Characterization of Biofilm Formation by the Dermatophyte Nannizzia gypsea
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism
2.2. Biofilm Formation
2.3. Biofilm Quantification
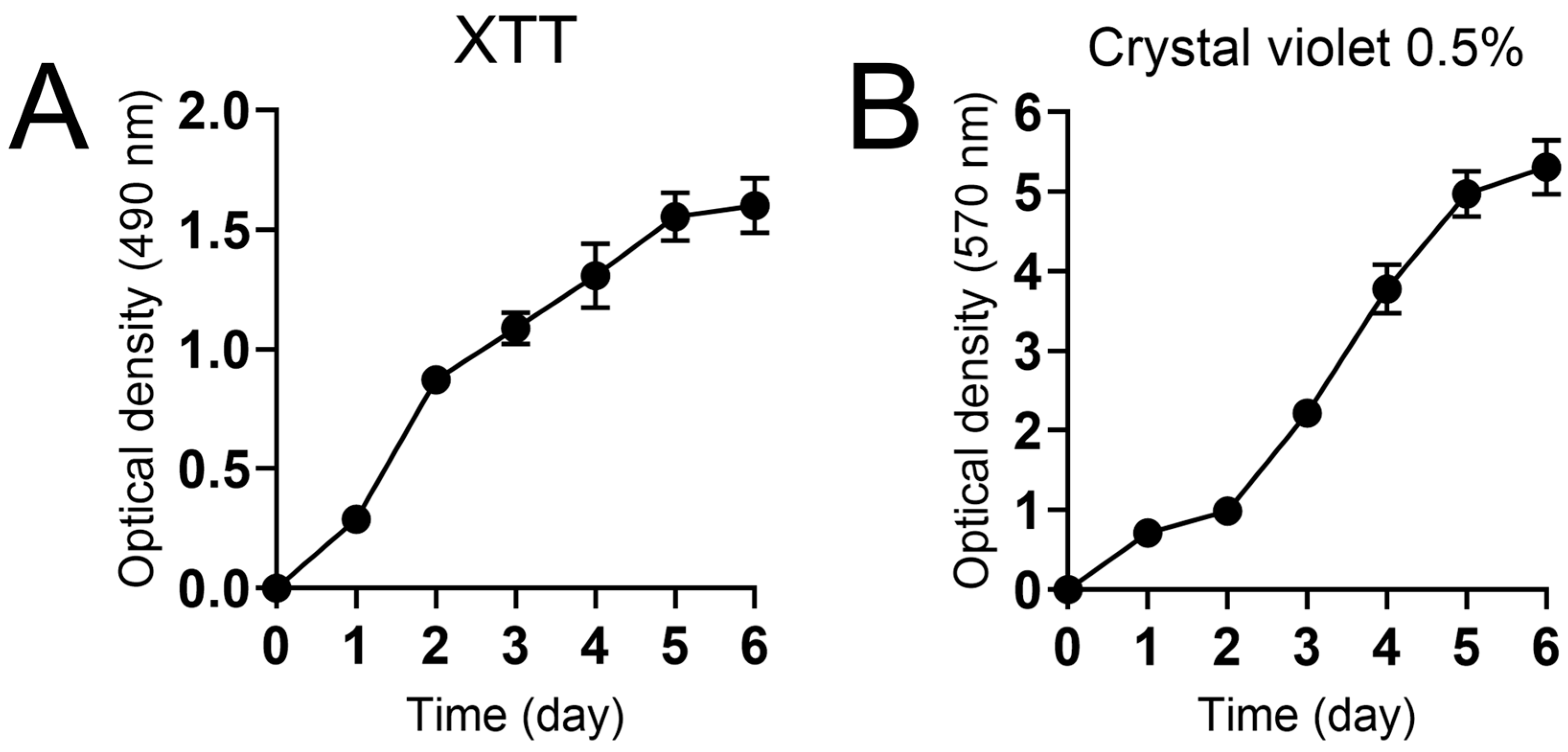
2.3.1. XTT Reduction Assay
2.3.2. Crystal Violet Staining
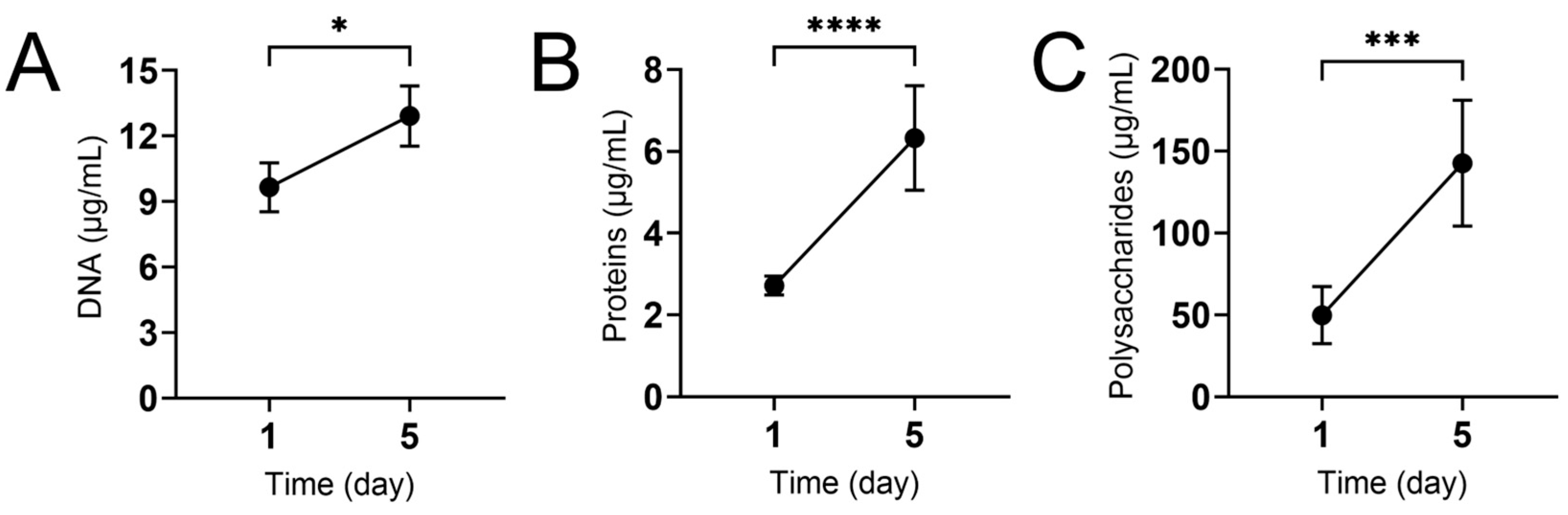
2.4. Quantification of Extracellular Matrix Components
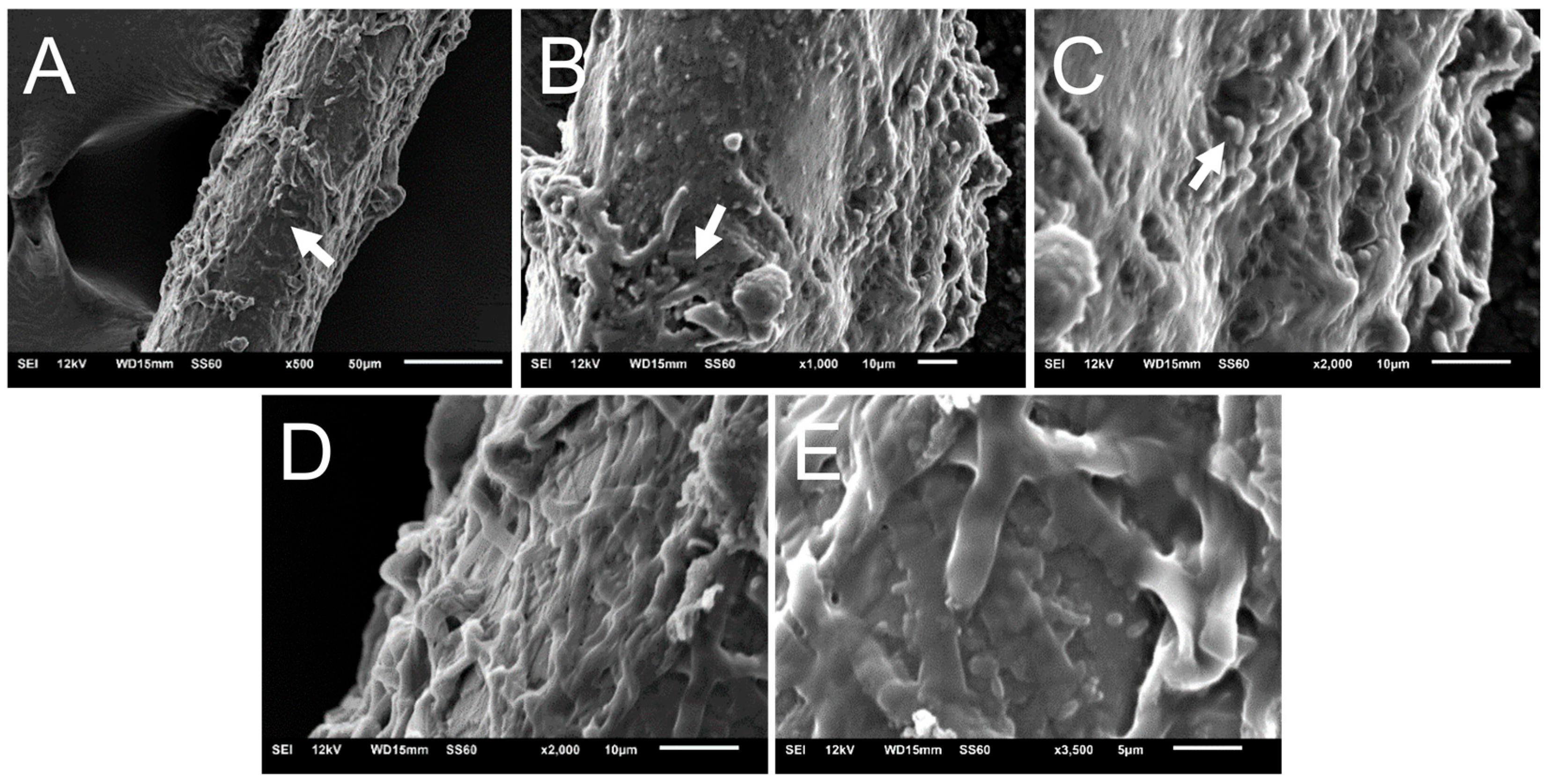
2.5. Structural Analysis of Biofilms In Vitro and Ex Vivo by Scanning Electron Microscopy
2.6. Hair Model Infection
2.7. Analysis of Virulence Gene Expression in N. gypsea
2.7.1. Primer Design
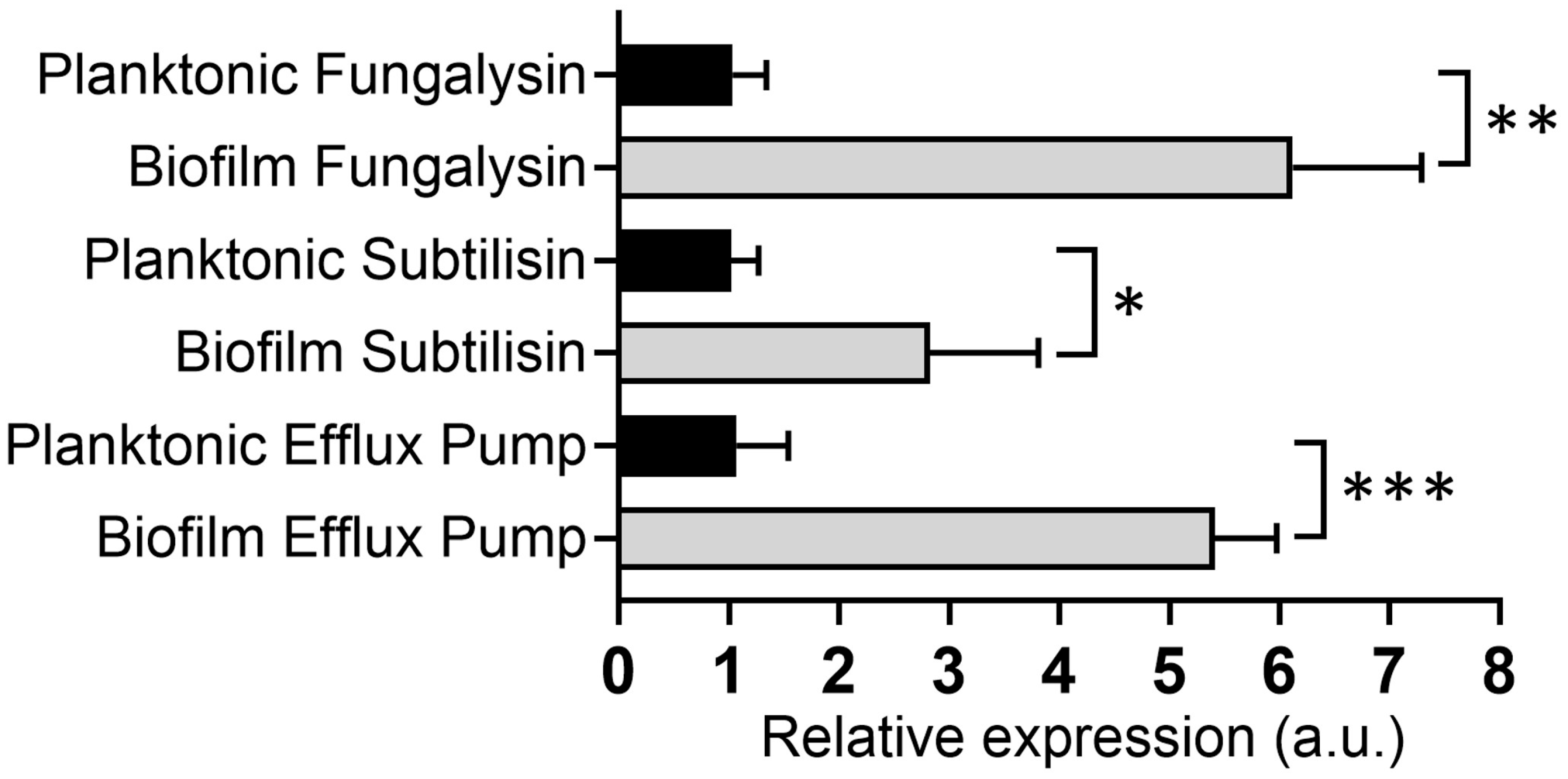
2.7.2. Evaluation of Genes Involved in Virulence and Pathogenicity by qPCR
2.8. Statistical Analysis
3. Results
3.1. N. gypsea Identification
3.2. Characterization of N. gypsea Biofilm Formation
3.3. The Macromolecular Composition of N. gypsea Biofilm Extracellular Matrix Increases over Time
3.4. Scanning Electron Microscopy of N. gypsea Showing Cells Adhering and Forming Biofilms In Vitro
3.5. N. gypsea Heavily Colonizes and Forms Biofilm on Hair Ex Vivo
3.6. Assessment of Nucleic Acid Integrity and Purity
3.7. N. gypsea-Biofilm-Derived Cells Showed Higher Virulence Factor Expression than Planktonic Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhan, P.; Liu, W. The changing face of dermatophytic infections worldwide. Mycopathologia 2017, 182, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Peres, N.T.; Bitencourt, T.A.; Martins, M.P.; Rossi, A. State-of-the-art dermatophyte infections: Epidemiology aspects, pathophysiology, and resistance mechanisms. J. Fungi 2021, 7, 629. [Google Scholar] [CrossRef] [PubMed]
