The First Whole Genome Sequence and Methylation Profile of Gerronema lapidescens QL01
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Material and Nucleic Acid Extraction
2.2. Genome Assembly and Chromosome Assembly
2.3. Genome Annotation
2.4. SNP Site Detection
2.5. Reconstruction of the Phylogenetic Tree
2.6. Gene Family Expansion and Contraction Analysis
2.7. BGC Analysis and Visualisation
2.8. Partial Core Gene Methylation Prediction
2.9. Data Availability
3. Results
3.1. Collection and Domestication Cultivation of Wild Macrofungi Resources in the Qinling Mountains
3.2. Genome Sequencing, De Novo Assembly, and Annotation
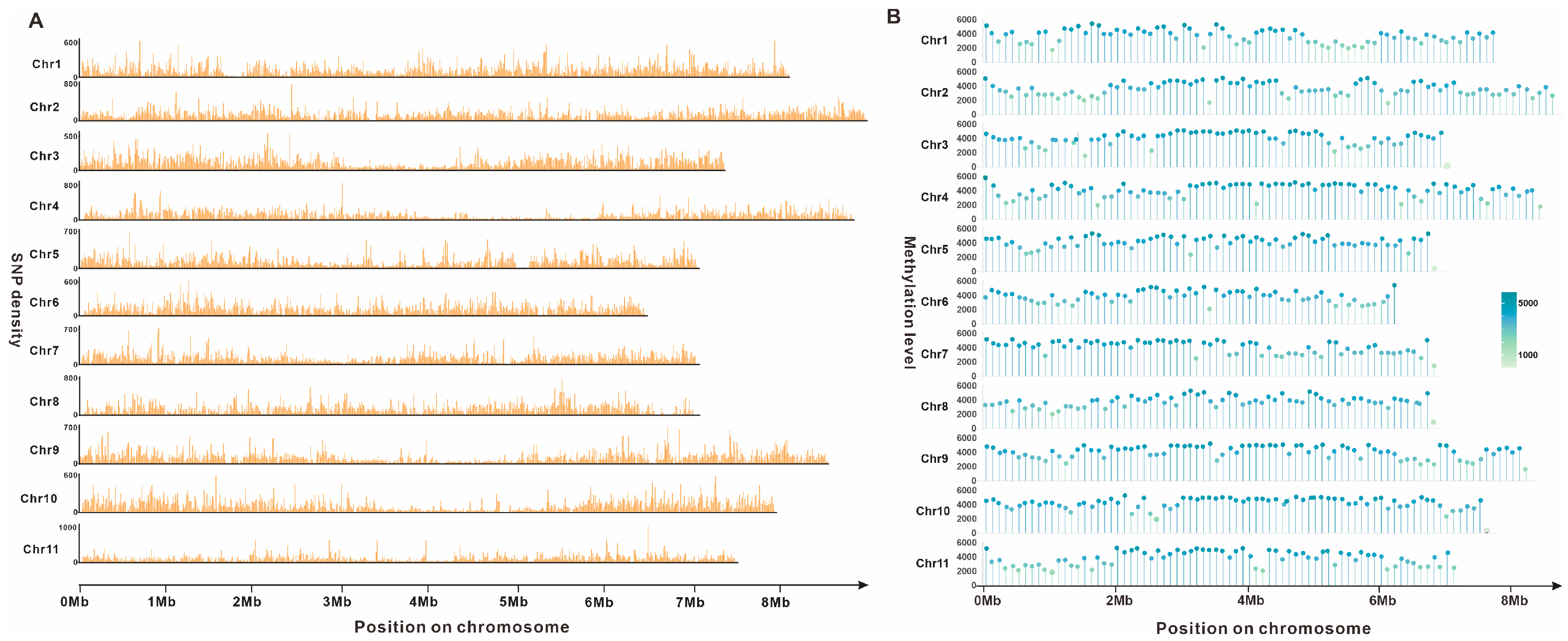
3.3. SNP and Methylation Analysis of Gerronema lapidescens QL01
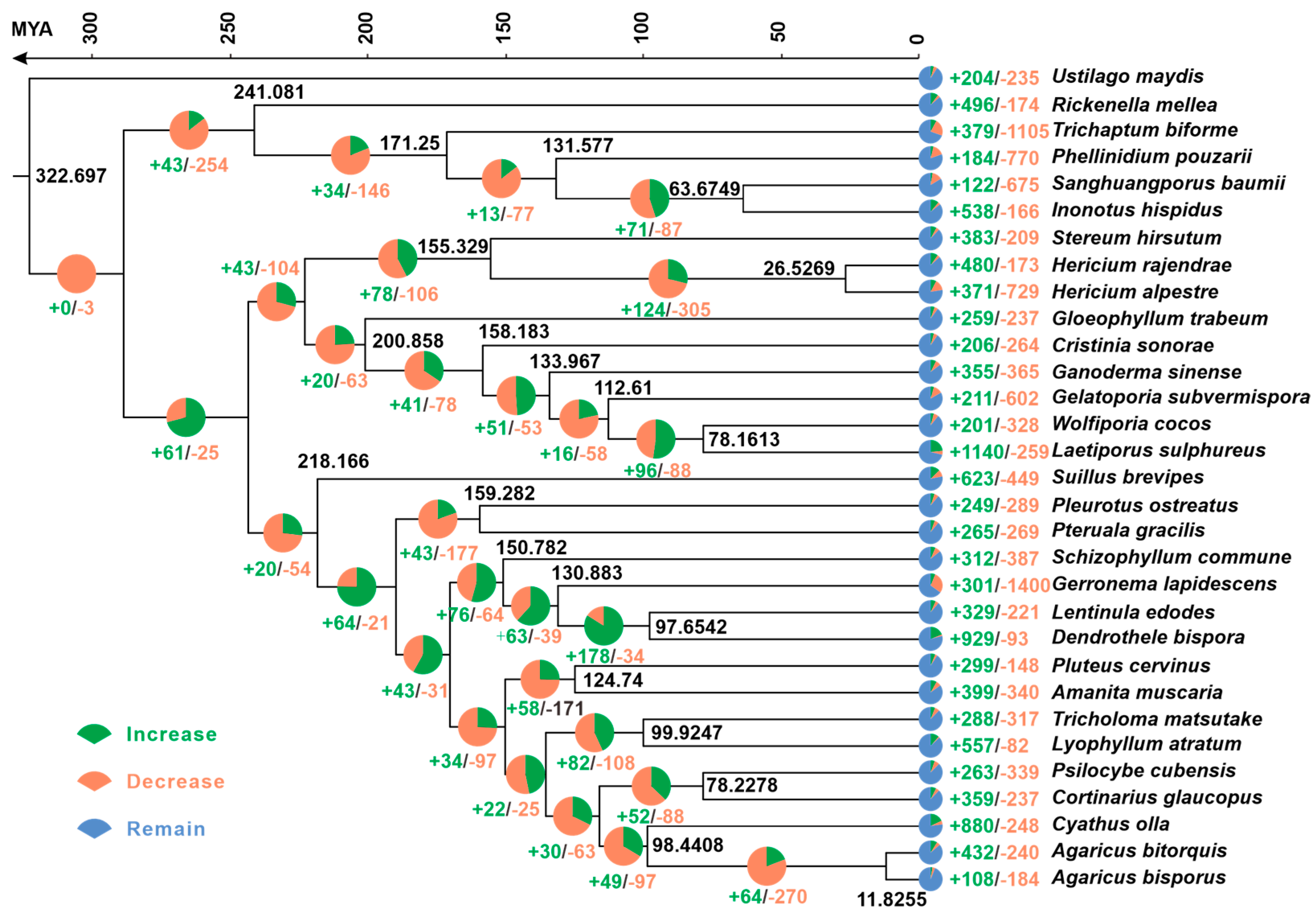
3.4. Phylogenetic and Gene Family Variation Analysis
3.5. CAZyme Analysis and Developing SSR Markers
3.6. Search and Analysis of Secondary Metabolite-Related Genes
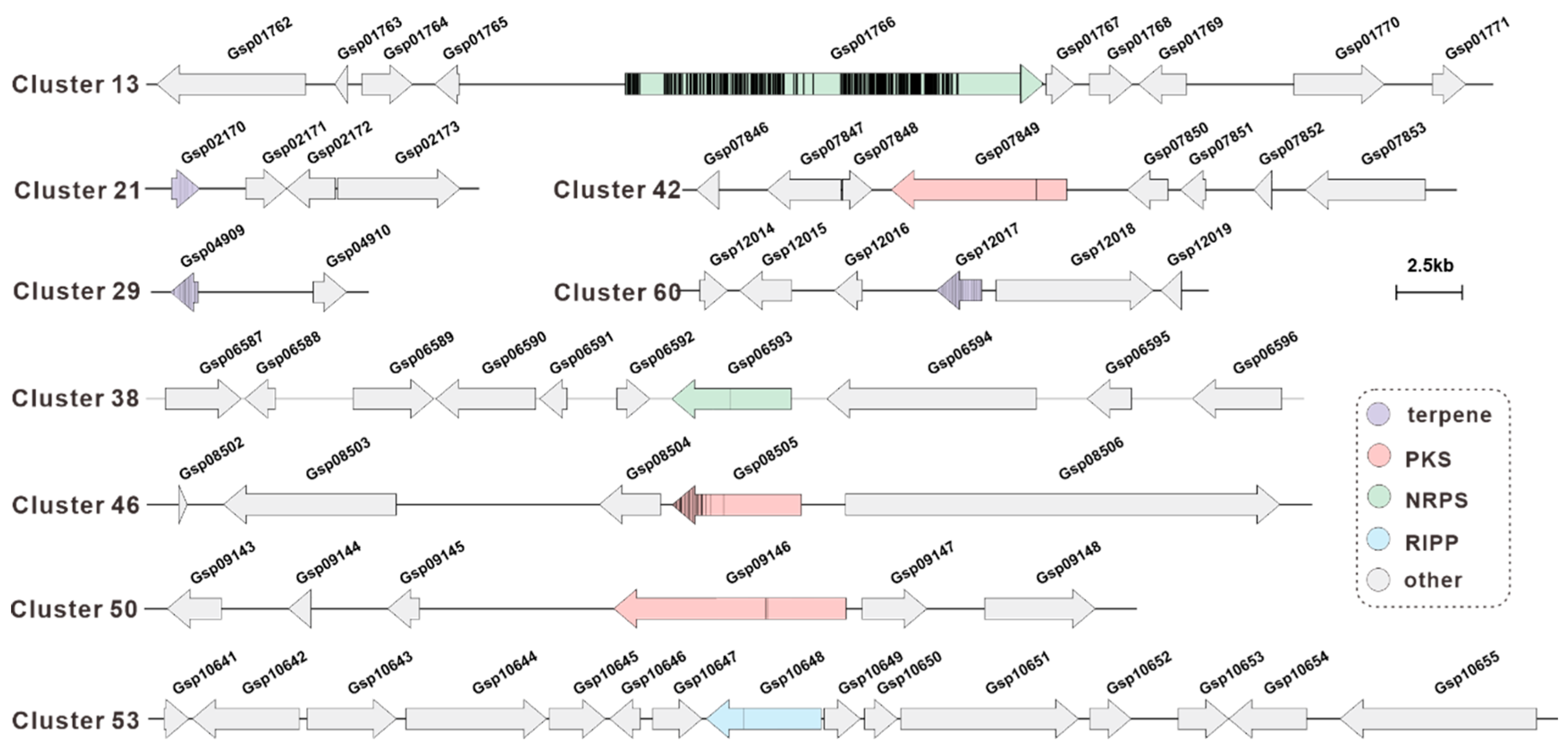
3.7. Methylation Analysis of the BGCs for Secondary Metabolite
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Wang, C.; Deng, W.; Chang, C.; Zhang, M. A Taxonomic Revision of Rare Medicinal Fungus Leiwan. Acta Edulis Fungi 2024, 31, 90–99. [Google Scholar] [CrossRef]
- Li, N.; Zhuo, W.; Yu, Z.; Zhao, X.; Wang, H.; Lu, S.; Cao, W. Developmental characteristics and domestication of Gerronema lapidescens. Mycosystema 2025, 44, 84–96. [Google Scholar] [CrossRef]
- Mueller, G.M.; Wu, Q.; Field Museum of Natural History. Mycological Contributions of Rolf Singer: Field Itinerary, Index to New Taxa, and List of Publications; Field Museum of Natural History: Chicago, IL, USA, 1997. [Google Scholar]
- Wei, G. A review of the clinical applications of Leiwan in ancient and modern times. Liaoning J. Tradit. Chin. Med. 2014, 41, 1866–1867. [Google Scholar] [CrossRef]
