Seven New Series and Four New Species in Sections Subinflati and Trachyspermi of Talaromyces (Trichocomaceae, Eurotiales)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Materials
2.2. Morphological Observations
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Phylogenetic Analyses
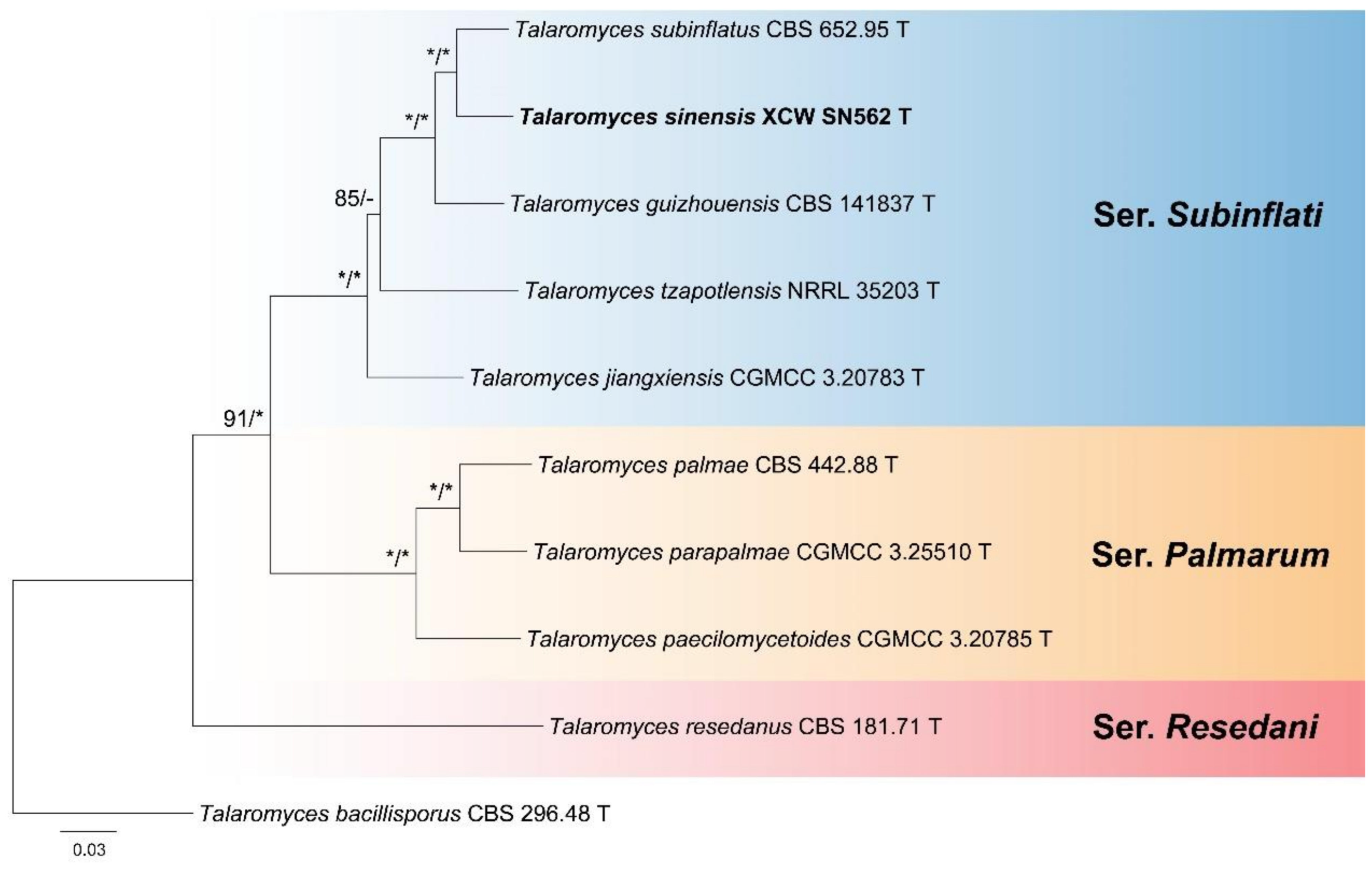
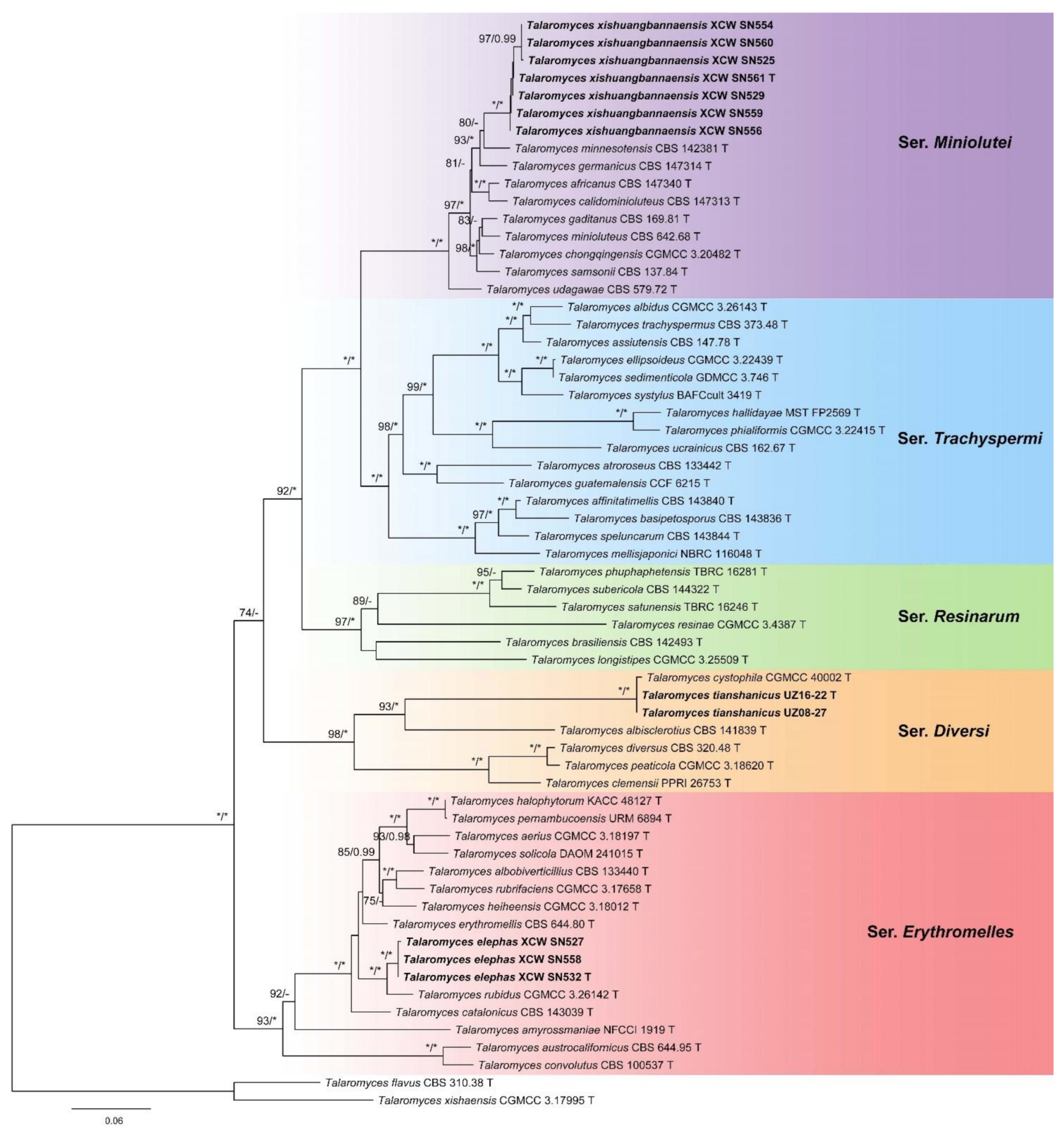
3. Results
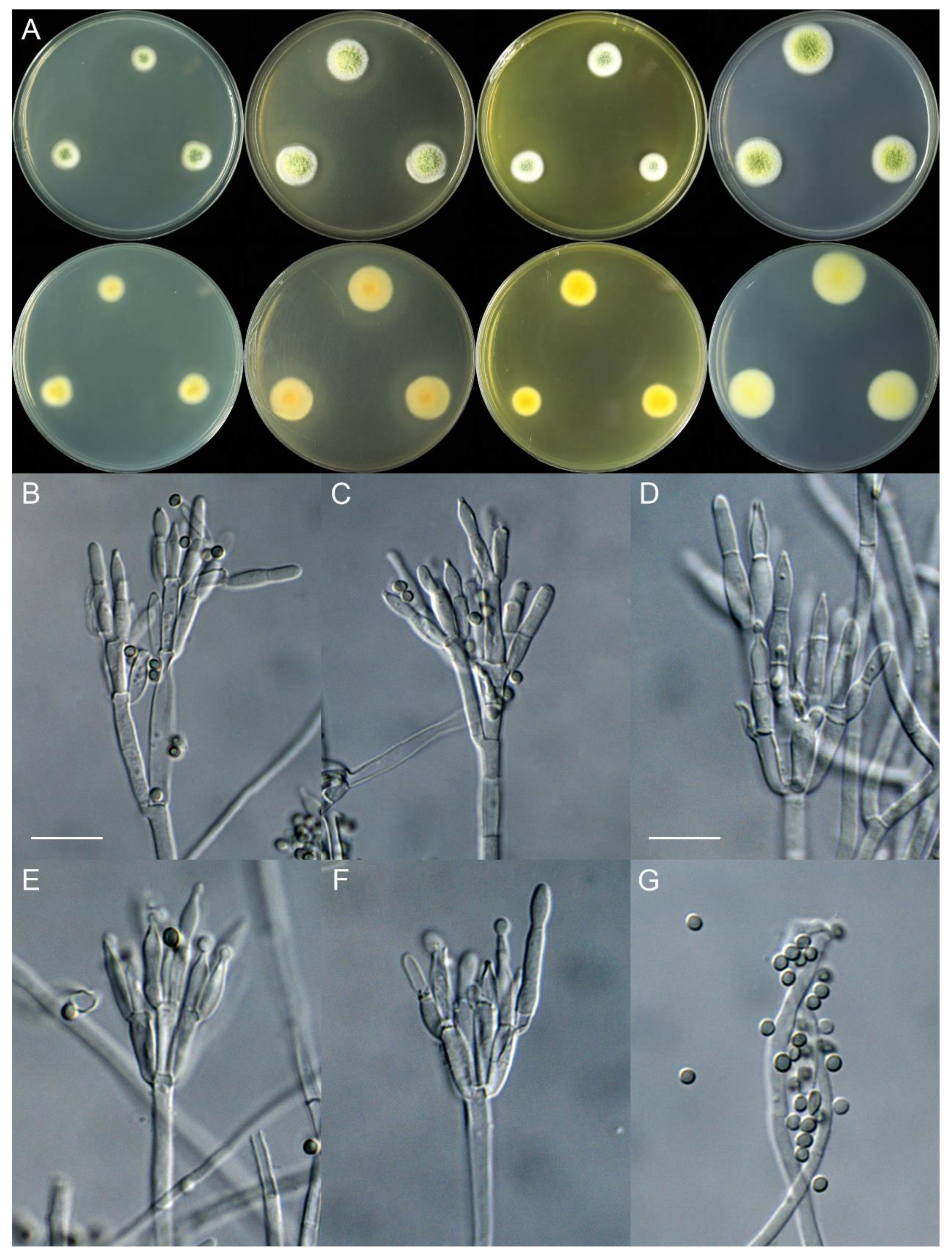
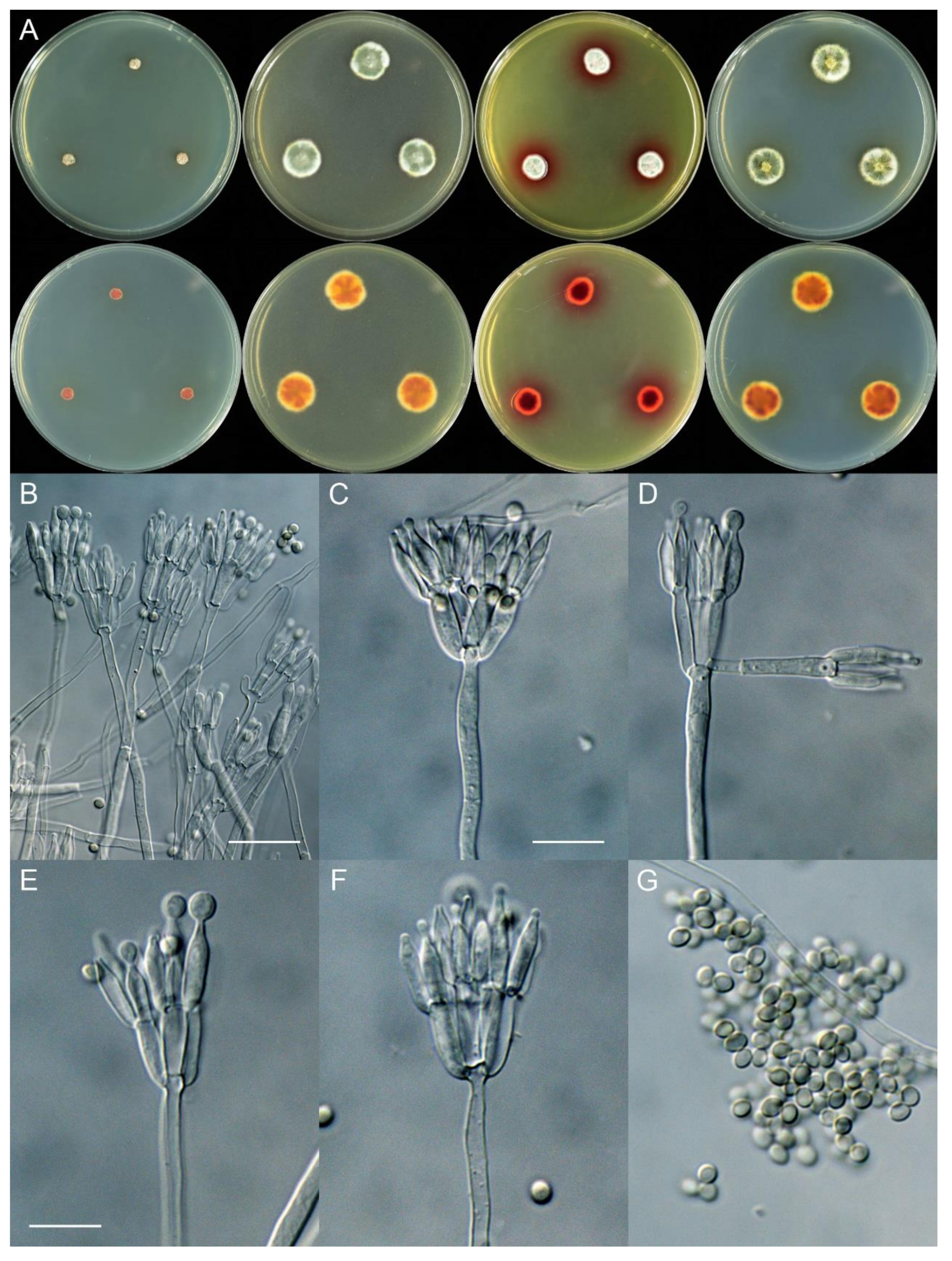
4. Taxonomy
4.1. New Series
4.2. Emended Series
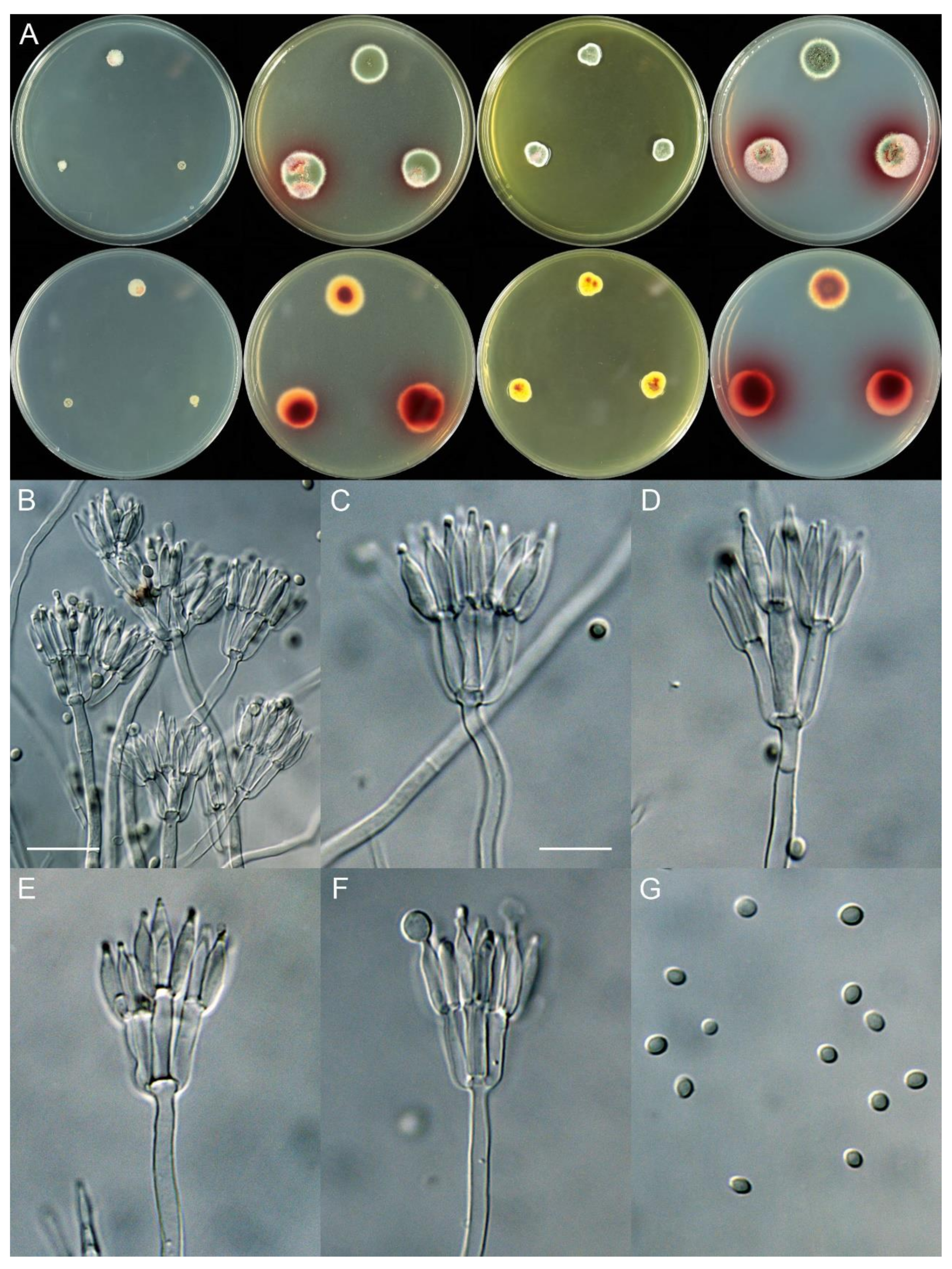
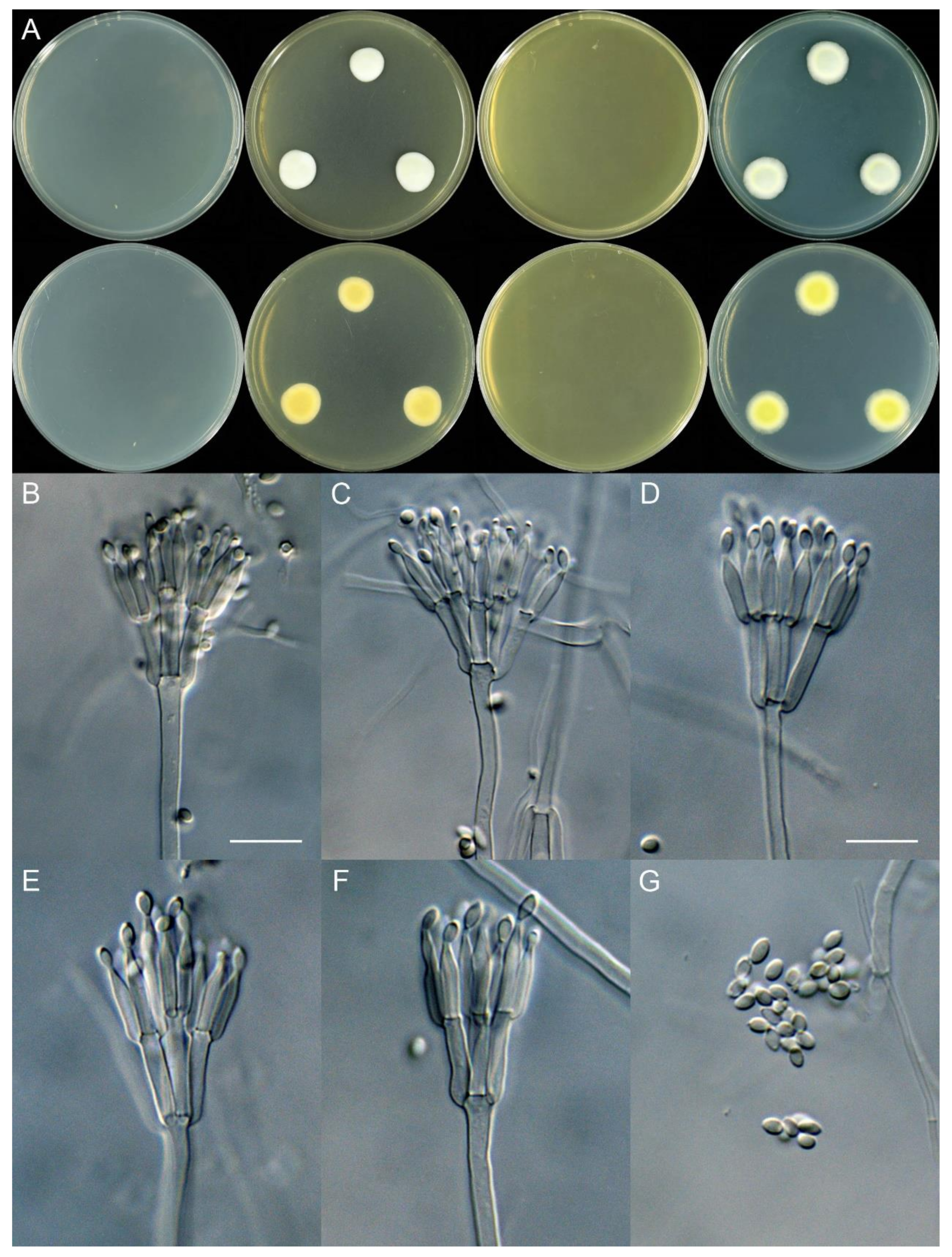
4.3. New Species
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Z.; Chen, J.; Ni, D.; Xu, W.; Zhang, W.; Mu, W. Microbial dextran-hydrolyzing enzyme: Properties, structural features, and versatile applications. Food Chem. 2024, 437, 137951. [Google Scholar] [CrossRef]
- Zhgun, A.A. Industrial Production of Antibiotics in Fungi: Current state, deciphering the molecular basis of classical strain improvement and increasing the production of high-yielding strains by the addition of low-molecular weight inducers. Fermentation 2023, 9, 1027. [Google Scholar] [CrossRef]
- Lacey, A.E.; Minns, S.A.; Chen, R.; Vuong, D.; Lacey, E.; Kalaitzis, J.A.; Tan, Y.P.; Shivas, R.G.; Butler, M.S.; Piggott, A.M. Talcarpones A and B: Bisnaphthazarin-derived metabolites from the Australian fungus Talaromyces johnpittii sp. nov. MST-FP2594. J. Antibiot. 2024, 77, 147–155. [Google Scholar] [CrossRef]
- Umar, A.; Darwish, D.B.E.; Alenezi, M.A. Fungal pigments: Secondary metabolites and their application. In Fungal Secondary Metabolites: Synthesis and Applications in Agroecosystem; Abd-Elsalam, K.A., Mohamed, H.I., Eds.; Elsevier: Amsterdam, The Netherlands, 2024; pp. 173–195. [Google Scholar]
- Wang, F.; Han, R.; Chen, S. An overlooked and underrated endemic mycosis—Talaromycosis and the pathogenic fungus Talaromyces marneffei. Clin. Microbiol. Rev. 2023, 36, e0005122. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsube, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar]
- Wang, X.C.; Zhuang, W.Y. New species of Talaromyces (Trichocomaceae, Eurotiales) from southwestern China. J. Fungi 2022, 8, 647. [Google Scholar] [CrossRef]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef]
