3.3. Taxonomy
Two new species and a new record are described in this study.
Leptobacillium Zare & W. Gams, Mycol. Progr. 15: 1001 (2016).
Systematic position: Fungi, Dikarya, Ascomycota, Pezizomycotina, Sordariomycetes, Hypocreomycetidae, Hypocreales, Cordycipitaceae
Type species: Leptobacillium leptobactrum (W. Gams) Zare & W. Gams, Mycol. Progr. 15: 1003 (2016).
Table 2 lists all hosts, substrates, and geographical locations of
Leptobacillium species.
Table 3 reveals differences with asexual morphs, including the conidial shapes and conidiogenous structures of known species in
Leptobacillium.
Table 2.
Leptobacillium hosts, substrates, and geographical location.
Table 2.
Leptobacillium hosts, substrates, and geographical location.
| No. | Species | Host/Substrate | Countries Found | References |
|---|
| 1 | L. cavernicola | Air | France | [20] |
| 2 | L. chinense | Wood submerged in freshwater, environmental microorganisms | China | [18] |
| 3 | L. coffeanum | Coffea arabica (branches), Ophiocordyceps nutans, soil | China, Brazil | [21,64] |
| 4 | L. filiforme | Endophyte from Citrullus lanatus, Thozetella pindobacuensis | Brazil | [37] |
| 5 | L. gasaense | Gibellula sp. | China | This study |
| 6 | L. latisporum | Soil | Thailand | [23] |
| 7 | L. leptobactrum | Decaying wood, Lactarius rufus, Phlebia tremellosa, human nail, sandy soil, unknown ascomycete | Poland, France, Iran, Netherlands | [6] |
| 8 | L. leptobactrum var. calidius | Living lepidopterous larva, cyst of Heterodera glycines, Hemileia vastatrix on Coffea | Ghana, USA, Brazil, France, Netherlands | [6] |
| 9 | L. leptobactrum var. leptobactrum | Lepidoptera larva | Ghana, Poland, France, Iran, Netherlands | [6] |
| 10 | L. muralicola | On acrylic varnish coatings of murals | China | [22] |
| 11 | L. symbioticum | From sori of soybean rust fungus | Japan | [19] |
| 12 | L. xianyushanense | Camellia oleifera rhizosphere soil | China | [24] |
Table 3.
Morphological comparisons of asexual morphs in Leptobacillium.
Table 3.
Morphological comparisons of asexual morphs in Leptobacillium.
| No. | Species | Colony | Phialides (µm) | Conidia (µm) | References |
|---|
| 1 | L. cavernicola | White, reverse dark brown | Solitary, 5.1–27.2 × 1.2–1.7 | Narrowly cylindrical to slightly fusiform, 3.1–6.9 × 0.9–1.5 | [20] |
| 2 | L. chinense | White, reverse cream-colored to light yellow | Solitary, (6.0–) 15–30 (–68.0) × 1.5 | Mostly oval, ellipsoidal or cylindrical, 3.5–5.0 × 1.0–1.5; the apical conidia of the conidial chains subglobose to obovoid, 1.5–2.5 × 1.5–2.0 | [18] |
| 3 | L. coffeanum | White, reverse cream-colored | Solitary, rarely in whorls of 2–3, 11–44 (–70) × 1.0–2.4 | Macroconidia spindle-shaped, 5.3–8.8 × 1.0–1.6 µm; Microconidia ellipsoidal to fusiform, 2.2–3.8 × 0.8–1.5 | [21] |
| 3 | L. coffeanum | White to cream, reverse orange-yellow | Solitary or in whorls of 2–3, 13.7–81.1 × 1.8–2.9 | Macroconidia spindle-shaped, 3.3–6.2 × 1.2–3.2; Microconidia ellipsoidal to fusiform, 2.9–4.1 × 1.4–2.2 | [64] |
| 4 | L. filiforme | White, reverse white to yellowish cream | Solitary, 9–18 × 1 | Fusoid to filiform, catenulate, sometimes forming zigzag chains, 7.2–12.5 × 1 | [37] |
| 5 | L. gasaense | White to cream, reverse pale luteous, yellow to yellowish brown | Solitary, rarely in whorls of 2–3, 12.2–30.3 × 1.3–2.5 | Bacilliform or narrowly cylindrical (rod-shaped), 3.9–6.8 × 0.9–2.5; the apical conidia of the conidial chains, fusiform, subglobose to obovoid, 2.7–3.8 × 1.8–2.6 | This study |
| 6 | L. latisporum | White, reverse grayish orange to orange-white at the margin | Solitary or in whorls of 2–3, cylindrical, 13.2–40.8 × 3–4.8 | Slightly fusoid to narrowly cylindrical, 4–6.3 × 1.9–3.8 | [23] |
| 7 | L. leptobactrum var. calidius | White to cream, reverse pale yellow to brown | Solitary, rarely in whorls of 1–2, 18.4–60.0 × 0.7–2.0 | Narrowly cylindrical (rod-shaped) to slightly fusiform, 3.0–5.7 × 0.7–1.7 | [64] |