- Li, J.W.; Li, Z.J. Compendium of Materia Medica Annotated; Liaohai Publishing House: Shenyang, China, 2001. [Google Scholar]
- China Pharmacopoeia Committee. Pharmacopoeia of China: Dai Medicinal Herbs Volume; Shanghai Scientific and Technical Publishers: Shanghai, China, 2005. [Google Scholar]
- Yu, Y.; Gong, J.; Yu, M. Studies on the purification and properties of Omphalia lapidescens lectin. Syst. Mycol. 2000, 19, 278–282. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.; Yang, H.; Yang, R.; Xiao, X. Research progress in the chemical composition, pharmacological effects, and quality control of Omphalia lapidescens. Chin. J. New Drugs 2024, 33, 1002–1008. [Google Scholar]
- Silberborth, S.; Stumpf, A.; Erkel, G.; Anke, T.; Sterner, O. Gerronemins A-F, cytotoxic biscatechols from a Gerronema species. Phytochemistry 2002, 59, 643–648. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Wei, Z.Q.; Hu, Z.K. The Impact of the training materials with different percentage of carbon and nitrogen to the mycelia growthin cultivated species of Omphaolia lapidescens Schroet. In Proceedings of the Fourth International Conference on Traditional Chinese Medicine Modernization, Chengdu, China, 26–27 September 2013; pp. 1–2. [Google Scholar]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Chen, Y.; Nie, F.; Xie, S.Q.; Zheng, Y.F.; Dai, Q.; Bray, T.; Wang, Y.X.; Xing, J.F.; Huang, Z.J.; Wang, D.P.; et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021, 12, 60. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Li, H. compleasm: A faster and more accurate reimplementation of BUSCO. Bioinformatics 2023, 39, btad595. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Zhao, Q.; Ming, R.; Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 2019, 5, 833–845. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Nachtweide, S.; Stanke, M. Multi-Genome Annotation with AUGUSTUS. Methods Mol. Biol. 2019, 1962, 139–160. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wood, W.I. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics 2003, 19, 307–308. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021, 50, D571–D577. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Heldenbrand, J.R.; Baheti, S.; Bockol, M.; Drucker, T.M.; Hart, S.N.; Hudson, M.E.; Iyer, R.K.; Kalmbach, M.T.; Klee, E.W.; Wieben, E.D.; et al. Performance benchmarking of GATK3.8 and GATK4. bioRxiv 2018. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef]
- Mendes, F.K.; Vanderpool, D.; Fulton, B.; Hahn, M.W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 2020, 36, 5516–5518. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.H. Synthaser: A CD-Search enabled Python toolkit for analysing domain architecture of fungal secondary metabolite megasynth(et)ases. Fungal Biol. Biotechnol. 2021, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Czajka, K.; Mehes-Smith, M.; Nkongolo, K. DNA methylation and histone modifications induced by abiotic stressors in plants. Genes Genom. 2022, 44, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, Y.-C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef]
- He, X.L.; Huo, W.Y.; Zhang, L.G.; Dai, L.; Qi, P.; Liu, Y.; Li, J.Z. A New Species of Hygrophoropsis (Boletales, Basidiomycota) from Qinling Mountains in China. J. Fungal Res. 2024, 22, 135–141. [Google Scholar] [CrossRef]
- He, X.L.; Huo, W.Y.; Zhang, L.G.; Dai, L.; Liu, Y.; Li, J.Z. Two New Species of Helvella (Pezizales, Ascomycota) Collected from the Qinling Mountains of China. J. Fungal Res. 2024, 22, 226–235. [Google Scholar] [CrossRef]