- Petrucelli, M.F.; de Araujo Filho, J.A.; Romagnolo, V.; Queiroz-Telles, F. Epidemiology and diagnostic perspectives of dermatophytoses. J. Fungi 2020, 6, 310. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Lim, B.S.; Lim, K.P.; Ab Malik, N.; Abd Wahab, M.A.; Baharudin, R. Detection of emerging genotypes in Trichophyton mentagrophytes species complex: A proposal for handling biodiversity in dermatophytes. Front. Microbiol. 2022, 13, 960190. [Google Scholar] [CrossRef]
- Kidd, S.E.; Abdolrasouli, A.; Hagen, F. Fungal nomenclature: Managing change is the name of the game. Open Forum Infect. Dis. 2023, 10, ofac559. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Dukik, K.; Monod, M.; Packeu, A.; Stubbe, D.; Hendrickx, M.; Kupsch, C.; Stielow, J.B.; Freeke, J.; Göker, M.; et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia 2017, 182, 5–31. [Google Scholar] [CrossRef]
- Conceição ABda, S.; Mendes, N.S.; Vieira, M.M.; Ferreira Filho, S.P.; Santos, M. Avaliação da presença de fungos dermatófitos e resistência antimicrobiana a partir de amostras ambientais de comunidades ribeirinhas da Região Amazônica: Uma questão de vigilância em saúde. Res. Soc. Dev. 2024, 13, e9313445590. [Google Scholar] [CrossRef]
- Mercer, D.K.; Stewart, C.S. Keratin hydrolysis by dermatophytes. Med. Mycol. 2019, 57, 13–22. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Sardi, J.C.; Santos, C.T.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. In vitro characterization of Trichophyton rubrum and T. mentagrophytes biofilms. Biofouling 2014, 30, 719–727. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; de Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal biofilms and polymicrobial diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Serafim-Pinto, A.; da Silva, P.B.; Bila, N.M.; Bonatti, J.L.C.; Scorzoni, L.; Singulani, J.L.; Dos Santos, C.T.; Nazaré, A.C.; Chorilli, M.; et al. Incorporation of nonyl 3,4-dihydroxybenzoate into nanostructured lipid systems: Effective alternative for maintaining anti-dermatophytic and antibiofilm activities and reducing toxicity at high concentrations. Front. Microbiol. 2020, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Monod, M. Secreted proteases from dermatophytes. Mycopathologia 2008, 166, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Peres, N.T.; Rossi, A. Dermatophyte resistance to antifungal drugs: Mechanisms and prospectus. Front. Microbiol. 2018, 9, 1108. [Google Scholar] [CrossRef] [PubMed]
- Sardi, J.D.C.O.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. Highlights in pathogenic fungal biofilms. Rev. Iberoam. Micol. 2014, 31, 22–29. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Brilhante, R.S.N.; Correia, E.E.M.; Guedes, G.M.M.; Pereira, V.S.; Oliveira, J.S.; Bandeira, S.P.; Alencar, L.P.; Andrade, A.R.C.; Castelo-Branco, D.S.C.M.; Cordeiro, R.A.; et al. Quantitative and structural analyses of the in vitro and ex vivo biofilm-forming ability of dermatophytes. J. Med. Microbiol. 2017, 66, 1045–1052. [Google Scholar] [CrossRef]
- Kaplan, E.; Eshetu, D.A.; Yeshanew, A.G.; Bauer, J.; Gräser, Y. Genes encoding proteolytic enzymes fungalysin and subtilisin in dermatophytes of human and animal origin: A comparative study. Mycopathologia 2019, 184, 673–683. [Google Scholar] [CrossRef]
- Chen, J.; Gao, Y.; Xiong, S.; Peng, Z.; Zhan, P. Expression profiles of protease in onychomycosis-related pathogenic Trichophyton rubrum and Tinea capitis-related pathogenic Trichophyton violaceum. Mycopathologia 2024, 189, 14. [Google Scholar] [CrossRef]