- Sun, B.D.; Chen, A.J.; Houbraken, J.; Frisvad, J.C.; Wu, W.P.; Wei, H.L.; Zhou, Y.G.; Jiang, X.Z.; Samson, R.A. New section and species in Talaromyces. MycoKeys 2020, 68, 75–113. [Google Scholar] [CrossRef]
- Stolk, A.C.; Samson, R.A. The genus Talaromyces. Studies on Talaromyces and related genera. II. Stud. Mycol. 1972, 2, 1–65. [Google Scholar]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Wang, X.C.; Zhuang, W.Y.; Cheng, X.H.; Zhao, P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology 2021, 10, 745. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhuang, W.Y. New species of Aspergillus (Aspergillaceae) from tropical islands of China. J. Fungi 2022, 8, 225. [Google Scholar] [CrossRef]
- Wang, X.C.; Zhang, Z.K.; Zhuang, W.Y. Species diversity of Penicillium in Southwest China with discovery of forty-three new species. J. Fungi 2023, 9, 1150. [Google Scholar] [CrossRef]
- Song, H.; Ding, Y.J.; Zhuang, W.Y.; Ding, G.Z.; Wang, X.C. Three new species of Penicillium from East and Northeast China. J. Fungi 2024, 10, 342. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Hong, S.B.; Cho, H.S.; Shin, H.D.; Frisvad, J.C.; Samson, R.A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 477–486. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Guevara-Suarez, M.; Sutton, D.A.; Gene, J.; Garcia, D.; Wiederhold, N.; Guarro, J.; Cano-Lira, J.F. Four new species of Talaromyces from clinical sources. Mycoses 2017, 60, 651–662. [Google Scholar] [CrossRef]
- Pyrri, I.; Visagie, C.M.; Soccio, P.; Houbraken, J. Re-evaluation of the taxonomy of Talaromyces minioluteus. J. Fungi 2021, 7, 993. [Google Scholar] [CrossRef]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press Inc: London, UK, 1979; p. 634. [Google Scholar]
- Li, M.; Raza, M.; Song, S.; Hou, L.; Zhang, Z.F.; Gao, M.; Huang, J.E.; Liu, F.; Cai, L. Application of culturomics in fungal isolation from mangrove sediments. Microbiome 2023, 11, 272. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, L.; Liu, W.; Ta, K.; Wang, X.; Guo, J.; Luo, W.; Peng, Z.; Huang, Q.; Wang, Y. Talaromyces sedimenticola sp. nov., isolated from the Mariana Trench. Antonie Van Leeuwenhoek 2024, 117, 44. [Google Scholar] [CrossRef]
- Mo, Y.X.; Kan, Y.Z.; Jia, L.M.; Cao, X.T.; Sikandar, A.; Wu, H.Y. Characterization and effect of a nematophagous fungus Talaromyces cystophila sp. nov. for the biological control of corn cyst nematode. Phytopathology 2024, 114, 618–629. [Google Scholar] [CrossRef]