- Dong, W.G.; Wang, Z.X.; Feng, X.L.; Zhang, R.Q.; Shen, D.Y.; Du, S.; Gao, J.M.; Qi, J. Chromosome-Level Genome Sequences, Comparative Genomic Analyses, and Secondary-Metabolite Biosynthesis Evaluation of the Medicinal Edible Mushroom Laetiporus sulphureus. Microbiol. Spectr. 2022, 10, e0243922. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, M.; Liu, Y.Y.; Li, M.; Xie, X.; Qi, J. Haplotype-Phased Chromosome-Level Genome Assembly of Cryptoporus qinlingensis, a Typical Traditional Chinese Medicine Fungus. J. Fungi 2025, 11, 163. [Google Scholar] [CrossRef]
- Xie, X.; Zhao, L.; Song, Y.; Qiao, Y.; Wang, Z.X.; Qi, J. Genome-wide characterization and metabolite profiling of Cyathus olla: Insights into the biosynthesis of medicinal compounds. BMC Genom. 2024, 25, 618. [Google Scholar] [CrossRef]
- Wei, J.; Cheng, M.; Zhu, J.F.; Zhang, Y.; Cui, K.; Wang, X.; Qi, J. Comparative Genomic Analysis and Metabolic Potential Profiling of a Novel Culinary-Medicinal Mushroom, Hericium rajendrae (Basidiomycota). J. Fungi 2023, 9, 1018. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, H.; Wang, Y.; Liu, D.; Ma, A. Research Progress in the Biological Activities of Macrofungal Sclerotia. Sci. Technol. Food Ind. 2018, 39, 328–332. [Google Scholar] [CrossRef]
- China Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Zhao, K.; Xu, J. Genome-wide comparisons reveal broad variations in intraspecific SNP frequencies among species in Agaricomycetes, Basidiomycota. F1000Research 2023, 12, 200. [Google Scholar] [CrossRef]
- Zeng, Z.; Wu, J.; Kovalchuk, A.; Raffaello, T.; Wen, Z.; Liu, M.; Asiegbu, F.O. Genome-wide DNA methylation and transcriptomic profiles in the lifestyle strategies and asexual development of the forest fungal pathogen Heterobasidion parviporum. Epigenetics 2019, 14, 16–40. [Google Scholar] [CrossRef]
