of Fungi. 2025; 11(9):650. https://doi.org/10.3390/jof11090650
Chicago/Turabian StylePeng, Qingqing, Qifei Wei, and Xi Yu. 2025. "Harnessing Epigenetic Modifiers Reveals MAPK-Mediated Regulation Mechanisms in Hadal Fungi of Alternaria alternata Under High Hydrostatic Pressure" Journal of Fungi 11, no. 9: 650. https://doi.org/10.3390/jof11090650
APA StylePeng, Q., Wei, Q., & Yu, X. (2025). Harnessing Epigenetic Modifiers Reveals MAPK-Mediated Regulation Mechanisms in Hadal Fungi of Alternaria alternata Under High Hydrostatic Pressure. Journal of Fungi, 11(9), 650. https://doi.org/10.3390/jof11090650