- Elavarashi, E.; Kindo, A.J.; Rangarajan, S. Enzymatic and non-enzymatic virulence activities of dermatophytes on solid media. J. Clin. Diagn. Res. 2017, 11, DC23–DC25. [Google Scholar] [CrossRef]
- Nayak, A.P.; Green, B.J.; Beezhold, D.H. Fungal hemolysins. Med. Mycol. 2013, 51, 1–16. [Google Scholar] [CrossRef]
- Barman, A.; Ghosh, A.; Sengupta, M.; Chakraborty, A.; Datta, A.; Ghosh, A.K. Phospholipases play multiple cellular roles including growth, stress tolerance, sexual development, and virulence in fungi. Microbiol. Res. 2018, 209, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Sardana, K.; Gupta, A.; Mathachan, S.R. Immunopathogenesis of dermatophytoses and factors leading to recalcitrant infections. Indian. Dermatol. Online J. 2021, 12, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Dubljanin, E.; Zunic, J.; Vujcic, I.; Colovic Calovski, I.; Sipetic Grujicic, S.; Mijatovic, S.; Dzamic, A. Host-pathogen interaction and resistance mechanisms in dermatophytes. Pathogens 2024, 13, 657. [Google Scholar] [CrossRef] [PubMed]
- Baldo, A.; Monod, M.; Mathy, A.; Cambier, L.; Bagut, E.T.; Defaweux, V. Mechanisms of skin adherence and invasion by dermatophytes. Mycoses 2012, 55, 218–223. [Google Scholar] [CrossRef]
- Tabart, J.; Baldo, A.; Vermout, S.; Mignon, B.; Losson, B. Reconstructed interfollicular feline epidermis as a model for Microsporum canis dermatophytosis. J. Med. Microbiol. 2007, 56 Pt 7, 971–975. [Google Scholar] [CrossRef]
- Maciel Quatrin, P.; Roncada, T.M.; Martínez, M.E.; Fuentefria, A.M. Fungal infection models: Current progress of ex vivo methods. Mycoses 2019, 62, 860–873. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous, f.u.n.g.i.; approved standard CLSI document, M.3.8.-A.; CLSI: Wayne, PA, USA, 2002. [Google Scholar]
