Transfer of Downy Mildew Resistance Genes from Wild Cucumbers to Beit Alpha Types
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sources
2.2. Agronomy
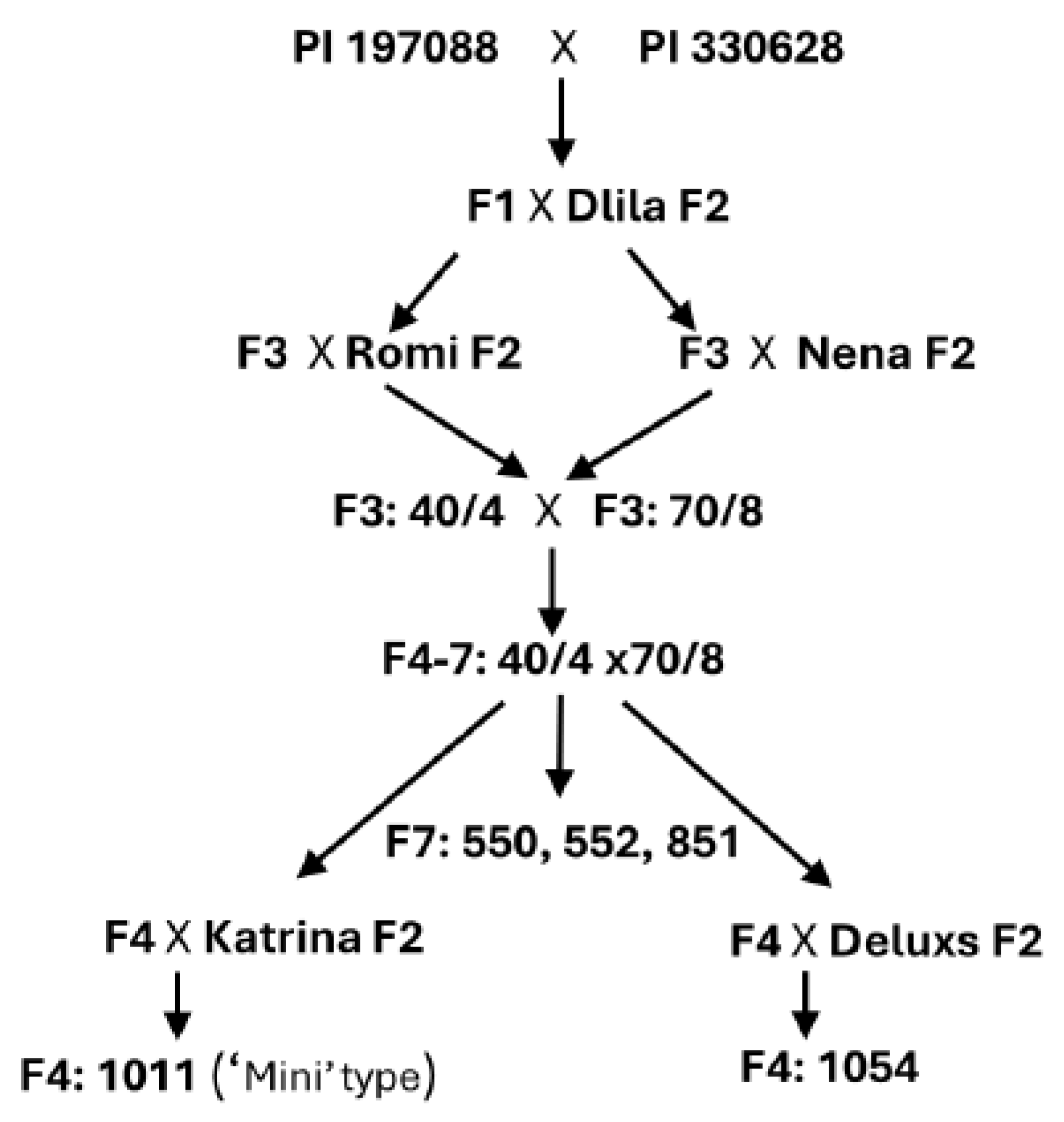
2.3. Breeding
2.4. Inoculation
2.5. Resistance Ratings
3. Data Analysis
4. Results
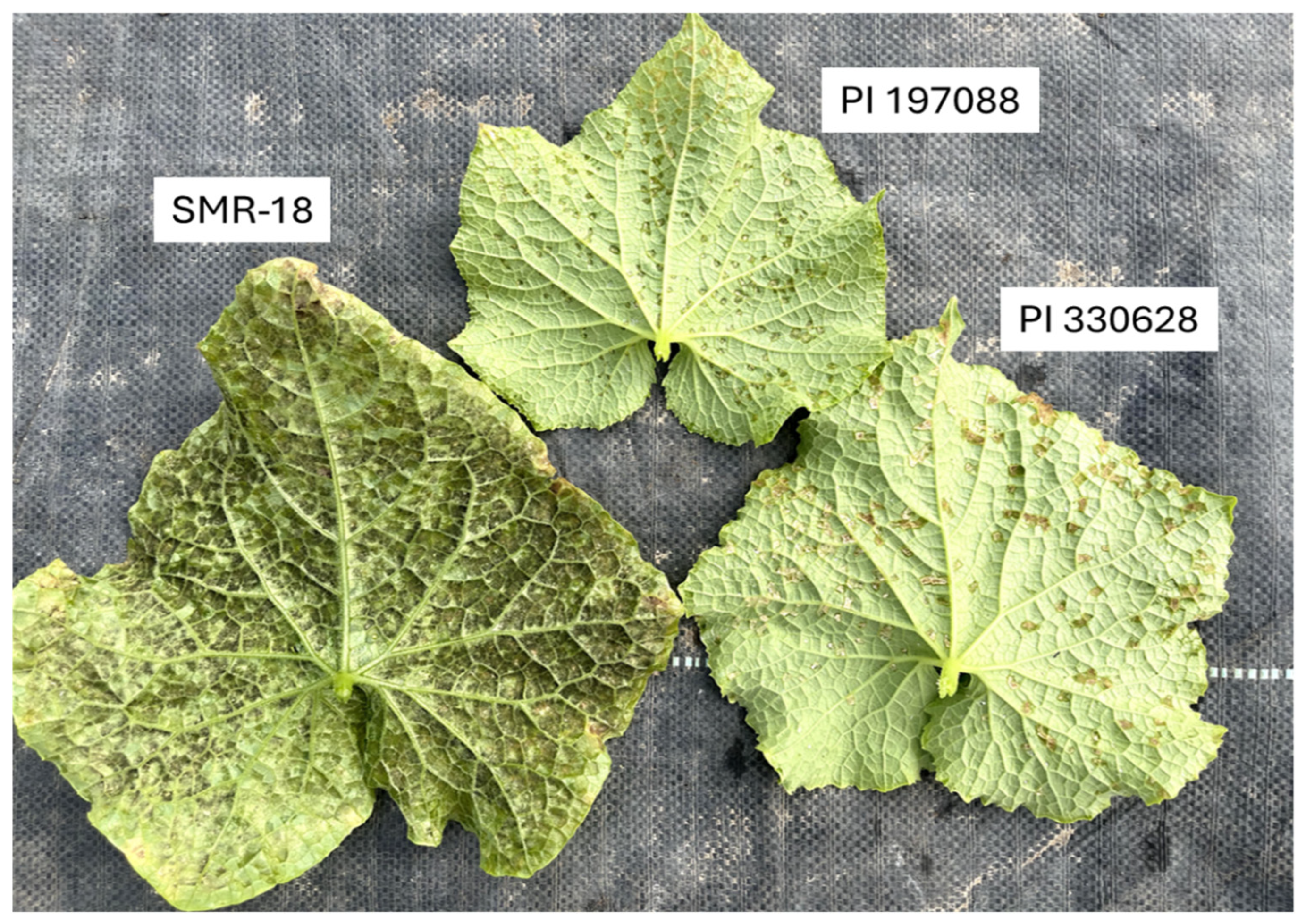
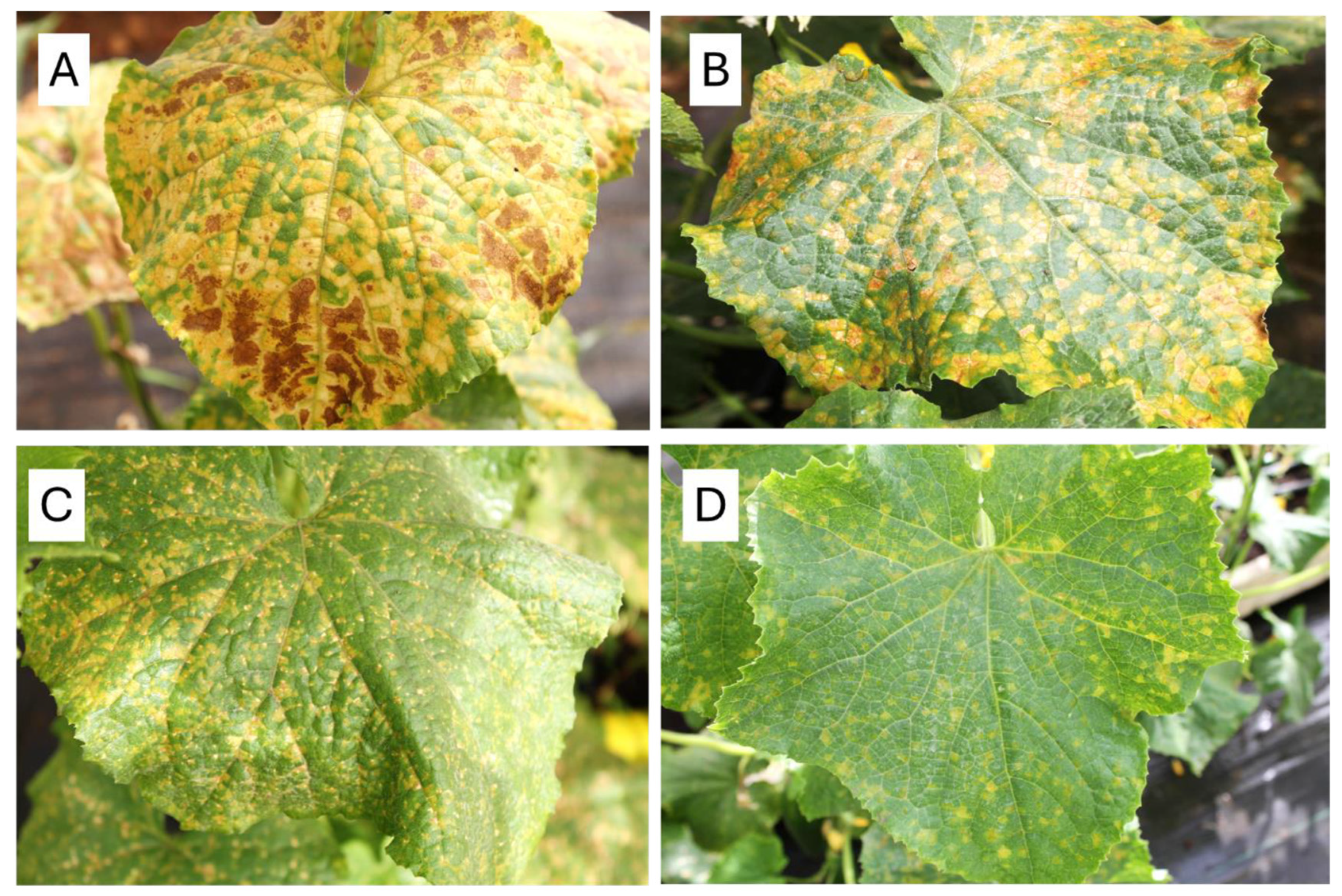
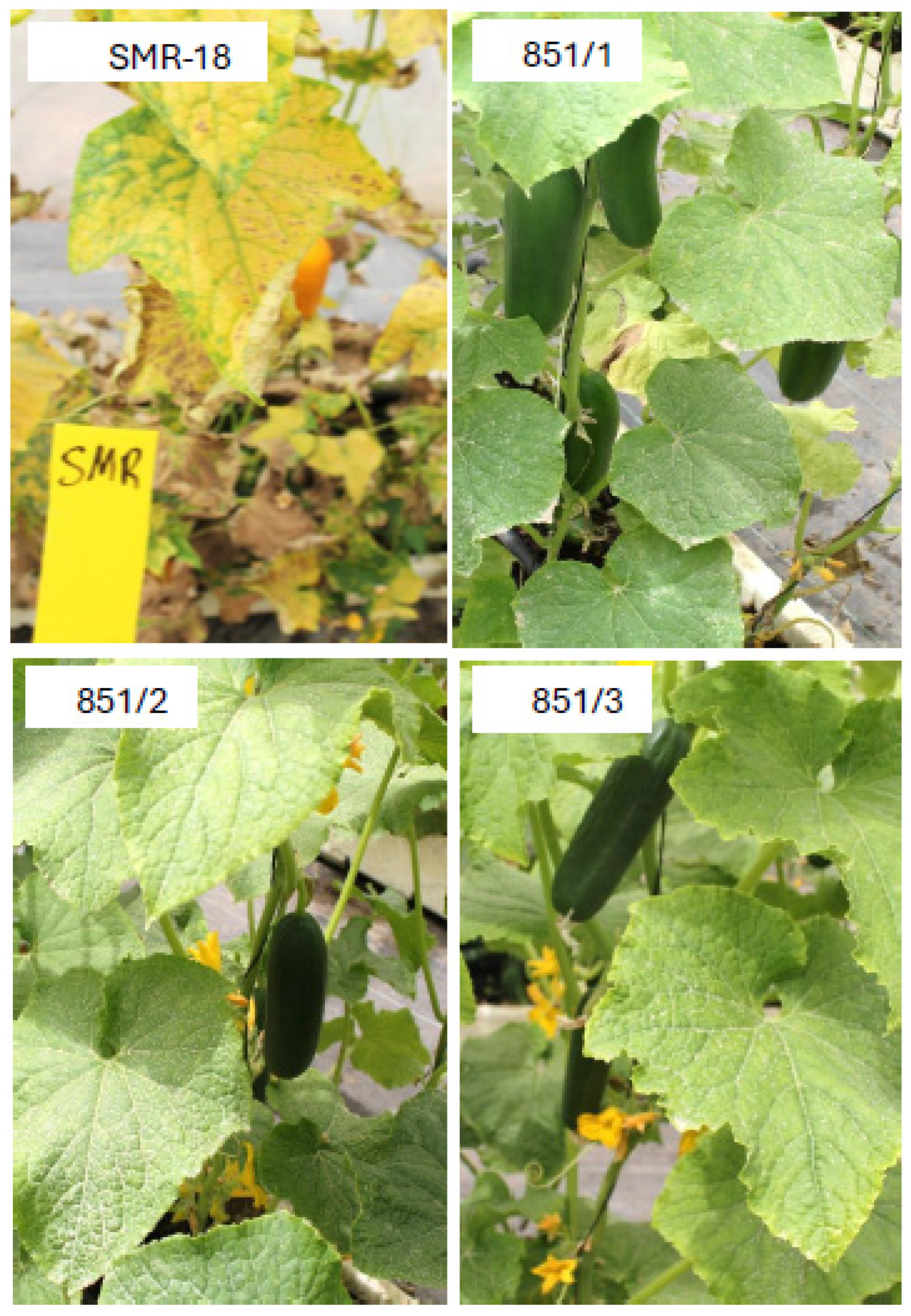
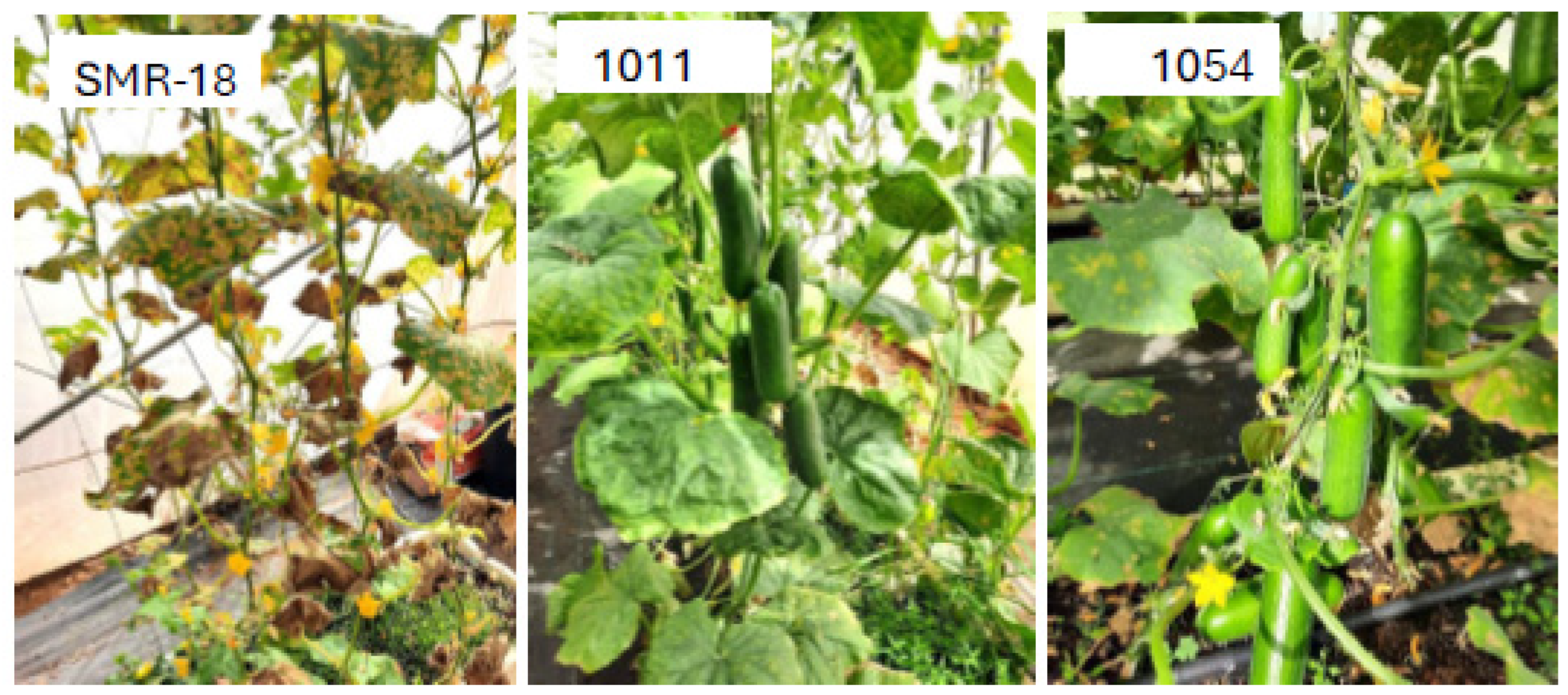
4.1. Disease Resistance
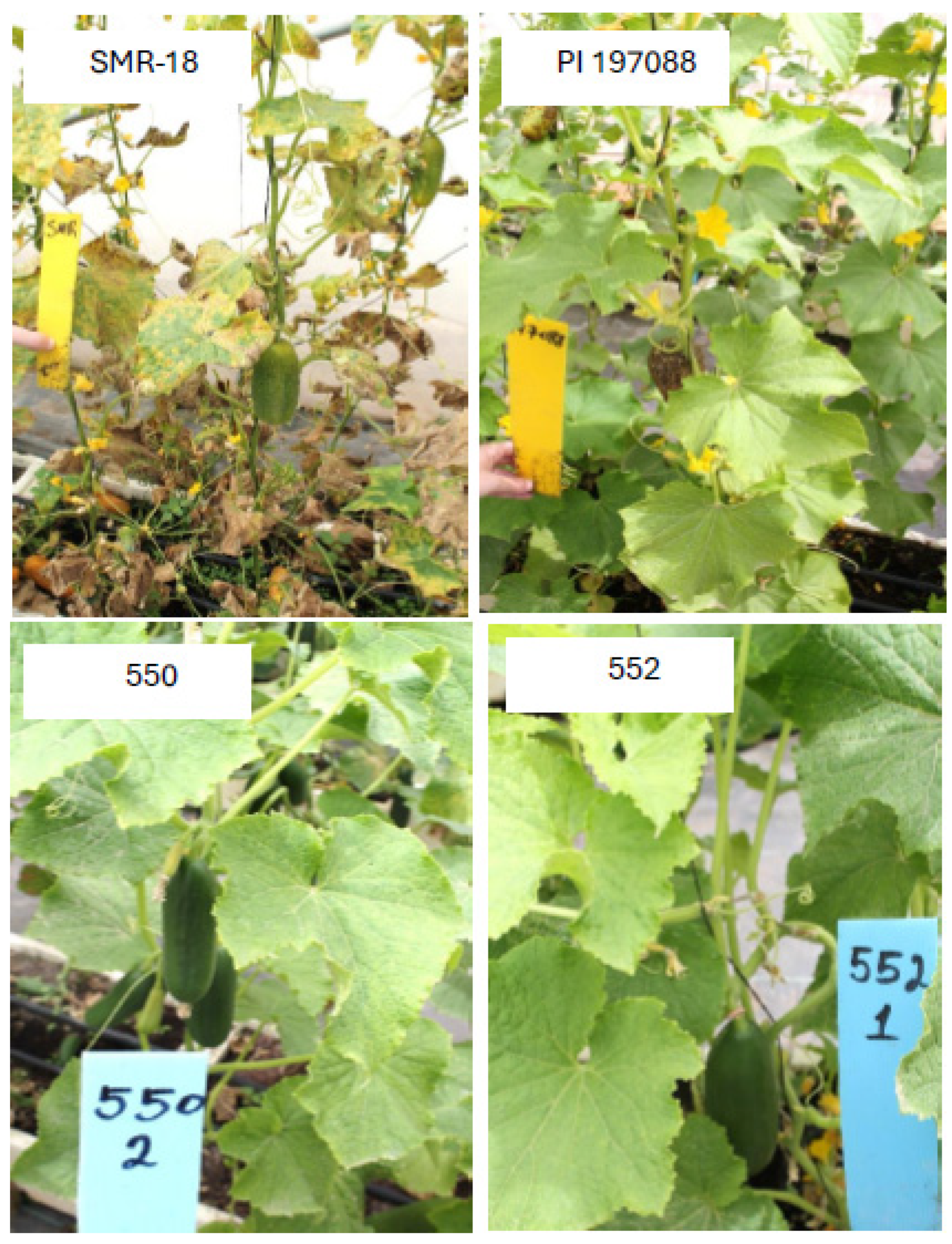
4.2. Performance of the Resistant Lines
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salcedo, A.; Hausbeck, M.; Pigg, S.; Quesada-Ocampo, L.M. Diagnostic Guide for Cucurbit Downy Mildew. Plant Health Prog. 2020, 21, 166–172. [Google Scholar] [CrossRef]