- Yuan, W.; Yuan, W.; Zhou, R.; Lv, G.; Sun, M.; Zhao, Y.; Zheng, W. Production of hispidin polyphenols from medicinal mushroom Sanghuangporus vaninii in submerged cultures. Chin. Herb. Med. 2023, 15, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Qi, J.; Lin, C.; Xu, Z.; Li, Z.; Liu, C. From natural laboratory to drug discovery: Chemical structures, bioactivities, and biosynthesis of meroterpenoids from Ganoderma species. Chin. Herb. Med. 2025. [Google Scholar] [CrossRef]
- Li, Y.; Lou, S.; Yang, R.; Zhang, L.; Zou, Q.; Shang, S.; Gao, L.; Wang, W. Cytotoxic sesquiterpene aryl esters from Armillaria gallica 012m. Chin. Herb. Med. 2023, 15, 343–346. [Google Scholar] [CrossRef]
- Wang, Z.-X.; Huang, K.; Pu, K.-L.; Li, L.; Jiang, W.-X.; Wu, J.; Kawagishi, H.; Li, M.; Qi, J. Naematelia aurantialba: A comprehensive review of its medicinal, nutritional, and cultivation aspects. Food Med. Homol. 2025, 2, 9420072. [Google Scholar] [CrossRef]
- Zhang, G.-P.; Pan, Y.-M.; Ye, S.-M.; Lu, Y.-C.; Fan, X.-J.; Zhang, A.-Q. Bioactive components of Ganoderma lucidum and their efficacy and application in cosmetics. Food Med. Homol. 2025, 2, 9420044. [Google Scholar] [CrossRef]


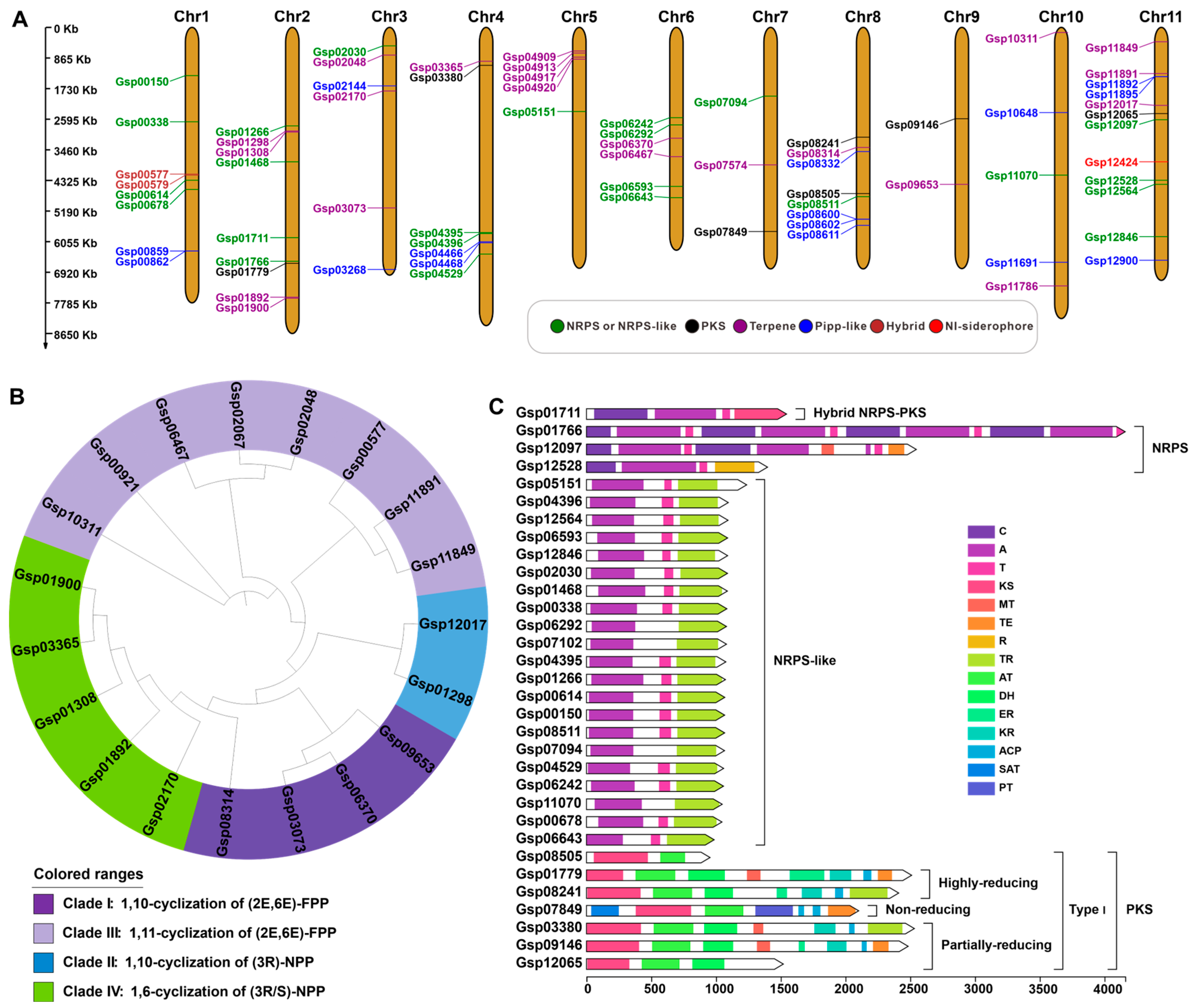
| Cluster. No | Location | Core Gene ID | Core Gene Type | ||
|---|---|---|---|---|---|
| Chromosome | Start (bp) | End (bp) | |||
| 1 | Chr1 | 1,333,163 | 1,362,998 | geneGsp00150 | NRPS-like |
| 2 | Chr1 | 2,660,560 | 2,701,903 | geneGsp00338 | NRPS-like |
| 3 * | Chr1 | 4,134,889 | 4,192,917 | geneGsp00577 | Terpene |
| geneGsp00579 | Ripp-like | ||||
| 4 | Chr1 | 4,285,042 | 4,327,761 | geneGsp00614 | NRPS-like |