- Zang, W.; Li, M.; Sun, J.; Gao, C.; Wang, L. Two new species of Talaromyces sect. Trachyspermi discovered in China. Mycopathologia 2023, 188, 793–804. [Google Scholar] [CrossRef]
- Stolk, A.C.; Samson, R.A. A new taxonomic scheme for Penicillium anamorphs. In Advances in Penicillium and Aspergillus Systematics; Samson, R.A., Pitt, J.I., Eds.; Springer: New York, NY, USA, 1986; pp. 163–192. [Google Scholar]
- Visagie, C.M.; Yilmaz, N.; Kocsubé, S.; Frisvad, J.C.; Hubka, V.; Samson, R.A.; Houbraken, J. A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Stud. Mycol. 2024, 107, 1–66. [Google Scholar] [CrossRef]
- Lima, J.M.S.; Barbosa, R.N.; Bento, D.M.; Barbier, E.; Bernard, E.; Bezerra, J.D.P.; Souza-Motta, C.M. Aspergillus, Penicillium, and Talaromyces (Eurotiales) in Brazilian caves, with the description of four new species. Fung. Syst. Evol. 2024, 14, 89–107. [Google Scholar] [CrossRef]
- Zárate-Ortiz, A.I.; Villarruel-Ordaz, J.L.; Sánchez-Espinosa, A.C.; Maldonado-Bonilla, L.D. Talaromyces oaxaquensis sp. nov. section Talaromyces (Trichocomaceae, Eurotiales) isolated from pseudostems of banana plants in Mexico. Sci. Fung. 2024, 55, e1457. [Google Scholar] [CrossRef]
| Species | Strain | Country | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. guizhouensis B.D. Sun et al. 2020 | CBS 141837 T | China: Guizhou | Soil | MN864277 | MN863346 | MN863323 | MN863335 |
| T. jiangxiensis Zhi Y. Zhang et al. 2023 | CGMCC 3.20783 T | China: Jiangxi | Soil | OL897029 | ON569044 | ON568888 | ON568963 |
| T. paecilomycetoides Zhi Y. Zhang et al. 2023 | CGMCC 3.20785 T | China: Yunnan | Soil | OL897033 | ON569040 | ON568890 | ON568959 |
| T. palmae (Samson et al.) Samson et al. 2011 | CBS 442.88 T | Netherlands | Seeds of Chrysalidocarpus lutescens | JN899396 | HQ156947 | KJ885291 | KM023300 |
| T. parapalmae Zhi Y. Zhang & Y.F. Han 2024 | CGMCC 3.25510 T | China: Guizhou | Soil | OR680520 | OR843225 | OR828456 | OR842937 |
| T. resedanus (McLennan & Ducker) A.J. Chen et al. 2020 | CBS 181.71 T | Australia | Acid, sandy soil | MN431413 | MN969436 | MN969355 | MN969214 |
| T. sinensis X.C. Wang, L.Y. Peng & W.Y. Zhuang, sp. nov. | XCW_SN 562 = CGMCC 3.28744 T | China: Yunnan | Rotten husk of an unknown fruit | PV085755 | PV102705 | PV102718 | PV102726 |
