Identification of Critical Candidate Genes Controlling Monokaryon Fruiting in Flammulina filiformis Using Genetic Population Construction and Bulked Segregant Analysis Sequencing
Abstract
1. Introduction
2. Materials and Methods
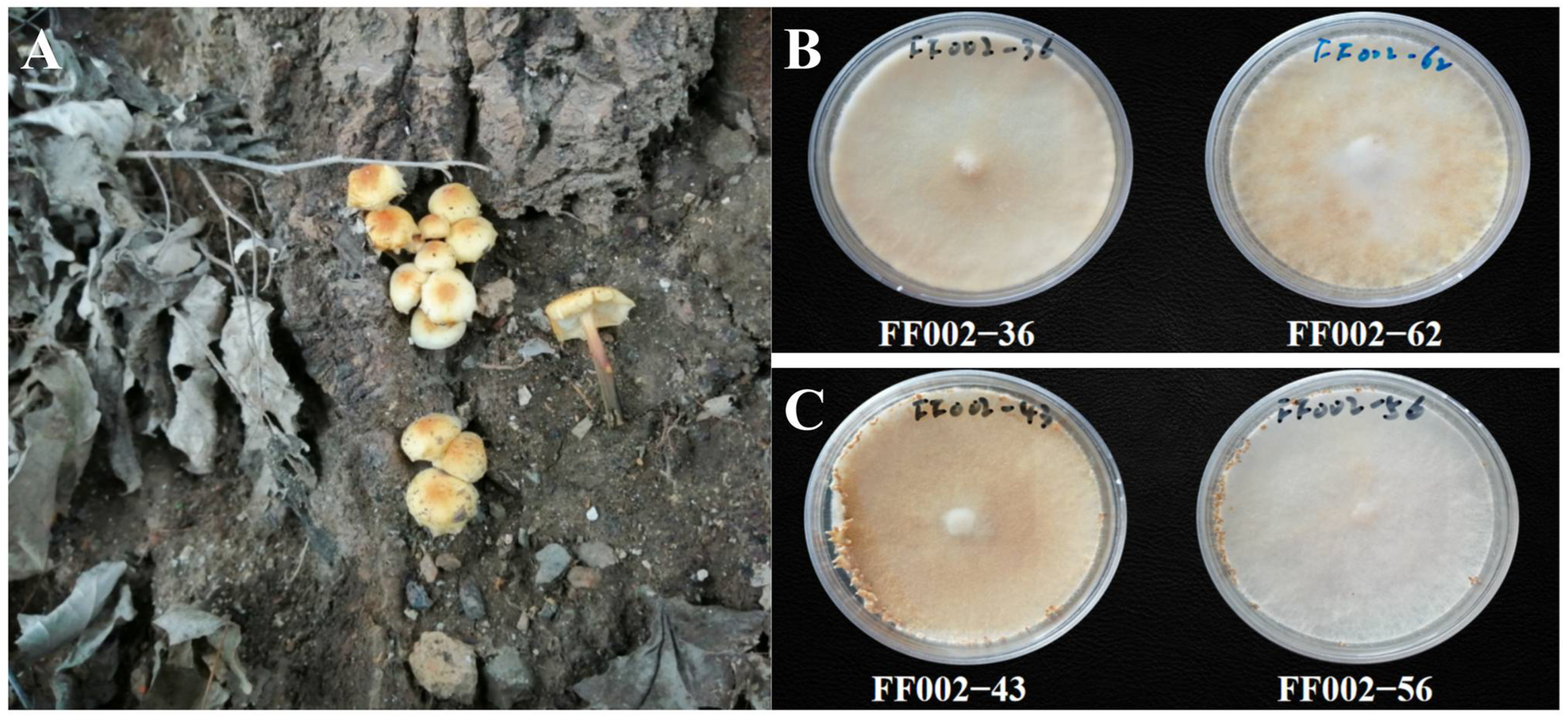
2.1. Strain Identification
2.2. Identification of Monokaryotic Fruiting Traits
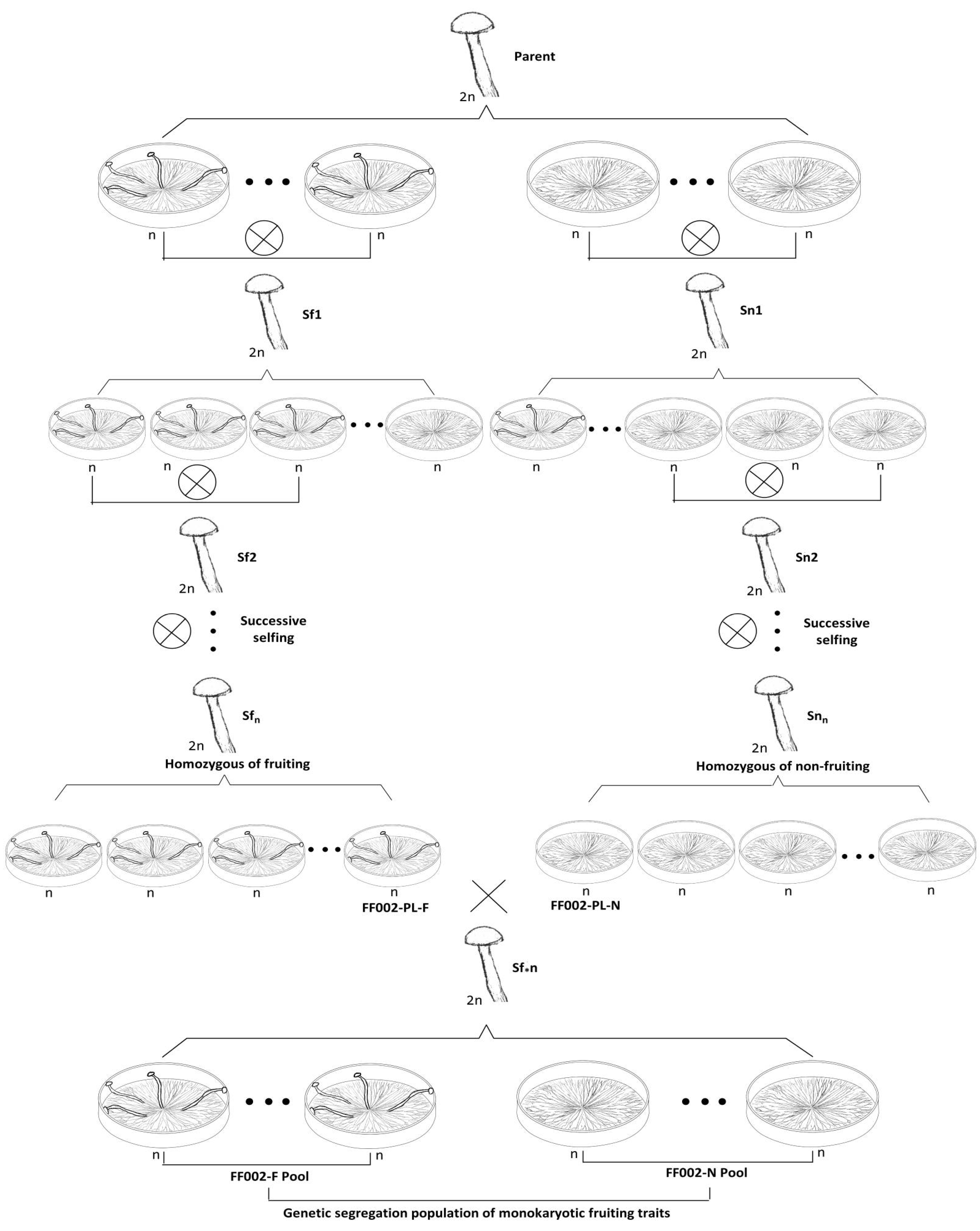
2.3. Genetic Population Construction
2.3.1. Preparation of Homozygous Strains with Monokaryotic Fruiting and Non-Fruiting Traits
2.3.2. Construction of a Genetic Segregating Population for Monokaryotic Fruiting Traits
2.4. Genomic DNA Extraction
2.5. BSA-Sequencing
2.6. Functional Annotation of Candidate Genes
2.7. Functional Validation and Phylogenetic Analysis of Target Genes
3. Results
3.1. Construction of a Genetic Segregation Population for Monokaryotic Fruiting Control Genes
3.2. Identification of Monokaryotic Fruiting Control Genes Through BSA-Seq Analysis
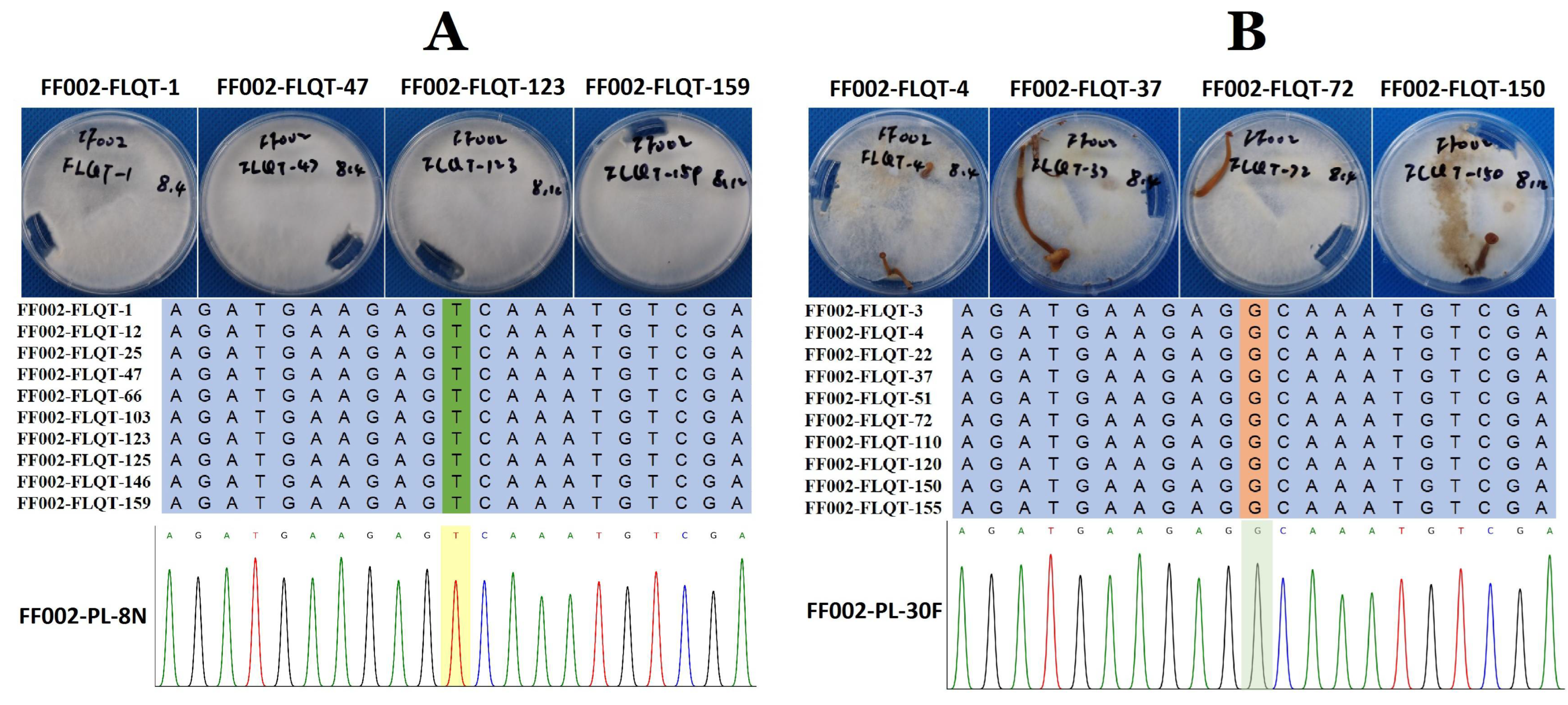
3.3. Validation of the Target Gene and Phylogenetics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Selfing Strain | Monokaryotic Fruiting Selfed Line of FF002 | Selfing Strain | Monokaryotic Non-Fruiting Selfed Line of FF002 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Selfing Generation | Selfing Combination | Segregation of Gamete Fruiting Traits | Homozygosity of Fruiting Traits | Selfing Generation | Selfing Combination | Segregation of Gamete Fruiting Traits | Homozygosity of Non-Fruiting Traits | ||
| (Fruiting: Non-Fruiting) | (Fruiting: Non-Fruiting) | ||||||||
| Sf1 | 43 × 56 | 47:0 | 100% | Sn1 | 36 × 62 | 15:38 | 71.69% | ||
| Sf2 | 1 × 4 | 39:0 | 100% | Sn2 | 5 × 14 | 0:33 | 100% | ||
| 3 × 12 | 33:0 | 100% | 10 × 11 | 8:19 | 70.37% | ||||
| 19 × 40 | 35:0 | 100% | 13 × 25 | Forming primordia but failing to form fruiting bodies | - | ||||
| Sf3 | - | - | - | Sn3 | 2 × 9 | 0:20 | 100% | ||
| 3 × 4 | 0:20 | 100% | |||||||
| 8 × 32 | 0:20 | 100% | |||||||
| SNP_id | FF002-F Type (and Read) | FF002-N_Type (and Read) | FF002-PL-8N_Type (and Read) | FF002-PL-30F_Type (and Read) | Gene Structure | Gene Function |
|---|---|---|---|---|---|---|