| 8 | L. leptobactrum | White, grayish white to pinkish white, reverse orange to orange-brown, ochraceous, pale luteous, milky white to dark buff | Solitary, rarely in whorls of 1–2, 20–45 µm long, 1–2 µm wide (base) to 0.5–0.7 µm wide (apex) | Narrowly cylindrical (rod-shaped) to slightly fusiform, 4.5–8 × 0.8–1.5 (–2) | [6] |
| 9 | L. Leptobactrum var. leptobactrum | White to cream, reverse light yellow to yellowish brown | Solitary, rarely in whorls of 2–3, 15.8–31.7 × 0.7–1.5 | Narrowly clavate or narrowly cylindrical (rod-shaped), 3.0–6.1 × 0.8–2.1 | [64] |
| 10 | L. muralicola | White, grayish white to greenish white, reverse pale luteous, milky white to dark buff, orange to orange-brown, ochraceous | Solitary, rarely in whorls of 1–2, 20–45 µm long, 1–2 µm wide (base) to 0.5–0.7 µm wide (apex) | Narrowly cylindrical (rod-shaped) to slightly fusiform, 4.5–6 × 1–2 | [22] |
| 11 | L. symbioticum | White, reverse orange-yellow to orange-brown | Solitary, rarely in whorls of 2–3, 7.1–30.6 × 1.6–3.5 | Slightly fusiform to narrowly cylindrical, 4.0–6.9 × 0.7–1.6 | [19] |
| 12 | L. xianyushanense | White, irregular floccose surface with divergent cracks, reverse orange to orange-brown, ochraceous, pale luteous | Solitary, rarely with branches of two, tapering 24.7–28.5 × 1.5–1.9 | Narrowly cylindrical (rod-shaped) to slightly fusiform. 3.8–5.6 × 0.5–1.2 | [24] |
Key to the species of
Leptobacillium| 1a. | Phialide simple, up to two whorls.....................………………………………………………………….. | 2 |
| 1b | Phialide simple, 2–3 whorls……….......................………………………………………………………….. | 6 |
| 2a | Conidia cylindrical (rod-shaped) to slightly fusiform…………………………………..................………………...……. | 3 |
| 2b | Conidia oval, ellipsoidal or filiform………………………………………………......................................……......……… | 5 |
| 3a | Phialide > 30 µm long; conidia relatively bigger (4.5–8 × 0.8–1.5(–2) µm)…….....……….…………… | L. leptobactrum |
| 3b | Phialide < 30 µm long; conidia relatively smaller……………………………….................……………………………… | 4 |
| 4a | Phialide relatively narrower, 1.2–1.7 µm………………………………………..……..…………....…...… | L. cavernicola |
| 4b | Phialide relatively wider, 1.5–1.9 µm………...……………………………................……………...…. | L. xianyushanense |
| 5a | Phialide relatively longer, (6.0–) 15–30 (–68.0) × 1.5 µm…………………….……………………………....… | L. chinense |
| 5b | Phialide relatively shorter, 9–18 × 1 µm…………………………………..……………………………..……… | L. filiforme |
| 6a | Phialide > 50 µm long……………………...........………………..…... ……………………………………………..………. | 7 |
| 6b | Phialide < 50 µm long……………………………......…………….………………….…………………………...…………. | 7 |
| 7a | Conidia spindle-shaped, 3.3–6.2 × 1.2–3.2 µm……...………….…………………………………….......…… | L. coffeanum |
| 7b | Conidia narrowly cylindrical (rod-shaped) to slightly fusiform, 3.0–5.7 × 0.7–1.7 µm……………………………......………………………………………………………….… | L. leptobactrum var. calidius |
| 8a | Isolated from inorganic substrate (acrylic varnish coatings of murals, soil)…....................................………….........… | 9 |
| 8b | Isolated on insecta (larvae, Lepidoptera) or fungi………………………………………………………….……..........… | 10 |
| 9a | Phialide relatively longer, 20–45 µm long; conidia relatively narrower, 1–2 µm ……..........................… | L. muralicola |
| 9b | Phialide relatively shorter, 13.2–40.8 µm long; conidia relatively wider, 1.9–3.8 µm……….…..…........ | L. latisporum |
| 10a | On Lepidoptera larva…...….………….……………………...………………………..…..………...… | L. var. leptobactrum |
| 10b | On fungi……………………....……………………………......……………………………......…………….……………… | 11 |
| 11a | Isolated from Gibellula sp., conidia relatively wider, 0.9–2.5 µm...….........................................................….. | L. gasaense |
| 11b | From sori of soybean rust fungus, conidia relatively narrower, 0.7–1.6 µm………………...........……..… | L. symbioticum |
Leptobacillium gasaense Q.Y. Dong sp. nov.
MycoBank: 857486
Etymology: The name reflects the location of Gasa County, where the species was isolated.
Holotype: China, Yunnan Province, Jinghong City, Gasa County, Anmalaozhai Village (22°08′53″ N, 100°40′01″ E, alt. 690 m), isolated from the Gibellula sp., 29 July 2024, Quanying Dong, dried culture on PDA (holotype CXTC 0003; ex-holotype living culture, CXCC 0003).
