Alternaria alternata botybirnavirus 1 (AaBRV1) Infection Affects the Biological Characteristics of Its Host Fungus Alternaria alternata
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Elimination of AaBRV1 from Strain SD-BZF-19
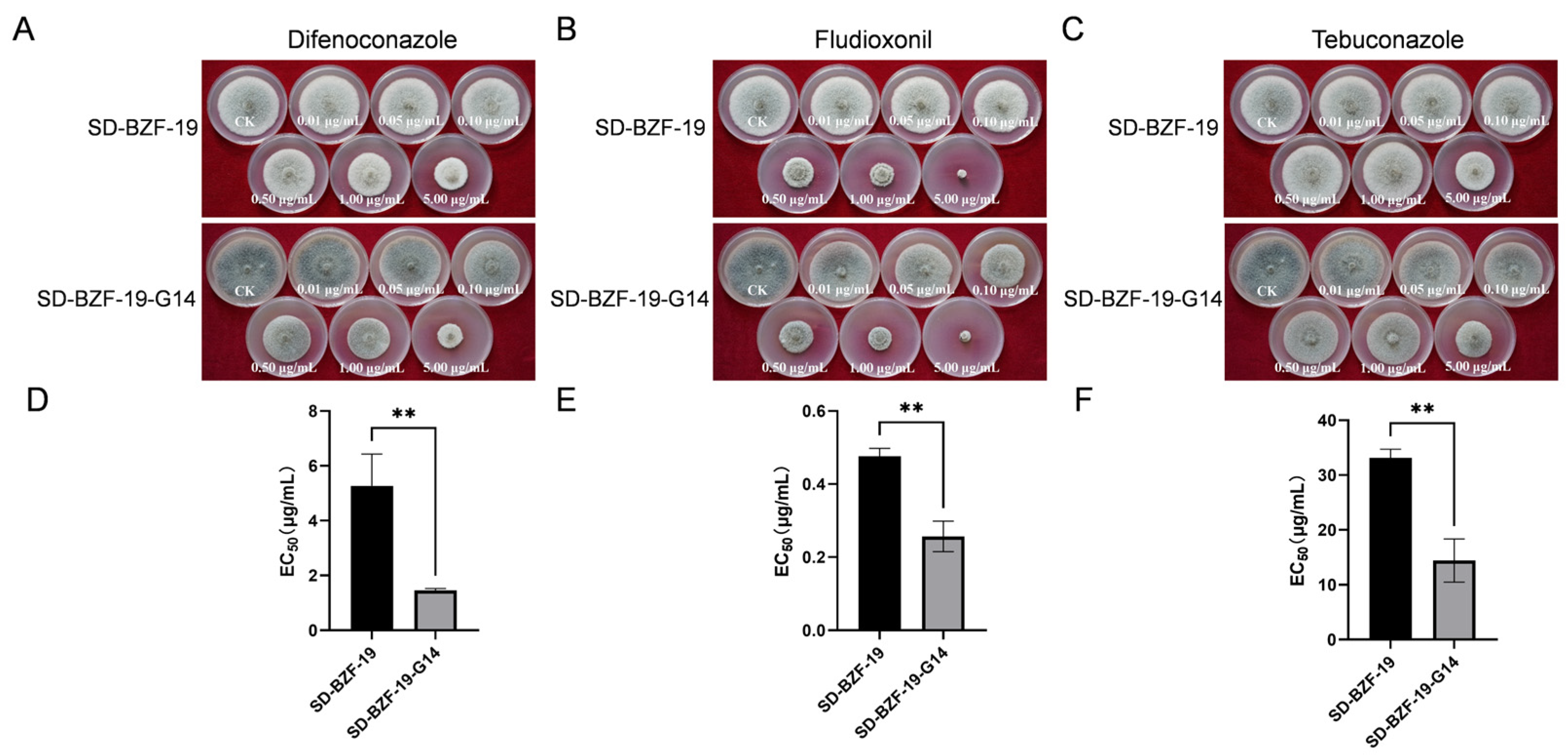
2.3. Observation of Colony Color and Measurements of Colony Growth Rate, Mycelial Biomass, and EC50 Value
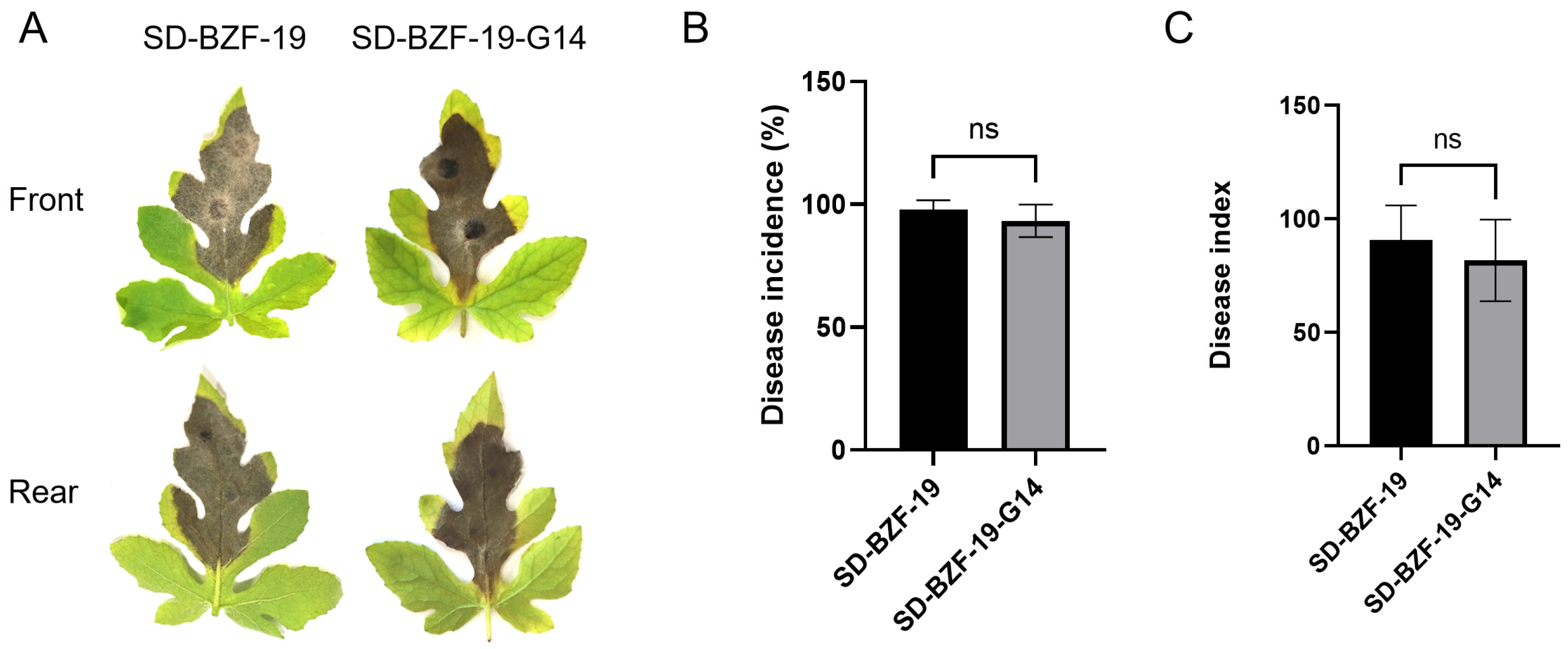