| scaffold19-46186-C-A | 1|1 (0|27) | 1|1 (0|45) | 1|1 (0|54) | 1|1 (0|35) | FV-L110034150 | mRNA-1:exon4:c.G863T:p.S288I |
| scaffold19-46310-T-C | 1|1 (0|25) | 1|1 (0|45) | 1|1 (0|40) | 1|1 (0|37) | FV-L110034150 | mRNA-1:exon4:c.A739G:p.N247D |
| scaffold19-46687-G-T | 1|1 (0|19) | 1|1 (0|41) | 1|1 (0|49) | 1|1 (0|35) | FV-L110034150 | mRNA-1:exon2:c.C455A:p.A152D |
| scaffold19-46698-T-G | 1|1 (0|23) | 1|1 (0|38) | 1|1 (0|49) | 1|1 (0|31) | FV-L110034150 | mRNA-1:exon2:c.A444C:p.E148D |
| scaffold19-46841-T-A | 1|1 (0|11) | 1|1 (0|20) | 1|1 (0|19) | 1|1 (0|7) | FV-L110034150 | mRNA-1:exon2:c.A301T:p.M101L |
| scaffold19-46863-G-T | 1|1 (0|5) | 1|1 (0|14) | 1|1 (0|14) | 1|1 (0|7) | FV-L110034150 | mRNA-1:exon2:c.C279A:p.S93R |
| scaffold19-46864-C-T | 1|1 (0|5) | 1|1 (0|14) | 1|1 (0|13) | 1|1 (0|7) | FV-L110034150 | mRNA-1:exon2:c.G278A:p.S93N |
| scaffold19-47775-T-G | 0|1 (21|13) | 0|1 (12|25) | 0|0 (0|63) | 1|1 (44|0) | FV-L110034160 | mRNA-1:exon3:c.T121G:p.S41A |
| scaffold19-47855-G-C | 1|1 (0|31) | 1|1 (0|32) | 1|1 (0|43) | 1|1 (0|37) | FV-L110034160 | mRNA-1:exon3:c.G201C:p.E67D |
| scaffold19-47922-T-G | 1|1 (0|19) | 1|1 (0|38) | 1|1 (1|44) | 1|1 (0|45) | FV-L110034160 | mRNA-1:exon4:c.T214G:p.L72V |
| scaffold19-51884-T-C | 1|1 (1|48) | 1|1 (0|75) | 1|1 (0|72) | 1|1 (0|72) | FV-L110034180 | mRNA-1:exon7:c.A604G:p.I202V |
| scaffold19-54000-G-C | 1|1 (0|19) | 1|1 (0|49) | 1|1 (0|43) | 1|1 (0|46) | FV-L110034190 | mRNA-1:exon6:c.C857G:p.A286G |
| Sample | Clean Reads | Clean Bases (bp) | Q20 (%) | Q30 (%) | Mapped (%) | GC (%) |
|---|---|---|---|---|---|---|
| FF002-F | 13628972 | 2044345800 | 97.19 | 92.37 | 30.54 | 49.10 |
| FF002-N | 20510422 | 3076563300 | 96.42 | 90.92 | 38.75 | 50.06 |
| FF002-PL-30F | 21785042 | 3267756300 | 96.91 | 92.89 | 37.07 | 49.70 |
| FF002-PL-8N | 17372780 | 2605917000 | 96.60 | 91.29 | 36.39 | 50.11 |
References
- Kües, U.; Liu, Y. Fruiting Body Production in Basidiomycetes. Appl. Microbiol. Biotechnol. 2000, 54, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Xiong, W.; Dong, C. Cloning and Analysis of the Oswc-1 Gene Encoding a Putative Blue Light Photoreceptor from Ophiocordyceps sinensis. Mycoscience 2014, 55, 241–245. [Google Scholar] [CrossRef]