Sexual morph: Undetermined.
Asexual morph: Colonies on PDA are moderately fast-growing, attaining a diameter of 34–37 mm in 21 days at 25 °C. Colonies cotton, fluffy, with high mycelial density, white to cream, reverse pale luteous, yellow to yellowish brown. Hyphae smooth-walled, branched, septate, hyaline, 1.3–2.5 µm wide. Cultures produce phialides and conidia on PDA after 10 days at room temperature. Phialides arising from aerial hyphae, usually solitary, rarely in whorls of two to three, 12.2–30.3 × 1.3–2.5 µm, 1.4–2.5 µm wide at the base, 0.9–1.3 µm wide at the top, long cylindrical, tapering gradually toward the apex. Conidia hyaline, one-celled, narrow clavate or narrowly cylindrical (rod-shaped), single or arranged in very long, slender chains at the apex of phialides, 3.9–6.8 × 0.9–2.5 µm. The first-formed conidium is usually shorter, ellipsoid, or with a rounded distal end, 2.7–3.8 × 1.8–2.6 µm. Chlamydospores not observed.
Substrate: Gibellula sp. (Cordycipitaceae)
Known distribution: Yunnan Province, China.
Additional specimens examined: China, Yunnan Province, Jinghong City, Gasa Country, Anmalaozhai Village (22°08′53″ N, 100°40′01″ E, alt. 690 m), isolated from the Gibellula sp., 29 July 2024, Quanying Dong, dried culture on PDA (paratype CXTC 0004; ex-paratype living culture CXCC 0004).
Commentary: Our phylogenetic analysis indicates that
Leptobacillium gasaense is closely related to
L. latisporum TBRC 16288 (BS = 70% and PP = 0.85), which was firstly isolated from soil in Thailand, mainly produces long solitary phialides, rarely in whorls of 2–3, 13.2–40.8 × 3–4.8 µm, conidia lisghtly fusoid to narrowly cylindrical, 4–6.3 × 1.9–3.8 µm [
23]. Morphologically,
L. gasaense differs from
L. latisporum in the following aspects. Relatively
, L. gasaense has thinner phialides (12.2–30.3 × 1.3–2.5 µm vs. 13.2–40.8 × 3–4.8 µm) and narrower conidia (3.9–6.8 × 0.9–2.5 µm vs. 4–6.3 × 1.9–3.8 µm). The molecular divergence between
L. gasaense and
L. latisporum, based on ITS and LSU sequence data.
Leptobacillium gasaense vs.
O. attenuatum: ITS: 85 bp differences. LSU: 17 bp differences.