| T. subinflatus Yaguchi & Udagawa 1993 | CBS 652.95 T | Japan | Soil | JN899397 | MK450890 | KJ885280 | KM023308 |
| T. tzapotlensis Jurjević & S.W. Peterson 2017 | NRRL 35203 T | Mexico | Hypothenemus hampei | KX946902 | KX946884 | KX946893 | KX946922 |
| T. bacillisporus (Swift) C.R. Benj. 1955 | CBS 296.48 T | North America | Leaves of Begonia | KM066182 | AY753368 | KJ885262 | JF417425 |
| Species | Strain | Country | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aerius A.J. Chen et al. 2016 | CGMCC 3.18197 T | China: Beijing | Indoor air | KU866647 | KU866835 | KU866731 | KU866991 |
| T. affinitatimellis Rodr.-Andr. et al. 2019 | CBS 143840 T | Spain | Honey | LT906543 | LT906552 | LT906549 | LT906546 |
| T. africanus Houbraken et al. 2021 | CBS 147340 T | South Africa | House dust | OK339610 | OK338782 | OK338808 | OK338833 |
| T. albidus L. Wang 2023 | CGMCC 3.26143 T | China: Shanghai | Soil | OQ746343 | OQ746324 | OQ746326 | OQ746328 |
| T. albisclerotius B.D. Sun et al. 2020 | CBS 141839 T | China: Xinjiang | Soil | MN864276 | MN863345 | MN863322 | MN863334 |
| T. albobiverticillius (H.M. Hsieh et al.) Samson et al. 2011 | CBS 133440 T | China: Taiwan | Fallen decaying leaves | HQ605705 | KF114778 | KJ885258 | KM023310 |
| T. amyrossmaniae Rajeshkumar et al. 2019 | NFCCI 1919 T | India | Decaying fruits of Terminalia bellerica | MH909062 | MH909064 | MH909068 | MH909066 |
| T. assiutensis Samson & Abdel-Fattah 1978 | CBS 147.78 T | Egypt | Soil | JN899323 | KJ865720 | KJ885260 | KM023305 |
| T. atroroseus N. Yilmaz et al. 2013 | CBS 133442 T | South Africa | House dust | KF114747 | KF114789 | KJ775418 | KM023288 |
| T. austrocalifornicus Yaguchi & Udagawa 1993 | CBS 644.95 T | USA | Soil | JN899357 | KJ865732 | KJ885261 | MN969147 |
| T. basipetosporus Stchigel et al. 2019 | CBS 143836 T | Argentina | Honey | LT906542 | LT906563 | n.a. | LT906545 |
| T. brasiliensis R.N. Barbosa et al. 2018 | CBS 142493 T | Brazil | Honey | MF278323 | LT855560 | LT855563 | MN969198 |
| T. calidominioluteus Houbraken & Pyrri 2021 | CBS 147313 T | Netherlands | Melon | OK339612 | OK338786 | OK338817 | OK338837 |
| T. catalonicus Guevara-Suarez et al. 2020 | CBS 143039 T | Spain | Herbivore dung | LT899793 | LT898318 | LT899775 | LT899811 |