- Eastwood, D.C.; Herman, B.; Noble, R.; Dobrovin-Pennington, A.; Sreenivasaprasad, S.; Burton, K.S. Environmental Regulation of Reproductive Phase Change in Agaricus bisporus by 1-Octen-3-ol, Temperature and CO2. Fungal Genet. Biol. 2013, 55, 54–66. [Google Scholar] [CrossRef]
- Muraguchi, H.; Kamada, T. A Mutation in the eln2 Gene Encoding a Cytochrome P450 of Coprinus cinereus Affects Mushroom Morphogenesis. Fungal Genet. Biol. 2000, 29, 49–59. [Google Scholar] [CrossRef]
- Ohm, R.A.; De Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; De Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome Sequence of the Model Mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome Sequence of the Button Mushroom Agaricus bisporus Reveals Mechanisms Governing Adaptation to a Humic-Rich Ecological Niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef] [PubMed]
- Muraguchi, H.; Umezawa, K.; Niikura, M.; Yoshida, M.; Kozaki, T.; Ishii, K.; Sakai, K.; Shimizu, M.; Nakahori, K.; Sakamoto, Y.; et al. Strand-Specific RNA-Seq Analyses of Fruiting Body Development in Coprinopsis cinerea. PLoS ONE 2015, 10, e0141586. [Google Scholar] [CrossRef]
- Yu, Y.H.; Wu, T.H.; Ye, Z.W.; Chen, B.X.; Guo, L.Q.; Lin, J.F. Transcriptional Profiling Reveals the Regulatory Network of Cold-Induced Primordium Formation in Flammulina velutipes. Acta Mycol. Sin. 2020, 39, 1065–1076. (In Chinese) [Google Scholar] [CrossRef]
- Liu, D.; Sun, X.; Diao, W.; Qi, X.; Bai, Y.; Yu, X.; Li, L.; Fang, H.; Chen, Z.; Liu, Q.; et al. Comparative Transcriptome Analysis Revealed Candidate Genes Involved in Fruiting Body Development and Sporulation in Ganoderma lucidum. Arch. Microbiol. 2022, 204, 514. [Google Scholar] [CrossRef]
- Zhang, L.; Gong, W.; Li, C.; Shi, N.; Yang, Y.; Zhao, Y.; Yuan, Y.; Qin, L.; Chen, S.; Zhou, Y. RNA-Seq-Based High-Resolution Linkage Map Reveals the Genetic Architecture of Fruiting Body Development in Shiitake Mushroom, Lentinula edodes. Comput. Struct. Biotechnol. J. 2021, 19, 1641–1653. [Google Scholar] [CrossRef]
- Quan, X.; Arshad, M.; Ya, L.; Wang, J.; Li, Y.; Song, C.; Zhang, D. Transcriptomic Profiling Revealed Important Roles of Amino Acid Metabolism in Fruiting Body Formation at Different Ripening Times in Hypsizygus marmoreus. Front. Microbiol. 2023, 14, 1169881. [Google Scholar] [CrossRef]