- Lebeda, A.; Cohen, Y. Cucurbit downy mildew (Pseudoperonospora cubensis)-biology, ecology, epidemiology, host-pathogen interaction and control. Eur. J. Plant Pathol. 2011, 129, 157–192. [Google Scholar] [CrossRef]
- Palti, J.; Cohen, Y. Downy mildew of Cucurbits (Pseudoperonospora cubensis): The Fungus and its hosts, distribution, epidemiology and control. Phytoparasitica 1980, 8, 109–147. [Google Scholar] [CrossRef]
- Savory, E.A.; Granke, L.L.; Quesada-Ocampo, L.M.; Varbanova, M.; Hausbeck, M.K.; Day, B. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol. Plant Pathol. 2011, 12, 217–226. [Google Scholar] [CrossRef]
- Granke, L.L.; Hausbeck, M.K. Dynamics of Pseudoperonospora cubensis Sporangia in Commercial Cucurbit Fields in Michigan. Plant Dis. 2011, 95, 1392–1400. [Google Scholar] [CrossRef]
- Ojiambo, P.S.; Holmes, G.J. Spatiotemporal Spread of Cucurbit Downy Mildew in the Eastern United States. Phytopathology 2011, 101, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Holmes, G.J.; Ojiambo, P.S.; Hausbeck, M.K.; Quesada-Ocampo, L.; Keinath, A.P. Resurgence of Cucurbit Downy Mildew in the United States: A Watershed Event for Research and Extension. Plant Dis. 2015, 99, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Rubin, A.E.; Galperin, M.; Ploch, S.; Runge, F.; Thines, M. Seed Transmission of Pseudoperonospora cubensis. PLoS ONE 2014, 9, e109766. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Rubin, A.E. Mating type and sexual reproduction of Pseudoperonospora cubensis, the downy mildew agent of cucurbits. Eur. J. Plant Pathol. 2012, 132, 577–592. [Google Scholar] [CrossRef]