| T. chongqingensis X.C. Wang & W.Y. Zhuang 2021 | CGMCC 3.20482 T | China: Chongqing | Soil | MZ358001 | MZ361343 | MZ361350 | MZ361357 |
| T. clemensii Visagie & N. Yilmaz 2019 | PPRI 26753 T | South Africa | Rotting wood in mine | MK951940 | MK951833 | MK951906 | MN418451 |
| T. convolutus Udawaga 1993 | CBS 100537 T | Nepal | Soil | JN899330 | KF114773 | MN969316 | JN121414 |
| T. cystophila Y.X. Mo & H.Y. Wu 2024 | CGMCC 40002 T | China: Guangxi | Heterodera zeae cyst | OM835900 | ON164851 | ON164853 | ON164852 |
| T. diversus (Raper & Fennell) Samson et al. 2011 | CBS 320.48 T | USA | Moldy leather | KJ865740 | KJ865723 | KJ885268 | KM023285 |
| T. elephas X.C. Wang, L.Y. Peng & W.Y. Zhuang, sp. nov. | XCW_SN 532 = CGMCC 3.28742 T | China: Yunnan | Rotten husk of an unknown fruit | PV085756 | PV102706 | PV102719 | PV102727 |
| XCW_SN 527 | China: Yunnan | Rotten husk of an unknown fruit | PV085757 | PV102707 | PV102720 | PV102728 | |
| XCW_SN 558 | China: Yunnan | Rotten husk of an unknown fruit | PV085758 | PV102708 | n.a. | PV102729 | |
| T. ellipsoideus M. Li & L. Cai 2023 | CGMCC 3.22439 T | China: Guangdong | Sediment | OQ798985 | OQ808981 | OQ808994 | OQ809036 |
| T. erythromellis (A.D. Hocking) Samson et al. 2011 | CBS 644.80 T | Australia | Soil | JN899383 | HQ156945 | KJ885270 | KM023290 |
| T. gaditanus (C. Ramírez & A.T. Martínez) Houbraken & Soccio 2021 | CBS 169.81 T | Spain | Air | MH861318 | OK338775 | OK338802 | OK338827 |
| T. germanicus Houbraken & Pyrri 2021 | CBS 147314 T | Germany | Wallboard | OK339619 | OK338799 | OK338812 | OK338845 |
| T. guatemalensis A. Nováková et al. 2019 | CCF 6215 T | Guatemala | Soil | MN322789 | MN329687 | MN329688 | MN329689 |
| T. hallidayae Y.P. Tan et al. 2024 | MST FP2569 T | Australia | Soil under turf grass | PP665728 | PP682580 | PP682551 | PP682567 |
| T. halophytorum Y.H. You & S.B. Hong 2020 | KACC 48127 T | South Korea | Roots of Limonium tetragonum | MH725786 | MH729367 | MK111426 | MK111427 |
| T. heiheensis X.C. Wang & W.Y. Zhuang 2017 | CGMCC 3.18012 T | China: Heilongjiang | Rotten wood | KX447526 | KX447525 | KX447532 | KX447529 |
| T. longistipes Zhi Y. Zhang & Y.F. Han 2024 | CGMCC 3.25509 T | China: Guizhou | Soil | OR680518 | OR843223 | OR828454 | OR842935 |
| T. mellisjaponici A. Okubo & D. Hirose 2024 | NBRC 116048 T | Japan | Honey | LC763421 | LC763430 | LC763439 | LC763448 |