- Belizario, J.A.; Bila, N.M.; Vaso, C.O.; Costa-Orlandi, C.B.; Mendonça, M.B.; Fusco-Almeida, A.M.; Pires, R.H.; Mendes-Giannini, M.J.S. Exploring the Complexity of the Interaction between T. rubrum and S. aureus/S. epidermidis in the Formation of Polymicrobial Biofilms. Microorganisms 2024, 12, 191. [Google Scholar] [CrossRef]
- Martins, M.; Henriques, M.; Azeredo, J.; Rocha, S.M.; Coimbra, M.A.; Oliveira, R. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010, 169, 323–331. [Google Scholar] [CrossRef]
- Oliveira, L.T.; Zambuzzi, W.F.; Fusco-Almeida, A.M.; Junqueira, J.C. Paracoccidioides spp.: The structural characterization of extracellular matrix, expression of glucan synthesis and associated genes and adhesins during biofilm formation. Front. Microbiol. 2024, 15, 1354140. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Oliveira, L.T.; Zambuzzi, W.F.; Junqueira, J.C.; Jorge, A.O.; Fusco-Almeida, A.M. Dynamics of mono- and dual-species biofilm formation and interactions between Paracoccidioides brasiliensis and Candida albicans. Front. Microbiol. 2020, 11, 551256. [Google Scholar] [CrossRef] [PubMed]
- Engle, K.; Kumar, G. Tackling multi-drug resistant fungi by efflux pump inhibitors. Biochem. Pharmacol. 2024, 226, 116400. [Google Scholar] [CrossRef] [PubMed]
- Barabote, R.D.; Thaminy, S.; Nagarajavel, V.R.; Clark, B.J.; Adams, M.W.; Saier, M.H., Jr. Xenobiotic efflux in bacteria and fungi: A genomics update. Adv. Enzym. Relat. Areas Mol. Biol. 2011, 77, 237–306. [Google Scholar]