| 5 | Chr1 | 4,548,301 | 4,587,172 | geneGsp00678 | NRPS-like |
| 6 * | Chr1 | 6,317,779 | 6,318,426 | geneGsp00859 | Ripp-like |
| geneGsp00862 | Ripp-like | ||||
| 7 | Chr1 | 6,967,152 | 6,971,998 | geneGsp00921 | Terpene |
| 8 | Chr2 | 2,763,751 | 2,792,130 | geneGsp01266 | Terpene |
| 9 | Chr2 | 2,908,572 | 2,926,762 | geneGsp01298 | Terpene |
| 10 | Chr2 | 2,936,974 | 2,952,672 | geneGsp01308 | Terpene |
| 11 | Chr2 | 3,789,149 | 3,818,583 | geneGsp01468 | NRPS-like |
| 12 | Chr2 | 5,934,048 | 5,954,781 | geneGsp01711 | NRPS |
| 13 | Chr2 | 6,591,072 | 6,638,658 | geneGsp01766 | NRPS |
| 14 | Chr2 | 6,650,704 | 6,697,700 | geneGsp01779 | T1PKS |
| 15 | Chr2 | 7,615,453 | 7,622,734 | geneGsp01892 | Terpene |
| 16 | Chr2 | 7,650,053 | 7,662,432 | geneGsp01900 | Terpene |
| 17 | Chr3 | 505,392 | 528,870 | geneGsp02030 | NRPS-like |
| 18 | Chr3 | 760,222 | 770,517 | geneGsp02048 | Terpene |
| 19 | Chr3 | 1,026,023 | 1,044,018 | geneGsp02067 | Terpene |
| 20 * | Chr3 | 1,645,178 | 1,724,472 | geneGsp02144 | Ripp-like |
| geneGsp02153 | Ripp-like | ||||
| 21 | Chr3 | 1,803,842 | 1,814,061 | geneGsp02170 | Terpene |
| 22 | Chr3 | 5,087,409 | 5,105,567 | geneGsp03073 | Terpene |
| 23 | Chr3 | 6,804,933 | 6,858,659 | geneGsp03268 | Ripp-like |
| 24 | Chr4 | 954,860 | 965,271 | geneGsp03365 | Terpene |
| 25 | Chr4 | 1,064,339 | 1,106,327 | geneGsp03380 | T1PKS |
| 26 * | Chr4 | 5,766,937 | 5,836,976 | geneGsp04395 | NRPS-like |
| geneGsp04396 | NRPS-like | ||||
| 27 * | Chr4 | 6,042,896 | 6,113,623 | geneGsp04466 | Ripp-like |
| geneGsp04468 | Ripp-like | ||||
| 28 | Chr4 | 6,394,666 | 6,423,026 | geneGsp04529 | NRPS-like |
| 29 | Chr5 | 674,185 | 680,885 | geneGsp04909 | Terpene |
| 30 | Chr5 | 721,838 | 723,100 | geneGsp04913 | Terpene |
| 31 | Chr5 | 826,981 | 832,702 | geneGsp04917 | Terpene |