| T. minioluteus (Dierckx) Samson et al. 2011 | CBS 642.68 T | Unknown | Unknown | JN899346 | MN969409 | KJ885273 | JF417443 |
| T. minnesotensis Guevara-Suarez et al. 2017 | CBS 142381 T | USA | Human ear | LT558966 | LT559083 | LT795604 | LT795605 |
| T. peaticola J.Q. Tian & J.Z. Sun 2021 | CGMCC 3.18620 T | China: Sichuan | Peaty soil of wetland | MF135613 | MF284705 | MF284703 | MF284704 |
| T. pernambucoensis R. Cruz et al. 2019 | URM 6894 T | Brazil | Soil | LR535947 | LR535945 | LR535946 | LR535948 |
| T. phialiformis M. Li & L. Cai 2023 | CGMCC 3.22415 T | China: Guangdong | Mangrove sediment | OQ798986 | OQ808982 | OQ808995 | n.a. |
| T. phuphaphetensis Nuankaew et al. 2022 | TBRC 16281 T | Thailand | Soil in cave | ON692803 | ON706960 | ON706962 | ON706964 |
| T. resinae (Z.T. Qi & H.Z. Kong) Houbraken & X.C. Wang 2020 | CGMCC 3.4387 T | China: Guizhou | Resin of Eucalyptus tereticornis | MT079858 | MN969442 | MT066184 | MN969221 |
| T. rubidus L. Wang 2023 | CGMCC 3.26142 T | China: Yunnan | Soil | OQ746342 | OQ746323 | OQ746325 | OQ746327 |
| T. rubrifaciens W.W. Gao 2016 | CGMCC 3.17658 T | China: Beijing | Hospital air | KR855658 | KR855648 | KR855653 | KR855663 |
| T. samsonii (Quintan.) Houbraken & Pyrri 2021 | CBS 137.84 T | Spain | Insect-damaged fruit of Malus pumila | MH861709 | OK338798 | OK338824 | OK338844 |
| T. satunensis Nuankaew et al. 2022 | TBRC 16246 T | Thailand | Soil in cave | ON692804 | ON706961 | ON706963 | n.a. |
| T. sedimenticola Y. Wang & H. Zhou 2024 | GDMCC 3.746 T | Mariana Trench | Sediment | ON000284 | ON384002 | ON326484 | ON000277 |
| T. solicola Visagie & K. Jacobs 2012 | DAOM 241015 T | South Africa | Fynbos soil | FJ160264 | GU385731 | KJ885279 | KM023295 |
| T. speluncarum Rodr.-Andr. et al. 2020 | CBS 143844 T | Spain | Sparkling wine | LT985890 | LT985901 | LT985906 | LT985911 |
| T. subericola Rodr.-Andr. et al. 2020 | CBS 144322 T | Spain | Sparkling wine | LT985888 | LT985899 | LT985904 | LT985909 |
| T. systylus S.M.Romero et al. 2016 | BAFCcult 3419 T | Argentina | Soil | KP026917 | KR233838 | KR233837 | n.a. |
| T. tianshanicus X.C. Wang, L.Y. Peng, Gafforov & W.Y. Zhuang, sp. nov. | UZ16-22 = CGMCC 3.28741 T | Uzbekistan | Soil | PV085759 | PV102709 | n.a. | PV102730 |
| UZ08-27 | Uzbekistan | Soil | PV085760 | PV102710 | PV102721 | n.a. | |