- Garcia, Í.R.; Silva, D.C.; Santos, N.C.; Pereira, M.F. Microbial resistance: The role of efflux pump superfamilies and their respective substrates. Life Sci. 2022, 295, 120391. [Google Scholar] [CrossRef]
- Brilhante, R.S.N.; Aguiar, L.; Sales, J.A.; Araújo, G.D.S.; Pereira, V.S.; Pereira-Neto, W.A.; Pinheiro, A.Q.; Paixão, G.C.; Cordeiro, R.A.; Sidrim, J.J.C.; et al. Ex vivo biofilm-forming ability of dermatophytes using dog and cat hair: An ethically viable approach for an infection model. Biofouling 2019, 35, 392–400. [Google Scholar] [CrossRef]
- Gupta, A.K.; Versteeg, S.G.; Shear, N.H. Global dermatophyte infections linked to human and animal health: A scoping review. Microorganisms 2025, 13, 575. [Google Scholar] [CrossRef]
- Sharifzadeh, A.; Shokri, H.; Khosravi, A.R. In vitro evaluation of antifungal susceptibility and keratinase, elastase, lipase and DNase activities of different dermatophyte species isolated from clinical specimens in Iran. Mycoses 2016, 59, 710–719. [Google Scholar] [CrossRef]
- Chen, B.; Sun, Y.; Zhang, J.; Chen, R.; Zhong, X.; Wu, X.; Zheng, L.; Zhao, J. In vitro evaluation of photodynamic effects against biofilms of dermatophytes involved in onychomycosis. Front. Microbiol. 2019, 10, 1228. [Google Scholar] [CrossRef]
- Garcia, L.M.; Costa-Orlandi, C.B.; Bila, N.M.; Vaso, C.O.; Gonçalves, L.N.C.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. A Two-Way Road: Antagonistic Interaction Between Dual-Species Biofilms Formed by Candida albicans/Candida parapsilosis and Trichophyton rubrum. Front. Microbiol. 2020, 11, 1980. [Google Scholar] [CrossRef]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. Fungal super glue: The biofilm matrix and its composition, assembly, and functions. PLoS Pathog. 2016, 12, e1005828. [Google Scholar] [CrossRef]