- Orban, A.; Weber, A.; Herzog, R.; Hennicke, F.; Rühl, M. Transcriptome of Different Fruiting Stages in the Cultivated Mushroom Cyclocybe aegerita Suggests a Complex Regulation of Fruiting and Reveals Enzymes Putatively Involved in Fungal Oxylipin Biosynthesis. BMC Genom. 2021, 22, 324. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of Markers Linked to Disease-Resistance Genes by Bulked Segregant Analysis: A Rapid Method to Detect Markers in Specific Genomic Regions by Using Segregating Populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Zou, C.; Wang, P.; Xu, Y. Bulked Sample Analysis in Genetics, Genomics and Crop Improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef]
- Pool, J.E. Genetic Mapping by Bulk Segregant Analysis in Drosophila: Experimental Design and Simulation-Based Inference. Genetics 2016, 204, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Wang, C.C.; Bian, Y.B.; Wang, Z.S.; Ma, X.L.; Zhou, Y. Advances in the Application of Bulked Segregant Analysis in Edible Fungi Genetic Research. Acta Edulis Fungi 2020, 27, 179–187. (In Chinese) [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid Mapping of Quantitative Trait Loci in Rice by Whole Genome Resequencing of DNA from Two Bulked Populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Wang, L.; Lu, Z.; Regulski, M.; Jiao, Y.; Chen, J.; Ware, D.; Xin, Z. BSA-seq: An Interactive and Integrated Web-Based Workflow for Identification of Causal Mutations in Bulked F2 Populations. Bioinformatics 2021, 37, 382–387. [Google Scholar] [CrossRef]
- Fang, X.; Sun, X.; Yang, X.; Li, Q.; Lin, C.; Xu, J.; Gong, W.; Wang, Y.; Liu, L.; Zhao LLiu, B.; et al. MS1 Is Essential for Male Fertility by Regulating the Microsporocyte Cell Plate Expansion in Soybean. Sci. China Life Sci 2021, 64, 1533–1545. [Google Scholar] [CrossRef]
- Song, Q.; Johnson, C.; Wilson, T.E.; Kumar, R.; Hyten, D.; Klepadlo, M.; Cregan, P.; Li, Z. Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth. PLoS Genet. 2014, 10, e1004570. [Google Scholar] [CrossRef]