| T. trachyspermus (Shear) Stolk & Samson 1972 | CBS 373.48 T | USA | Unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
| T. ucrainicus (Panas.) Udagawa 1966 | CBS 162.67 T | Japan | Soil | JN899394 | KF114771 | KJ885282 | KM023289 |
| T. udagawae Stolk & Samson 1972 | CBS 579.72 T | Japan | Soil | JN899350 | OK338783 | KX961260 | MN969148 |
| T. xishuangbannaensis X.C. Wang, L.Y. Peng & W.Y. Zhuang, sp. nov. | XCW_SN 561 = CGMCC 3.28743 T | China: Yunnan | Rotten husk of an unknown fruit | PV085761 | PV102711 | PV102722 | PV102731 |
| XCW_SN 525 | China: Yunnan | Rotten husk of an unknown fruit | PV085762 | PV102712 | n.a. | PV102732 | |
| XCW_SN 529 | China: Yunnan | Rotten husk of an unknown fruit | PV085763 | PV102713 | PV102723 | PV102733 | |
| XCW_SN 554 | China: Yunnan | Rotten husk of an unknown fruit | PV085764 | PV102714 | n.a. | PV102734 | |
| XCW_SN 556 | China: Yunnan | Rotten husk of an unknown fruit | PV085765 | PV102715 | PV102724 | PV102735 | |
| XCW_SN 559 | China: Yunnan | Rotten husk of an unknown fruit | PV085766 | PV102716 | PV102725 | PV102736 | |
| XCW_SN 560 | China: Yunnan | Rotten husk of an unknown fruit | PV085767 | PV102717 | n.a. | PV102737 | |
| T. flavus (Klöcker) Stolk & Samson 1972 | CBS 310.38 T | New Zealand | Unknown | JN899360 | JX494302 | KF741949 | JF417426 |
| T. xishaensis X.C. Wang et al. 2016 | CGMCC 3.17995 T | China: Hainan | Soil | KU644580 | KU644581 | KU644582 | MZ361364 |
| Dataset | Gene Fragment | No. of Seq. | Length of Alignment (bp) | No. of Variable Sites | No. of Parsimony-Informative Sites | Model for BI |
|---|---|---|---|---|---|---|
| Subinflati | ITS | 10 | 518 | 129 | 68 | |
| BenA | 10 | 383 | 131 | 65 | ||
| CaM | 10 | 532 | 234 | 141 | ||
| RPB2 | 10 | 1008 | 287 | 166 | ||
| BenA + CaM + RPB2 | 10 | 1923 | 652 | 372 | TrNef + I + G | |
| Trachyspermi | ITS | 62 | 626 | 218 | 162 | |
| BenA | 62 | 590 | 290 | 221 | ||
| CaM | 56 | 658 | 398 | 311 | ||
| RPB2 | 58 | 917 | 349 | 314 | ||
| BenA + CaM + RPB2 | 62 | 2165 | 1037 | 846 | GTR + I + G |
| Series | Species | CYA 25 °C (mm) | CYA 37 °C (mm) | MEA (mm) | YES (mm) | Conidiophore | Conidia Shape | Conidia Wall | Conidia Size (µm) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Diversi | T. tianshanicus | 16–19 | 10–14 | 21–23 | 11–15 | Irregularly biverticillate to quaterverticillate | Subglobose to ellipsoidal | Smooth | 2.0–2.5 × 1.7–2.0 | This study |