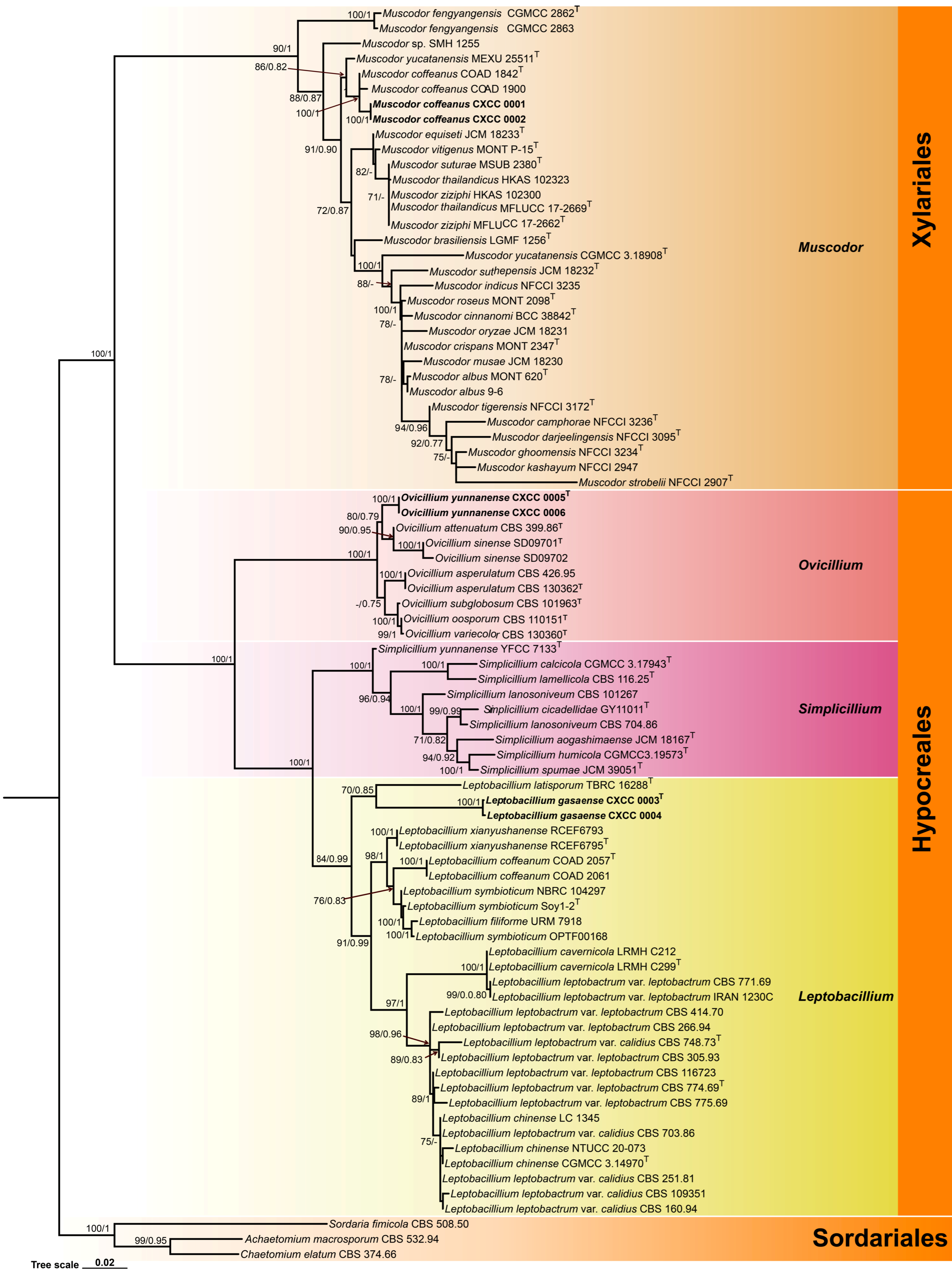
Figure 2.
Morphology of Leptobacillium gasaense. (A,B) Colonies on PDA after 21 days ((A) obverse; (B) reverse). (C–J) Solitary phialides with conidia in chains are produced on prostrate aerial hyphae. Scale bars: (A,B) = 10 mm; (C–G) = 20 µm; (H–J) = 10 µm.
Figure 2.
Morphology of Leptobacillium gasaense. (A,B) Colonies on PDA after 21 days ((A) obverse; (B) reverse). (C–J) Solitary phialides with conidia in chains are produced on prostrate aerial hyphae. Scale bars: (A,B) = 10 mm; (C–G) = 20 µm; (H–J) = 10 µm.
Muscodor Worapong, Strobel & W.M. Hess, Mycotaxon 79: 71 (2001)
Systematic position: Fungi, Dikarya, Ascomycota, Pezizomycotina, Sordariomycetes, Xylariomycetidae, Xylariales
Type species: Muscodor albus Worapong, Strobel & W.M. Hess 2001
Muscodor coffeanus A.A.M. Gomes, Pinho & O.L. Pereira [as ‘coffeanum’], Cryptog. Mycol. 36(3): 368 (2015)
Sexual morph: For detailed descriptions and images of
M. coffeanus, see Li and Kang [
65].
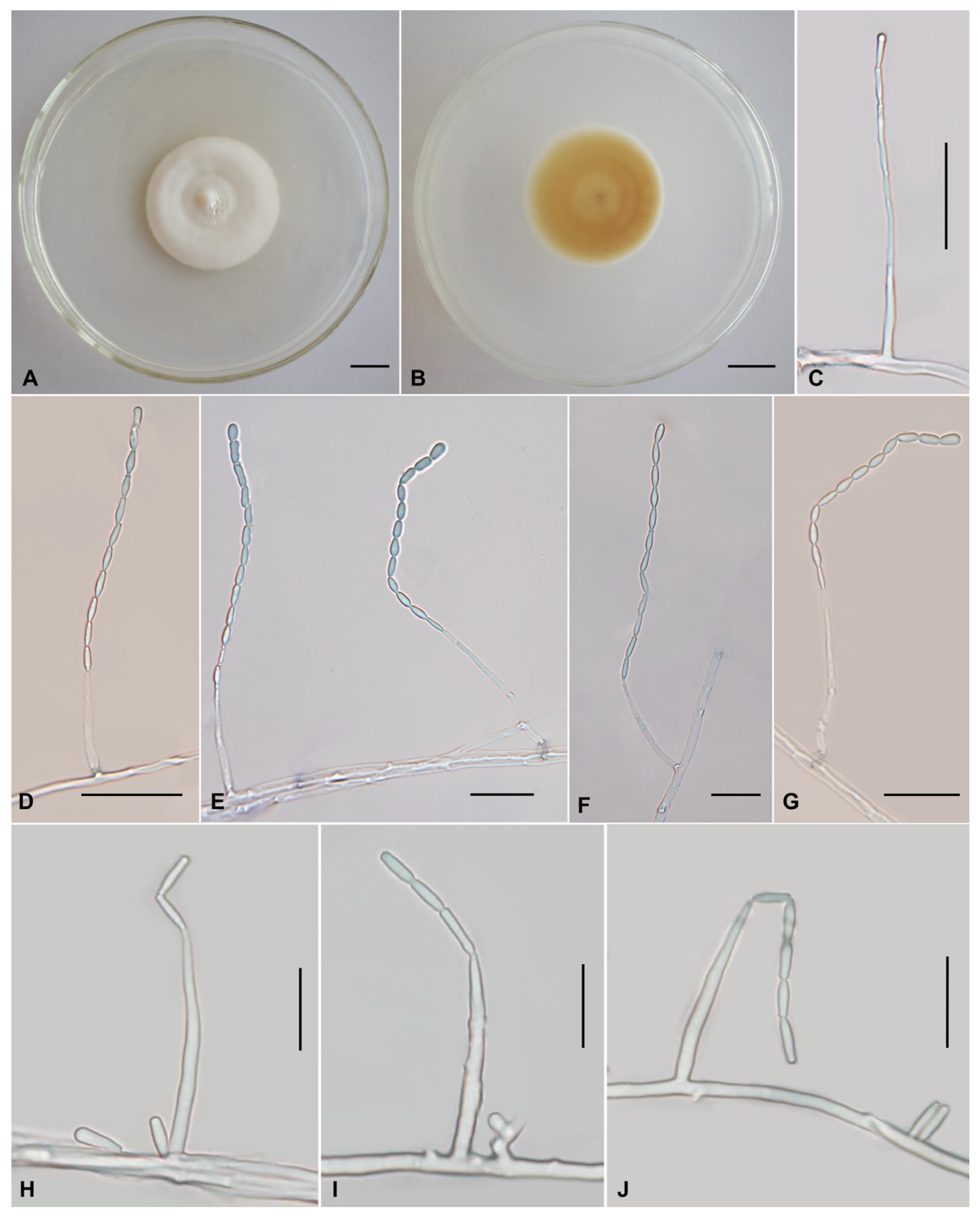
Asexual morph: Colonies on PDA are slowly growing, attaining a diameter of 37–40 mm after 21 days at 25 °C. Colonies white to pale yellow, with high mycelial density, flocculose. Reverse yellow to brown. Hyphae hyaline, branched, smooth-walled, septate, 1.2–2.4 µm wide. Cultures readily produced phialides and conidia on PDA after four months at 25 °C. Conidiophores cylindrical, hyaline, smooth-walled, solitary or verticillate, 15.4–55.8 × 1.1–2.6 µm, 1.4–2.4 µm wide (apex), 1.1–2.2 µm wide (base). Phialides from aerial mycelium, straight to slightly flexuose, solitary or in whorls of two to five on each branch, cylindrical, usually with a slightly swollen basal part, tapering into the apex form a long neck, 6.9–34.7 × 1.6–2.5 µm, 0.3–1.5 µm wide (apex), 1.4–2.5 µm wide (base). Conidia one-celled, hyaline, smooth, ovoid to ellipsoidal, globose to subglobose; the conidia exhibit catenulate arrangement, producing distinct chains, 2.0–3.2 × 1.5–2.5 µm. Chlamydospores not observed.
Known distribution: Brazil, China, Thailand [
42,
65].