| 32 | Chr5 | 897,400 | 908,671 | geneGsp04920 | Terpene |
| 33 | Chr5 | 2,345,287 | 2,387,459 | geneGsp05151 | NRPS-like |
| 34 | Chr6 | 2,525,938 | 2,563,354 | geneGsp06242 | NRPS-like |
| 35 | Chr6 | 2,721,335 | 2,761,718 | geneGsp06292 | NRPS-like |
| 36 | Chr6 | 3,126,334 | 3,144,062 | geneGsp06370 | Terpene |
| 37 | Chr6 | 3,648,047 | 3,664,942 | geneGsp06467 | Terpene |
| 38 | Chr6 | 4,484,271 | 4,526,656 | geneGsp06593 | NRPS-like |
| 39 | Chr6 | 4,802,482 | 4,842,126 | geneGsp06643 | NRPS-like |
| 40 * | Chr7 | 1,909,694 | 1,988,097 | geneGsp07094 | NRPS-like |
| geneGsp07102 | NRPS-like | ||||
| 41 | Chr7 | 3,890,379 | 3,906,718 | geneGsp07574 | Terpene |
| 42 | Chr7 | 5,762,576 | 5,790,301 | geneGsp07849 | T1PKS |
| 43 | Chr8 | 3,083,461 | 3,128,991 | geneGsp08241 | T1PKS |
| 44 | Chr8 | 3,391,092 | 3,400,084 | geneGsp08314 | Terpene |
| 45 | Chr8 | 3,505,329 | 3,516,333 | geneGsp08332 | Ripp-like |
| 46 | Chr8 | 4,667,825 | 4,708,615 | geneGsp08505 | T1PKS |
| 47 | Chr8 | 4,749,215 | 4,786,027 | geneGsp08511 | NRPS-like |
| 48 * | Chr8 | 5,397,733 | 5,458,632 | geneGsp08600 | Ripp-like |
| geneGsp08602 | Ripp-like | ||||
| 49 * | Chr8 | 5,581,739 | 5,639,225 | geneGsp08611 | Ripp-like |
| geneGsp08613 | Ripp-like | ||||
| 50 | Chr9 | 2,552,571 | 2,584,816 | geneGsp09146 | T1PKS |
| 51 | Chr9 | 4,433,774 | 4,453,262 | geneGsp09653 | Terpene |
| 52 | Chr10 | 122,324 | 141,261 | geneGsp10311 | Terpene |
| 53 | Chr10 | 2,391,981 | 2,444,186 | geneGsp10648 | Ripp-like |
| 54 | Chr10 | 4,164,792 | 4,201,924 | geneGsp11070 | NRPS-like |
| 55 | Chr10 | 6,619,032 | 6,669,729 | geneGsp11691 | Ripp-like |
| 56 | Chr10 | 7,300,583 | 7,307,881 | geneGsp11786 | Terpene |
| 57 | Chr11 | 396,663 | 397,903 | geneGsp11849 | Terpene |