| T. cystophila | 21–26 | 18–21 | 32–33 | 19–24 | Biverticillate | Subglobose to ellipsoidal | Smooth | 2.6–3.0 × 2.5–3.0 | [31] | |
| Erythromelles | T. elephas | 3–7 | No growth | 15–19 | 8–11 | Biverticillate | Ellipsoidal | Smooth | 3.0–3.5 × 2.0–3.0 | This study |
| T. rubidus | 5–6 | No growth | 17–18 | 6–7 | Biverticillate | Ovoid to spheroidal | Echinulate | 2.5–3.0 | [32] | |
| Miniolutei | T. xishuangbannaensis | 4–5 | No growth | 15–16 | 10–12 | Biverticillate | Subglobose to ellipsoidal | Smooth | 3.0–4.0 × 2.0–3.0 | This study |
| T. germanicus | 20–22 | No growth | 23–25 | 22–24 | Biverticillate | Narrow ellipsoidal to fusiform | Smooth | 2.5–3.5 × 1.5–2.5 | [27] | |
| T. minnesotensis | 24–26 | No growth | 13–15 | 21–24 | Biverticillate | Ellipsoidal | Smooth | 2.5–3.5 × 2.0–3.0 | [26] | |
| Subinflati | T. sinensis | No growth | No growth | 13–16 | no growth | Biverticillate | Ellipsoidal | Smooth | 3.0–4.0 × 2.0–2.5 | This study |
| T. subinflatus | 3–4 | No growth | 14–15 | 3–4 | Biverticillate | Ellipsoidal to fusiform | Smooth | 2.5–4.0 × 1.5–2.0 | [8] | |
| T. guizhouensis | 8–9 | No growth | 24–27 | 12–13 | Biverticillate | Subglobose to fusiform | Finely rough | 2.5–4.5 × 2.5–3.0 | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, L.-Y.; Wang, X.-C.; Gafforov, Y.; Zhuang, W.-Y. Seven New Series and Four New Species in Sections Subinflati and Trachyspermi of Talaromyces (Trichocomaceae, Eurotiales). J. Fungi 2025, 11, 508. https://doi.org/10.3390/jof11070508
Peng L-Y, Wang X-C, Gafforov Y, Zhuang W-Y. Seven New Series and Four New Species in Sections Subinflati and Trachyspermi of Talaromyces (Trichocomaceae, Eurotiales). Journal of Fungi. 2025; 11(7):508. https://doi.org/10.3390/jof11070508
Chicago/Turabian StylePeng, Lu-Yao, Xin-Cun Wang, Yusufjon Gafforov, and Wen-Ying Zhuang. 2025. "Seven New Series and Four New Species in Sections Subinflati and Trachyspermi of Talaromyces (Trichocomaceae, Eurotiales)" Journal of Fungi 11, no. 7: 508. https://doi.org/10.3390/jof11070508
APA StylePeng, L.-Y., Wang, X.-C., Gafforov, Y., & Zhuang, W.-Y. (2025). Seven New Series and Four New Species in Sections Subinflati and Trachyspermi of Talaromyces (Trichocomaceae, Eurotiales). Journal of Fungi, 11(7), 508. https://doi.org/10.3390/jof11070508