Specimens examined: China, Sichuan Province, Dujiangyan City, Qingcheng Hou Mountain, Youyi Village (30°56′10″ N, 103°28′19″ E, alt. 1270 m), isolated from the Ophiocordyceps sp., 19 August 2023, Quanying Dong, dried culture on PDA (CXTC 0001, living culture CXCC 0001; CXTC 0002, living culture CXCC 0002).
Commentary:
Muscodor coffeanus as an endophytic fungus with a full description and illustration of the sterile mycelium described from Brazil by Hongsanan et al. [
42], and subsequently collected from Thailand, is isolated from a deadwood piece of an unidentified plant with sexual morphological characteristics and phylogenetic analyses results [
65]. VOCs, including cyclosativene compound and phytase, have been detected in the culture of
M.
coffeanus [
66].
Muscodor coffeanus produces VOCs with antifungal activity against
Botrytis cinerea Pers. and antibacterial activity against
Staphylococcus aureus Rosenbach,
Enterococcus faecalis (Andrewes and Horder) Schleifer and Kilpper-Bälz, and
E. faecium (Orla-Jensen) Schleifer and Kilpper-Bälz; moreover, anti-nematode activity against
Meloidogyne incognita (Kofoid & White) Chitwood [
15,
66,
67].
In our two-locus (ITS and nrLSU) phylogenetic analysis, M. coffeanus is, with strong support (BS = 86% and PP = 0.82), related to M. yucatanensis (M.C. González, A.L. Anaya, Glenn & Hanlin) Samarak. et al.; the four strains (COAD 1842, COAD 1900, CXCC 0001 and CXCC 0002) formed a distinct lineage. COAD 1842 as the type material of M. coffeanus from Brazil. Since no significant ITS sequence differences were found between the Chinese collections and that of Brazil, we treated CXCC 0001 and CXCC 0002 as Muscodor coffeanus, a new record for China based on its mycelial characteristics, asexual morphology, and phylogenetic analyses. Muscodor coffeanus is easily distinguished by its rope-like hyphal bodies and coil-like structure; solitary or verticillate, cylindrical conidiophores; solitary or in whorls of two to five, cylindrical phialides and ovoid to ellipsoidal, globose to subglobose conidia. Moreover, this report is the first study on asexual characters of Muscodor genus.