- Cohen, Y.; Van Den Langenberg, K.M.; Wehner, T.C.; Ojiambo, P.S.; Hausbeck, M.; Quesada-Ocampo, L.M.; Lebeda, A.; Sierotzki, H.; Gisi, U. Resurgence of Pseudoperonospora cubensis: The causal agent of Cucurbit downy mildew. Phytopathology 2015, 105, 998–1012. [Google Scholar] [CrossRef]
- Cohen, Y.; Rubin, A.E.; Galperin, M. Host Preference of Mating Type in Pseudoperonospora cubensis, the Downy Mildew Causal Agent of Cucurbits. Plant Dis. 2013, 97, 292. [Google Scholar] [CrossRef]
- Rani, R.; Negi, P.; Sharma, S.; Jain, S. Occurrence of oosporic stage of Pseudoperonospora cubensis on cucumber, in Punjab, India: A first report. Crop Prot. 2022, 155, 105939. [Google Scholar] [CrossRef]
- Kikway, I.; Keinath, A.P.; Ojiambo, P.S. Field Occurrence and Overwintering of Oospores of Pseudoperonospora cubensis in the Southeastern United States. Phytopathology 2022, 112, 1946–1955. [Google Scholar] [CrossRef] [PubMed]
- Runge, F.; Choi, Y.J.; Thines, M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur. J. Plant Pathol. 2011, 129, 135–146. [Google Scholar] [CrossRef]
- Wallace, E.C.; D’Arcangelo, K.N.; Quesada-Ocampo, L.M. Population analyses reveal two host-adapted clades of pseudoperonospora cubensis, the causal agent of cucurbit downy mildew, on commercial and wild cucurbits. Phytopathology 2020, 110, 1578–1587. [Google Scholar] [CrossRef]
- Cohen, Y.; Meron, I.; Mor, N.; Zuriel, S. A New Pathotype of Pseudoperonospora cubensis Causing Downy Mildew in Cucurbits in Israel. Phytoparasitica 2003, 31, 458–466. [Google Scholar] [CrossRef]
- Cohen, Y.; Reuveni, M.; Gur, L.; Ovadia, S. Survival in the field of Pseudoperonospora cubensis and Plasmopara viticola after extreme hot and dry weather conditions in Israel. Phytoparasitica 2020, 48, 699–703. [Google Scholar] [CrossRef]
- Keinath, A.P. Congruent and Differential Responses of Pseudoperonospora cubensis Clades 1 and 2 to Downy Mildew Fungicides. Plant Health Prog. 2023, 24, 405–410. [Google Scholar] [CrossRef]
- Kenigsbuch, D.; Cohen, Y. Inheritance of Resistance to Downy Mildew in Cucumis melo PI 124112 and Commonality of Resistance Genes with PI 124111F. 1992. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19922320262 (accessed on 1 August 2025).