- Wenger, J.W.; Schwartz, K.; Sherlock, G. Bulk Segregant Analysis by High-Throughput Sequencing Reveals a Novel Xylose Utilization Gene from Saccharomyces cerevisiae. PLoS Genet. 2010, 6, e1000942. [Google Scholar] [CrossRef] [PubMed]
- Swart, K.; Van der Vondervoort, P.J.I.; De Vries, R.P.; Hondel, C.A.M.J.J.; Visser, J. Parasexual Crossings for Bulk Segregant Analysis in Aspergillus niger to Facilitate Mutant Identification via Whole Genome Sequencing. Fungal Genet. Biol. 2017, 103, 1–10. [Google Scholar] [CrossRef]
- Dai, Y.C.; Yang, Z.L. New Annotations on the Scientific Names of Five Important Edible Fungi in China. Acta Mycol. Sin 2018, 37, 1572–1577. (In Chinese) [Google Scholar] [CrossRef]
- Tan, Y.; Wang, B.; Zhao, R.L. Nuclear Phase Changes in the Life Cycle of Flammulina velutipes. Acta Edulis Fungi 2015, 22, 13–19. (In Chinese) [Google Scholar] [CrossRef]
- Yi, R.R.; Cao, H.; Pan, Y.J. Monokaryotic Fruiting and Color of the Fruiting Body of Flammulina velutipes. Acta Edulis Fungi 2007, 14, 33–36. (In Chinese) [Google Scholar] [CrossRef]
- Pan, B.H.; Li, C.P. Research and Application of the Monospore Fruiting of Flammulina velutipes. China Edible Fungi 1994, 13, 21–22. (In Chinese) [Google Scholar]
- Zhang, P.; Zhang, Z.G. Research on Monokaryotic Fruiting Bodies of Flammulina velutipes. Edible Fungi China 1998, 17, 17. (In Chinese) [Google Scholar]
- Kang, Z.S.; Li, Z.Q.; Shang, H.S.; Ma, Q.; Wang, H.W. Technique of Double Fluorescence Staining for Nuclei and Septa of Plant Pathogenic Fungi. Acta Mycol. Sin. 1993, 12, 174–176. (In Chinese) [Google Scholar]
- Liu, F. Preliminary Study of Flammulina velutipes Genome and Transcriptome. Ph.D. Dissertation, Fujian Agriculture and Forestry University, Fuzhou, China, 2014. (In Chinese). [Google Scholar]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Mitsuoka, C.; Tamiru, M.; Innan, H.; et al. Genome Sequencing Reveals Agronomically Important Loci in Rice Using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The Statistics of Bulk Segregant Analysis Using Next Generation Sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Bao, D.; Gong, M.; Zheng, H.; Chen, M.; Zhang, L.; Wang, H.; Jiang, J.; Wu, L.; Zhu, Y.; Zhu, G.; et al. Transcriptomic Analysis of Flammulina velutipes Reveals Differentially Expressed Genes during Development Stages. BMC Genom. 2013, 14, 413. [Google Scholar] [CrossRef]