- Lakhani, P.; Patil, A.; Majumdar, S. Challenges in the polyene- and azole-based pharmacotherapy of ocular fungal infections. J. Ocul. Pharmacol. Ther. 2019, 35, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Howell, P.L. Biofilm exopolysaccharides of pathogenic fungi: Lessons from bacteria. J. Biol. Chem. 2016, 291, 12529–12537. [Google Scholar] [CrossRef] [PubMed]
- Dario Junior, C.; Ramos, M.L.M.; Almeida-Paes, R.; Frases, S. New insights in dermatophytes: Microsporum spp. and Nannizzia spp. Curr. Trop. Med. Rep. 2022, 9, 15–27. [Google Scholar] [CrossRef]
- Dukik, K.; de Hoog, G.S.; Stielow, J.B.; Freeke, J.; van den Ende, B.G.; Vicente, V.A.; Menken, S.B.J.; Ahmed, S.A. Molecular and phenotypic characterization of Nannizzia (Arthrodermataceae). Mycopathologia 2020, 185, 9–35. [Google Scholar] [CrossRef]
- Aneke, C.I.; Rhimi, W.; Hubka, V.; Otranto, D.; Cafarchia, C. Virulence and antifungal susceptibility of Microsporum canis strains from animals and humans. Antibiotics 2021, 10, 296. [Google Scholar] [CrossRef]
- Kunert, J.; Kushwaha, R.; Guarro, J. Physiology of keratinophilic fungi. Rev. Iberoam. Micol. 2000, 17, 77–85. [Google Scholar]
- Kaatz, G.W.; Mcaleese, F.; Seo, S.M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005, 49, 1857–1864. [Google Scholar] [CrossRef]

| Gene | Sequence | GenBank |
|---|---|---|
| GAPDH | F 5′ GCCGTGTTGTTGACCTCATC 3′ R 5′ GCGATGTAGGCTGTGAGAGA 3′ | NW_003345199.1 |
| Subtilisin (Sub7) | F 5′ ACTGTCGCCGGTACCAAAT 3′ R 5′ GGCGTCGTTGGTAGCAAAT 3′ | OM397965.1 |
| Multidrug and Toxin Extrusion Protein 2 (Mate2) | F 5′ GCCATCACCCAGCTCTTCTA 3′ R 5′ CCCTTGCCGTTGTTGTTCAT 3′ | NW_003345198.1 |