- Taler, D.; Galperin, M.; Benjamin, I.; Cohen, Y.; Kenigsbuch, D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 2004, 16, 172–184. [Google Scholar] [CrossRef]
- Barnes, W.C.; Epps, W.M. An unreported type of resistance to cucumber downy mildew. Plant Rep. 1954, 38, 620. [Google Scholar]
- Fanourakis, N.E.; Simon, P.W. Analysis of genetic linkage in the cucumber. J. Hered. 1987, 78, 238–242. [Google Scholar] [CrossRef]
- Horejsi, T.; Staub, J.E.; Thomas, C. Linkage of random amplified polymorphic DNA markers to downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 2000, 115, 105–113. [Google Scholar] [CrossRef]
- Thomas, A.; Carbone, I.; Choe, K.; Quesada-Ocampo, L.M.; Ojiambo, P.S. Resurgence of cucurbit downy mildew in the United States: Insights from comparative genomic analysis of Pseudoperonospora cubensis. Ecol. Evol. 2017, 7, 6231–6246. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tan, J.; Wu, Z.; VandenLangenberg, K.; Wehner, T.C.; Wen, C.; Zheng, X.; Owens, K.; Thornton, A.; Bang, H.H.; et al. STAYGREEN, STAY HEALTHY: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Lebeda, A.; Křístková, E.; Sedláková, B.; McCreight, J.D.; Coffey, M.D. Cucurbit powdery mildews: Methodology for objective determination and denomination of races. Eur. J. Plant Pathol. 2016, 144, 399–410. [Google Scholar] [CrossRef]
- Wang, Y.; VandenLangenberg, K.; Wehner, T.C.; Kraan, P.A.G.; Suelmann, J.; Zheng, X.; Owens, K.; Weng, Y. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor. Appl. Genet. 2016, 129, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; VandenLangenberg, K.; Wen, C.; Wehner, T.C.; Weng, Y. QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by-sequencing in RIL population. Theor. Appl. Genet. 2018, 131, 597–611. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhu, W.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef]
- Chen, T.; Katz, D.; Ben Naim, Y.; Hammer, R.; Ben Daniel, B.H.; Rubin, A.E.; Cohen, Y. Isolate-Dependent Inheritance of Resistance Against Pseudoperonospora cubensis in Cucumber. Agronomy 2020, 10, 1086. [Google Scholar] [CrossRef]
- Caldwell, D.; Chan, E.; de Vries, J.; Joobeur, T.; King, J.; Reina, A.; Shetty, N.V. Methods and Compositions for Identifying Downy Mildew Resistant Cucumber Plants. 2011. Available online: https://patents.google.com/patent/WO2011050296A1/en (accessed on 28 April 2011).
- Li, L.; He, H.; Zou, Z.; Li, Y. QTL Analysis for Downy Mildew Resistance in Cucumber Inbred Line PI 197088. Plant Dis. 2018, 102, 1240–1245. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Sakata, Y.; Sugiyama, M.; Fukino, N. Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 2014, 198, 265–276. [Google Scholar] [CrossRef]
- Holdsworth, W.L.; Summers, C.F.; Glos, M.; Smart, C.D.; Mazourek, M. Development of downy mildew-resistant cucumbers for late-season production in the northeastern United States. HortScience 2014, 49, 10–17. [Google Scholar] [CrossRef]
- Brzozowski, L.; Holdsworth, W.L.; Mazourek, M. ‘DMR-NY401’: A New Downy Mildew–resistant Slicing Cucumber. HortScience 2016, 51, 1294–1296. [Google Scholar] [CrossRef]
- Evaluation of Cucumber Varieties Resistant to Downy Mildew, 2021 Vegetable Pathology—Long Island Horticultural Research & Extension Center. Available online: https://blogs.cornell.edu/livegpath/research/cucurbit-downy-mildew/evaluation-of-cucumber-varieties-resistant-to-downy-mildew2/ (accessed on 7 July 2025).