- Ohm, R.A.; Aerts, D.; Han, A.B.; Wösten, H.A.B. The Blue Light Receptor Complex WC-1/2 of Schizophyllum commune Is Involved in Mushroom Formation and Protection against Phototoxicity. Environ. Microbiol. 2013, 15, 943–955. [Google Scholar] [CrossRef]
- Nakazawa, T.; Tatsuta, Y.; Fujita, T.; Nakahori, K.; Kamada, T. Mutations in the Cc.rmt1 Gene Encoding a Putative Protein Arginine Methyltransferase Alter Developmental Programs in the Basidiomycete Coprinopsis cinerea. Curr. Genet. 2010, 56, 361–367. [Google Scholar] [CrossRef]
- Ando, Y.; Nakazawa, T.; Oka, K.; Nakahori, K.; Kamada, T. Cc.snf5, a Gene Encoding a Putative Component of the SWI/SNF Chromatin Remodeling Complex, Is Essential for Sexual Development in the Agaricomycete Coprinopsis cinerea. Fungal Genet. Biol. 2013, 50, 82–89. [Google Scholar] [CrossRef]
- Inada, K.; Morimoto, Y.; Arima, T.; Murata, Y.; Kamada, T. The clp1 Gene of the Mushroom Coprinus cinereus Is Essential for A-Regulated Sexual Development. Genetics 2001, 157, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Boontawon, T.; Nakazawa, T.; Horii, M.; Ohnuki, S.; Nakamoto, M.; Sakamoto, M.; Inoue, Y.; Izumitsu, K.; Shigenobu, S.; Kaneko, S.; et al. Functional Analyses of Pleurotus ostreatus pcc1 and clp1 Using CRISPR/Cas9. Fungal Genet. Biol. 2021, 154, 103599. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Jiang, S.; Wang, L.; Chou, T.; Wang, Q.; Meng, L.; Mukhtar, I.; Xie, B.; Wang, W. The Fvclp1 Gene Regulates Mycelial Growth and Fruiting Body Development in Edible Mushroom Flammulina velutipes. Arch. Microbiol. 2021, 203, 5373–5380. [Google Scholar] [CrossRef]
- Muraguchi, H.; Kamada, T. The ich1 Gene of the Mushroom Coprinus cinereus Is Essential for Pileus Formation in Fruiting. In Proceedings of the Annual Meeting of the Japanese Society for Molecular Biology, Nagoya, Japan, 18 December 1998; p. 3133. [Google Scholar]
- Schuren, F.H.J. Atypical Interactions between thn and Wild-Type Mycelia of Schizophyllum commune. Mycol. Res. 1999, 103, 1540–1544. [Google Scholar] [CrossRef]