Figure 3.
Morphology of Muscodor coffeanus. (A,B) Colonies on PDA after 1 month ((A) obverse; (B) reverse). (C–N) Phialides and conidia. Scale bars: (A,B) = 10 mm; (C,D,G,H,J) = 20 µm; (E,F,I,K–N) = 10 µm.
Figure 3.
Morphology of Muscodor coffeanus. (A,B) Colonies on PDA after 1 month ((A) obverse; (B) reverse). (C–N) Phialides and conidia. Scale bars: (A,B) = 10 mm; (C,D,G,H,J) = 20 µm; (E,F,I,K–N) = 10 µm.
Ovicillium Zare & W. Gams, Mycol. Progr. 15: 1020 (2016)
Systematic position: Fungi, Dikarya, Ascomycota, Pezizomycotina, Sordariomycetes, Hypocreomycetidae, Hypocreales, Bionectriaceae
Type species: Ovicillium attenuatum Zare & W. Gams, Mycol. Progr. 15: 1021 (2016)
Table 4 lists the hosts, substrates, and geographical locations of all
Ovicillium species.
Table 5 shows the differences between the asexual morphs, including conidiogenous structures, conidial shapes, and chlamydospores of known species in
Ovicillium.
Table 4.
Hosts, substrates, and geographic distribution of Ovicillium.
Table 4.
Hosts, substrates, and geographic distribution of Ovicillium.
| No. | Species | Host/Substrate | Countries Found | References |
|---|
| 1 | O. asperulatum | Forest soil | Spain | [26] |
| 2 | O. attenuatum | Auricularia sp. | Cuba, Ecuador, Papua New Guinea | [6] |
| 3 | O. oosporum | Theobroma gileri, Grandinia pallidula, Theobroma sp., human, Hypholoma sp, Leptomitus lacteus, Fomitopsis pinicola, soil under Elaeis guineënsis, Xylaria sp. on log | South America (Brazil?), Belgium, Ecuador, France, Netherlands, Poland, Surinam, USA | [6] |
| 4 | O. sinense | Lepidoptera pupa | China | [27] |
| 5 | O. subglobosum | Soil, Theobroma gileri | China, Brazil, Puerto Rico | [6] |
| 6 | O. variecolor | Forest soil | Spain | [26] |
| 7 | O. yunnanense | Dead insect on leaf | China | This study |
Table 5.
Morphological comparisons of asexual morphs in Ovicillium.
Table 5.
Morphological comparisons of asexual morphs in Ovicillium.
| No. | Species | Colony | Conidiophores | Phialides (µm) | Conidia (µm) | Chlamydospores | References |
|---|
| 1 | O. asperulatum | Dark, white or yellowish-white, reverse yellowish or amber yellow | Solitary or in whorls of 2–4, up to 105 µm long | Acicular, 28–68 µm long, 1–2 µm wide at the base | Globose, 3–4 (–5) µm diam, chromophilic, arranged in slimy heads | Present | [26] |
| 2 | O. attenuatum | White, dirty white to pinkish, ochraceous to pale ochraceous, light hazel to buff, reverse pinkish to pale white | Solitary and verticillate | Aculeate, 25–50 × 1.7–3.3 | Oval to subglobose, strongly cyanophilic, 3.5–5 × 2.5–3.8, aggregated in large globose to subglobose heads | Absent | [6] |
| 3 | O. oosporum | Grayish to dark buff, light honey to hazel, reverse white, pale brown, gray to grayish white, pale yellow to brown | Solitary or in whorls of 2–5, 20–50 × 1.2–2.2 | – | Subglobose, oval to broadly oval, cyanophilic, 4–6 × 2.5–4, aggregated in large globose heads | Present or absent | [6] |
| 4 | O. sinense | White, reverse yellowish | Solitary or in whorls of 2–5, 17.0–21.7 × 2.3–3.0 | Cylindrical, 16.2–25.8 × 1.7–2.4 | Globose to ovoid, 2.1–2.9 × 1.1–1.7, aggregated in large globose to subglobose heads | – | [27] |
| 5 | O. subglobosum | Grayish buff to dark buff, light smoke-gray to light hazel, reverse pale gray, dirty white to grayish cream-colored | Solitary or in whorls of 2–4 | 25–55 × 1.5–2.2 | Subglobose (or nearly globose), rather cyanophilic, 3.5–5.5 × 3.5–4.5 | Absent | [6] |