- Zhang, S.P.; Liu, M.M.; Miao, H.; Zhang, S.Q.; Yang, Y.H.; Xie, B.Y.; Wehner, T.C.; Gu, X.F. Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis. 2013, 97, 245–251. [Google Scholar] [CrossRef]
- Win, K.T.; Vegas, J.; Zhang, C.; Song, K.; Lee, S. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor. Appl. Genet. 2017, 130, 199–211. [Google Scholar] [CrossRef]
- Hammer, R.S.; Cohen, Y. Non-Sikkim Cucumber Accessions Resistant to Downy Mildew (Pseudoperonospora cubensis). Seeds 2025, 4, 8. [Google Scholar] [CrossRef]
- Mirzwa-Mróz, E.; Zieniuk, B.; Yin, Z.; Pawełkowicz, M. Genetic Insights and Molecular Breeding Approaches for Downy Mildew Resistance in Cucumber (Cucumis sativus L.): Current Progress and Future Prospects. Int. J. Mol. Sci. 2024, 25, 12726. [Google Scholar] [CrossRef] [PubMed]
- Performance of Beit Alpha Cucumbers in the Negev, Israel 2022–2023 (in Hebrew). Available online: https://www.moprn.org/node/42 (accessed on 10 April 2025).
- Thaxton, B.; Brecke, B.; Byars, A.; Smith, M. High Tunnel Beit Alpha Cucumber Variety Study. 2015. Available online: https://wfrec.ifas.ufl.edu/variety-testing/specialty-crops/ (accessed on 7 July 2015).
- Shaw, N.L.; Cantliffe, D.J.; Stoffella, P.J. A new crop for north american greenhouse growers: Beit alpha cucumber—Progress of production technology through university research trials. Acta Hortic. 2007, 731, 251–258. [Google Scholar] [CrossRef]


| AUDPC | ||||||
|---|---|---|---|---|---|---|
| Pedigree (Code) | Fruit Type | Replicates | Mean | Std Dev | Dunn’s Test | % Reduction |
| Ten commercial cultivars | Various | 104 | 3269 | 776 | a | 0 |
| [(40/4 × 70/8 #5) × Katrina F2] F4 (1054) | Beit Alpha | 32 | 875 | 261 | b | 73.2 |
| [(40/4 × 70/8 #5) × Deluxs F2] F4 (1011) | Beit Alpha | 32 | 855 | 255 | b | 73.8 |
| 40/4 × 70/8 #3 (550) | Beit Alpha | 32 | 830 | 244 | b | 74.6 |
| DMR 401 F2 | Slicer | 16 | 536 | 145 | bc | 83.6 |
| [(40/4 × 70/8 #3) × DMR 401 F2] F1 | Slicer | 16 | 279 | 306 | c | 91.5 |
| PI 330628 | Sikkim | 32 | 272 | 46 | c | 91.7 |
| [(40/4 × 70/8 #3) × PI 330628] F1 | Sikkim | 8 | 256 | 274 | bc | 92.2 |
| 40/4 × 70/8 #1 (851) | Beit Alpha | 24 | 249 | 199 | c | 92.4 |
| 40/4 × 70/8 #5 (552) | Beit Alpha | 24 | 236 | 190 | c | 92.8 |
| PI 197088 | Sikkim | 16 | 92 | 96 | c | 97.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammer, R.S.; Ben Naim, Y.; Brand, A.; Cohen, Y. Transfer of Downy Mildew Resistance Genes from Wild Cucumbers to Beit Alpha Types. J. Fungi 2025, 11, 597. https://doi.org/10.3390/jof11080597
Hammer RS, Ben Naim Y, Brand A, Cohen Y. Transfer of Downy Mildew Resistance Genes from Wild Cucumbers to Beit Alpha Types. Journal of Fungi. 2025; 11(8):597. https://doi.org/10.3390/jof11080597
Chicago/Turabian StyleHammer, Rivka S., Yariv Ben Naim, Arnon Brand, and Yigal Cohen. 2025. "Transfer of Downy Mildew Resistance Genes from Wild Cucumbers to Beit Alpha Types" Journal of Fungi 11, no. 8: 597. https://doi.org/10.3390/jof11080597
APA StyleHammer, R. S., Ben Naim, Y., Brand, A., & Cohen, Y. (2025). Transfer of Downy Mildew Resistance Genes from Wild Cucumbers to Beit Alpha Types. Journal of Fungi, 11(8), 597. https://doi.org/10.3390/jof11080597