- Stahl, U.; Esser, K. Genetics of Fruit Body Production in Higher Basidiomycetes. I. Monokaryotic Fruiting and Its Correlation with Dikaryotic Fruiting in Polyporus ciliatus. Mol. Gen. Genet. 1976, 148, 183–197. [Google Scholar] [CrossRef]
- Huang, Y.D. Research on the Polarity of Wood Ear Fungus. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2004. (In Chinese). [Google Scholar]
- Bao, D.P.; Wang, N.; Tan, Q.; Pan, Y.J. Research Progress on the Cultivation Physiology and Life Cycle of Pleurotus eryngii. Acta Edulis Fungi 1999, 6, 55–60. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, J.J.; Liu, J.J.; Liu, N.; Li, H. Research on the Morphology of Monokaryotic Mycelium and Fruiting of Pleurotus ostreatus. Acta Agric. Boreali-Sin. 2011, 26, 219–222. (In Chinese) [Google Scholar] [CrossRef]
- Peng, W.; Yao, F.J.; Lu, L.X.; Wang, Z.S.; Li, Y.; Wu, X.L. Map-Based Cloning of Genes Encoding Key Enzymes for Pigment Synthesis in Auricularia cornea. Fungal Biol. 2019, 123, 843–853. [Google Scholar] [CrossRef]
- Ma, X.X.; Lu, L.X.; Zhang, Y.M.; Yao, F.J.; Li, Y.; Wu, X.L. Velvet Family Members Regulate Pigment Synthesis of the Fruiting Bodies of Auricularia cornea. J. Fungi 2023, 9, 412. [Google Scholar] [CrossRef]
- Liu, F.; Ma, X.B.; Han, B.; Wang, B.; Xu, J.P.; Cao, B.; Ling, Z.L.; He, M.Q.; Zhu, X.Y.; Zhao, R.-L. Pan-genome analysis reveals genomic variations during enoki mushroom domestication, with emphasis on genetic signatures of cap color and stipe length. J. Adv. Res. 2024; in press. [Google Scholar] [CrossRef]
- van der Feltz, C.; Anthony, K.; Brilot, A.; Pomeranz Krummel, D.A. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 327–347. [Google Scholar] [CrossRef]
- Grützmann, K.; Sasso, S.; Deshpande, A.M.; Schöler, F.; Lohse, M.B.; Nowrousian, M. The occurrence and function of alternative splicing in fungi. Trends Genet. 2014, 30, 6–13. [Google Scholar] [CrossRef]
- Pelloth, C.; Schafer, K.; Sparber, F.; Převorovský, M.; Hogan, D.A.; Schüller, C. The spliceosome impacts morphogenesis in the human fungal pathogen Candida albicans. mBio 2023, 14, e00409-23. [Google Scholar] [CrossRef]
- Gottschalk, A.; Bartels, C.; Neubauer, G.; Luhrmann, R.; Fabrizio, P. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. MCB 2001, 21, 3037–3046. [Google Scholar] [CrossRef][Green Version]