| 6 | O. variecolor | Yellowish white to grayish yellow | Solitary or in whorls of 2–5, 290 µm long | Acicular, 18–95 µm long, 1–2 µm wide at the base | Subglobose or ovoid, 3–4 (–5) × 2–4, arranged in slimy heads; sessile conidia solitary, cylindrical or ellipsoidal, 5–7 (–9) × 2–3 (–4) | Absent | [26] |
| 7 | O. yunnanense | White to pale yellowish-orange, reverse yellow to brown | Solitary or in whorls of 2–5 | Cylindrical, 22.7–87.8 × 1.4–3.3 | Subglobose, ovoid to ellipsoidal, 2.1–4.4 × 1.9–3.8, aggregated in large globose to subglobose heads | Absent | This study |
Key to the species of
Ovicillium| 1a. | Conidia globose............................................................................................................................................... | O. asperulatum |
| 1b. | Conidia oval to subglobose...................................................................................................................................................... | 2 |
| 2a. | Isolated from dead insect on leaf.................................................................................................................... | O. yunnanense |
| 2b. | Isolated from other substrate or host...................................................................................................................................... | 3 |
| 3a. | Conidia > 3.0 µm long…………………………….................................................................…………….....................……. | 4 |
| 3b. | Conidia < 3.0 µm long ( 2.1–2.9 × 1.1–1.7 µm)……................................................................………..…………. | O. sinense |
| 4a. | Chlamydospores scarce if present, widespread geographical distribution………………..…..……….... | O. oosporum |
| 4b. | Chlamydospores absent, limited geographical distribution…….........................................................……........……. | 5 |
| 5a. | From soil, phialides relatively longer ……….....………...……..........................................................................……...…... | 6 |
| 5b. | From Auricularia sp, phialides relatively shorter (25–50 × 1.7–3.3 µm)..................................................... | O. attenuatum |
| 6a | Conidiophores solitary or in whorls of 2–4 phialides, phialides relatively shorter (25–55 µm).................................................................................................................................................................... | O. subglobosum |
| 6b | Conidiophores solitary or in whorls of 2–5 phialides, phialides relatively longer (18–95 µm)......................................................................................................................................................................... | O. variecolor |
Ovicillium yunnanense Q.Y. Dong, sp. nov.
MycoBank: 857488
Etymology: The name reflects the location of Yunan Province, where the species was isolated.
Holotype: China, Yunnan Province, Jinghong City, Gasa County, Huilaoxiaozhai Village (22°09′14″ N, 100°41′13″ E, alt. 670 m), isolated from dead insect on leaf, 29 July 2024, Quanying Dong, dried culture on PDA (holotype CXTC 0005; ex-holotype living culture CXCC 0005).
Sexual morph: Undetermined.
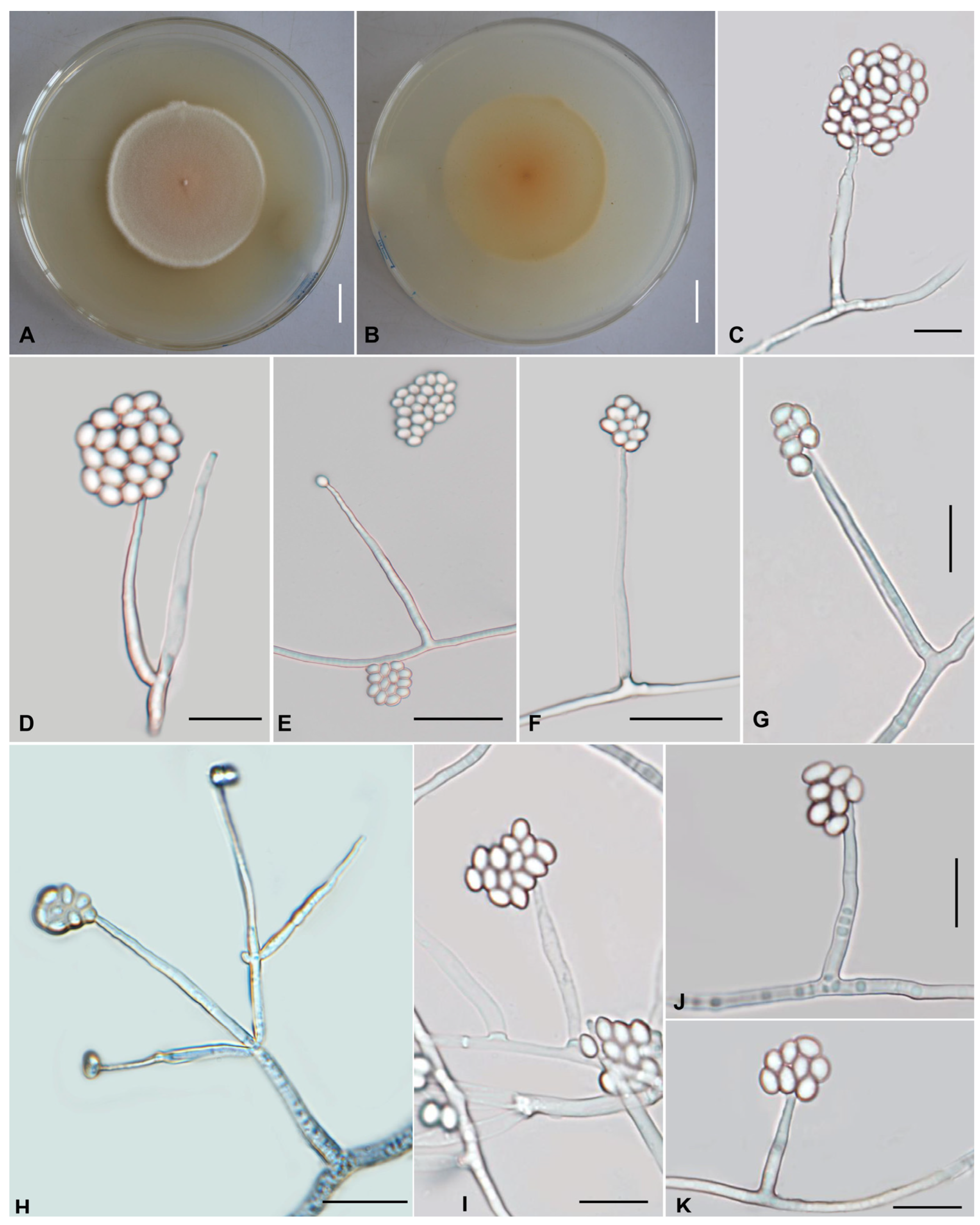