| 58 | Chr11 | 1,307,102 | 1,308,342 | geneGsp11891 | Terpene |
| 59 * | Chr11 | 1,386,710 | 1,394,778 | geneGsp11892 | Ripp-like |
| geneGsp11895 | Ripp-like | ||||
| 60 | Chr11 | 2,201,870 | 2,220,088 | geneGsp12017 | Terpene |
| 61 | Chr11 | 2,403,505 | 2,440,506 | geneGsp12065 | T1PKS |
| 62 | Chr11 | 2,594,189 | 2,636,978 | geneGsp12097 | NRPS |
| 63 | Chr11 | 3,787,596 | 3,815,670 | geneGsp12424 | NI-siderophore |
| 64 | Chr11 | 4,301,554 | 4,334,404 | geneGsp12528 | NRPS-like |
| 65 | Chr11 | 4,411,006 | 4,449,199 | geneGsp12564 | NRPS-like |
| 66 | Chr11 | 5,908,891 | 5,949,936 | geneGsp12846 | NRPS-like |
| 67 | Chr11 | 6,551,530 | 6,585,327 | geneGsp12900 | Ripp-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, Y.; Jia, Z.; Liu, Y.; Zhang, N.; Luo, C.; Meng, L.; Cheng, Y.; Li, M.; Xie, X.; Qi, J. The First Whole Genome Sequence and Methylation Profile of Gerronema lapidescens QL01. J. Fungi 2025, 11, 647. https://doi.org/10.3390/jof11090647
Qiao Y, Jia Z, Liu Y, Zhang N, Luo C, Meng L, Cheng Y, Li M, Xie X, Qi J. The First Whole Genome Sequence and Methylation Profile of Gerronema lapidescens QL01. Journal of Fungi. 2025; 11(9):647. https://doi.org/10.3390/jof11090647
Chicago/Turabian StyleQiao, Yanming, Zhiyuan Jia, Yuying Liu, Na Zhang, Chun Luo, Lina Meng, Yajie Cheng, Minglei Li, Xiuchao Xie, and Jianzhao Qi. 2025. "The First Whole Genome Sequence and Methylation Profile of Gerronema lapidescens QL01" Journal of Fungi 11, no. 9: 647. https://doi.org/10.3390/jof11090647
APA StyleQiao, Y., Jia, Z., Liu, Y., Zhang, N., Luo, C., Meng, L., Cheng, Y., Li, M., Xie, X., & Qi, J. (2025). The First Whole Genome Sequence and Methylation Profile of Gerronema lapidescens QL01. Journal of Fungi, 11(9), 647. https://doi.org/10.3390/jof11090647