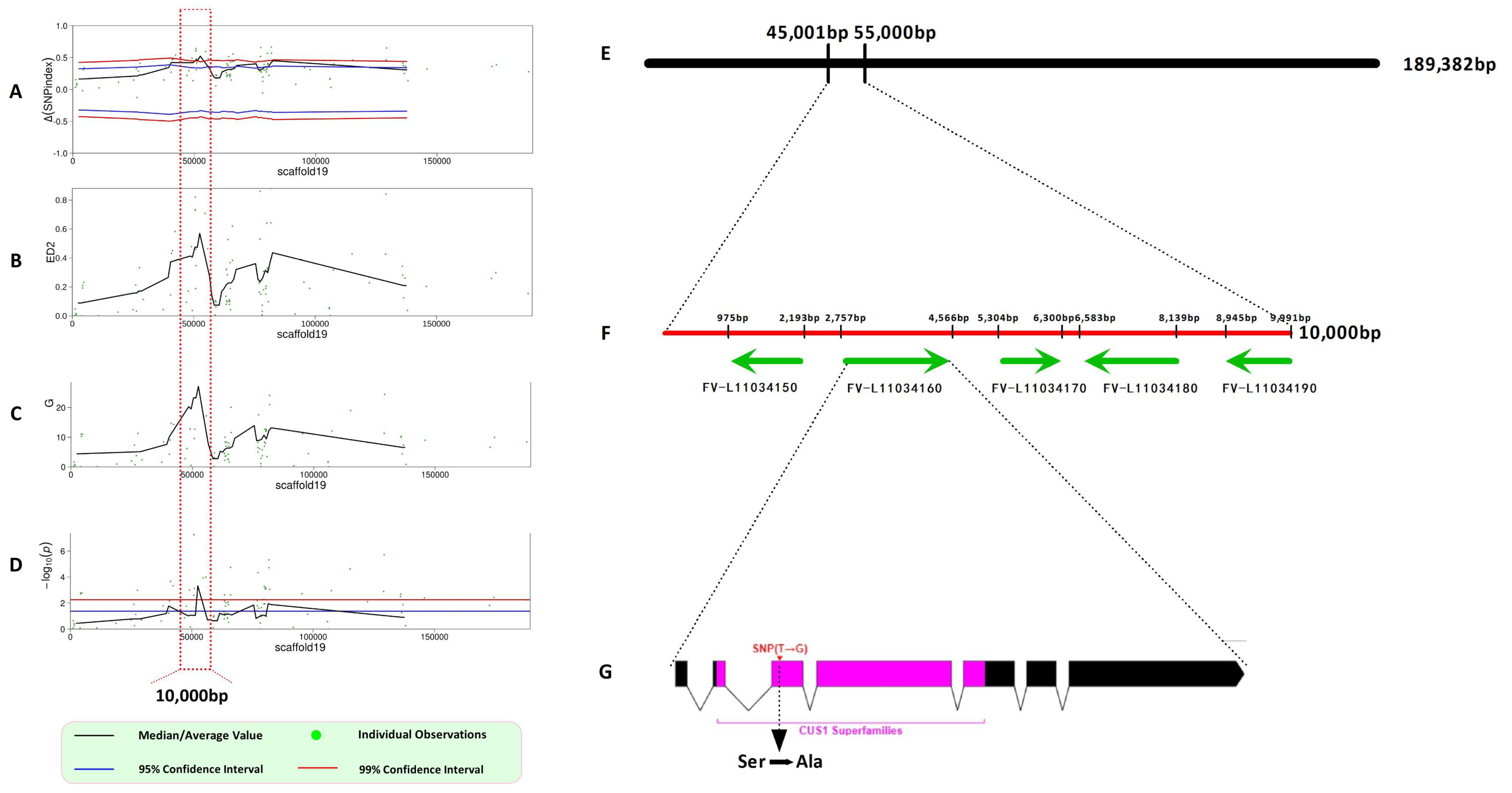

| Gene ID | Location | PFAMs | Pathway | Function Description |
|---|---|---|---|---|
| FV-L110034150 | Scaffold19 (45,969–47,338) | ASXH | - | Asx homology domain |
| FV-L110034160 | Scaffold19 (47,374–49,566) | DUF382 (CUS1) | ko03040 (pentose phosphate pathway) | DUF382-domain-containing protein |
| FV-L110034170 | Scaffold19 (49,778–51,300) | BCDHK_Adom3, HATPase_c | - | Dehydrogenase |
| FV-L110034180 | Scaffold19 (51,399–52,780) | 3Beta_HSD, Epimerase, NAD_binding_4 | - | Dihydrokaempferol 4-reductase activity |
| FV-L110034190 | Scaffold19 (53,799–55,299) | HAD_2 | ko00561 (glycerolipid metabolism), ko01100 (metabolic pathways) | HAD-hyrolase-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Yu, Y.; Xia, L.; Yan, Q.; Tan, X.; Wang, D.; Wang, X.; Zhang, Z.; Wen, J.; Huang, X. Identification of Critical Candidate Genes Controlling Monokaryon Fruiting in Flammulina filiformis Using Genetic Population Construction and Bulked Segregant Analysis Sequencing. J. Fungi 2025, 11, 512. https://doi.org/10.3390/jof11070512
Wang P, Yu Y, Xia L, Yan Q, Tan X, Wang D, Wang X, Zhang Z, Wen J, Huang X. Identification of Critical Candidate Genes Controlling Monokaryon Fruiting in Flammulina filiformis Using Genetic Population Construction and Bulked Segregant Analysis Sequencing. Journal of Fungi. 2025; 11(7):512. https://doi.org/10.3390/jof11070512
Chicago/Turabian StyleWang, Peng, Ya Yu, Lei Xia, Qi Yan, Xiao Tan, Dongyin Wang, Xue Wang, Zhibin Zhang, Jiawei Wen, and Xiao Huang. 2025. "Identification of Critical Candidate Genes Controlling Monokaryon Fruiting in Flammulina filiformis Using Genetic Population Construction and Bulked Segregant Analysis Sequencing" Journal of Fungi 11, no. 7: 512. https://doi.org/10.3390/jof11070512
APA StyleWang, P., Yu, Y., Xia, L., Yan, Q., Tan, X., Wang, D., Wang, X., Zhang, Z., Wen, J., & Huang, X. (2025). Identification of Critical Candidate Genes Controlling Monokaryon Fruiting in Flammulina filiformis Using Genetic Population Construction and Bulked Segregant Analysis Sequencing. Journal of Fungi, 11(7), 512. https://doi.org/10.3390/jof11070512