| Extracellular elastolytic metalloproteinase (Mmp12) | F 5′ GCCGTCAGCTTAGCCAGTAT 3′ R 5′ AGGCCGACTAGCTTCTTGTT 3′ | NW_003345200.1 |
| Biochemical Test | Nannizzia gypsea F35 LLH (Clinical Isolate) |
|---|---|
| Catalase | Positive |
| Lipase | Positive |
| Protease | Positive |
| Hemolytic activity | Negative |
| Urease | Positive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arantes, B.B.A.; Cabral, A.K.L.F.; dos Santos, K.S.; Mendonça, M.B.; dos Santos, R.C.; Bugalho, B.C.M.; Fernandes, L.D.S.; Martinez, L.R.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Characterization of Biofilm Formation by the Dermatophyte Nannizzia gypsea. J. Fungi 2025, 11, 455. https://doi.org/10.3390/jof11060455
Arantes BBA, Cabral AKLF, dos Santos KS, Mendonça MB, dos Santos RC, Bugalho BCM, Fernandes LDS, Martinez LR, Fusco-Almeida AM, Mendes-Giannini MJS. Characterization of Biofilm Formation by the Dermatophyte Nannizzia gypsea. Journal of Fungi. 2025; 11(6):455. https://doi.org/10.3390/jof11060455
Chicago/Turabian StyleArantes, Bruno B. A., Ana Karla L. F. Cabral, Kelvin S. dos Santos, Matheus B. Mendonça, Rafaela C. dos Santos, Beatriz C. M. Bugalho, Lígia De S. Fernandes, Luis R. Martinez, Ana Marisa Fusco-Almeida, and Maria José S. Mendes-Giannini. 2025. "Characterization of Biofilm Formation by the Dermatophyte Nannizzia gypsea" Journal of Fungi 11, no. 6: 455. https://doi.org/10.3390/jof11060455
APA StyleArantes, B. B. A., Cabral, A. K. L. F., dos Santos, K. S., Mendonça, M. B., dos Santos, R. C., Bugalho, B. C. M., Fernandes, L. D. S., Martinez, L. R., Fusco-Almeida, A. M., & Mendes-Giannini, M. J. S. (2025). Characterization of Biofilm Formation by the Dermatophyte Nannizzia gypsea. Journal of Fungi, 11(6), 455. https://doi.org/10.3390/jof11060455