Asexual morph: Colonies on PDA are fast-growing, attaining a diameter of 46–48 mm after 21 days at 25 °C. Colonies white to pale yellowish orange, margin thick, with high mycelial density, pulvinate, asperulate. Reverse yellow to brown. Hyphae hyaline, branched, smooth-walled, 1.4–3.1 µm wide. Cultures readily produced phialides and conidia on potato dextrose agar after 7 days at 25 °C. Conidiophores are cylindrical, hyaline, smooth-walled, simple to verticillate form 1–3. Phialides from aerial mycelium straight to slightly flexuose, solitary or in whorls of two to five on each branch, cylindrical, usually with a slightly swollen basal part, tapering into the apex from a long neck, 22.7–87.8 × 1.4–3.3 µm, 1.4–2.2 µm wide (apex), 1.8–2.3 µm wide (base). Conidia one-celled, hyaline, smooth, subglobose, ovoid to ellipsoidal, 2.1–4.4 × 1.9–3.8 µm, aggregated in large globose to subglobose heads. Chlamydospores not observed.
Other material examined: China, Yunnan Province, Jinghong City, Gasa County, Huilaoxiaozhai Village (22°09′14″ N, 100°41′13″ E, alt. 670 m), isolated from a dead insect on a leaf, 29 July 2024, Quanying Dong, dried culture on PDA (paratype CXTC 0006; ex-paratype living culture CXCC 0006).
Figure 4.
Morphology of Ovicillium yunnanense. (A,B) Colonies on PDA after 21 days ((A) obverse; (B) reverse). (C–K) Phialides and conidia. Scale bars: (A,B) = 10 mm; (C,D,G,I–K) = 10 µm; (E,F,H) = 20 µm.
Figure 4.
Morphology of Ovicillium yunnanense. (A,B) Colonies on PDA after 21 days ((A) obverse; (B) reverse). (C–K) Phialides and conidia. Scale bars: (A,B) = 10 mm; (C,D,G,I–K) = 10 µm; (E,F,H) = 20 µm.
Habitat: Dead insect on leaf.
Known Distribution: Yunnan Province, China.
Commentary:
Ovicillium yunnanense displays characteristic genus-level features consistent with other
Ovicillium species, including solitary or whorled phialides (2–5 per node) and conidia varying from subglobose to ovoid or ellipsoid in shape. The species can be distinguished by the following unique combination of morphological characteristics: solitary or in whorls of 2–5, cylindrical phialides, 22.7–87.8 × 1.4–3.3 µm; and mostly subglobose, ovoid to ellipsoidal conidia, 2.1–4.4 × 1.9–3.8 µm, conidia aggregated in large globose to subglobose heads. Furthermore, it is isolated from a dead insect on a leaf. It is phylogenetically, with high support (BS = 80%, PP = 0.79), clustered with
O. attenuatum and
O. sinense, but it is distinguished from the two latter species by forming a separate clade in this group (
Figure 1). Morphologically, these two species differ from
O. yunnanense in the following ways.
Ovicillium attenuatum, a species described from Cuba, Ecuador, and Papua, New Guinea, has an aculeate and relatively shorter phialides measuring 25–50 × 1.7–3.3 µm [
6].
Ovicillium sinense, a species described from Guizhou Province, Duyun City, and also similar to
O. yunnanense in appearance, has a relatively shorter phialide (16.2–25.8 × 1.7–2.4 µm vs. 22.7–87.8 × 1.4–3.3 µm), and relatively smaller conidia (2.1–2.9 × 1.1–1.7 µm vs. 2.1–4.4 × 1.9–3.8 µm) [
27]. The molecular divergence between
O. yunnanense,
O. attenuatum, and
O. sinense, based on ITS and LSU sequence data.
O. yunnanense vs.
O. attenuatum: ITS: 7 bp differences. LSU: 0 bp differences.
O. yunnanense vs. Ovicillium sinense: ITS: 13 bp differences. LSU: 2 bp differences.