2.4. Pathogenicity Assay
2.5. cDNA Library Preparation and Transcriptomic Analysis
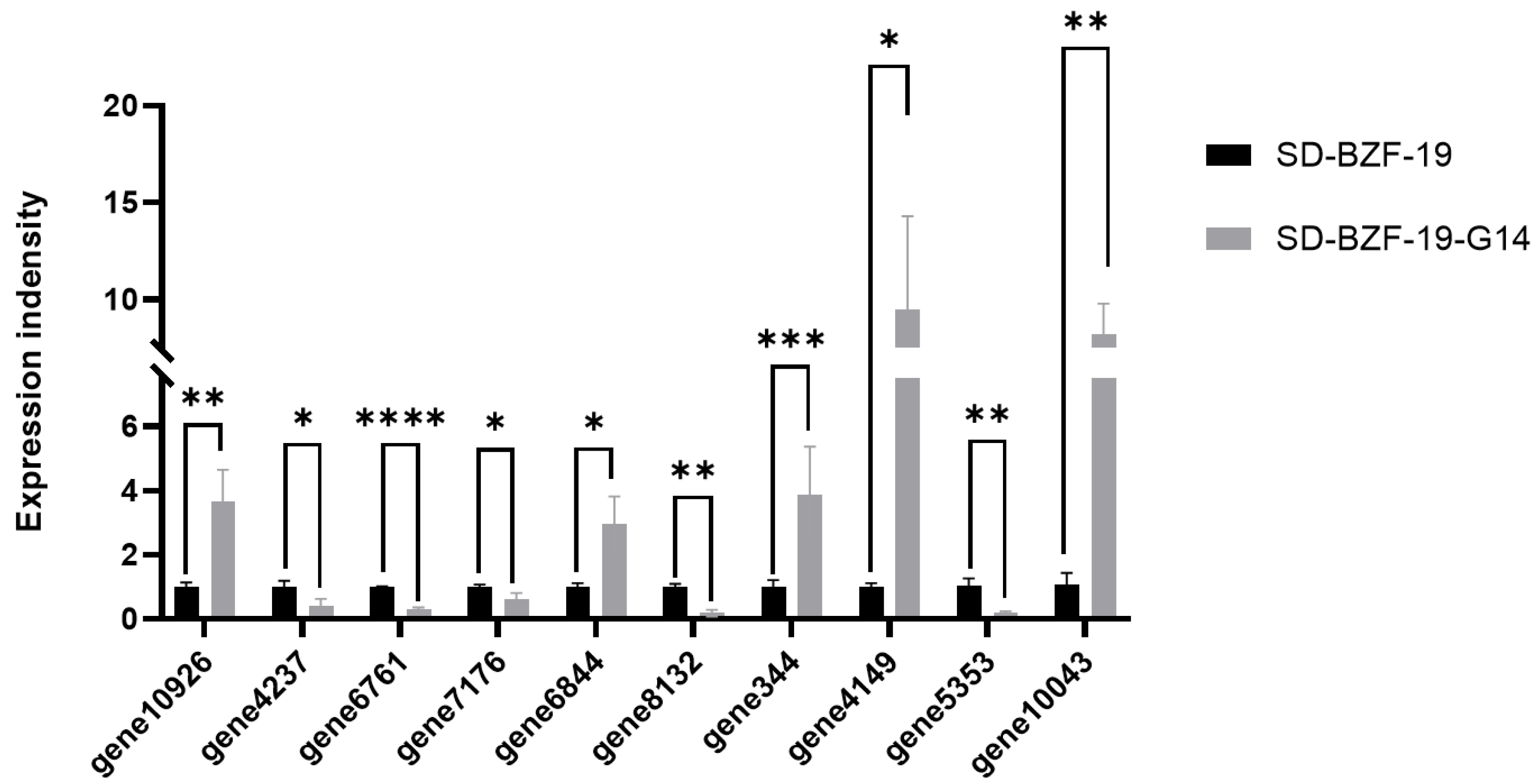
2.6. Validation of Transcriptome Data Using Reverse Transcription-Quantitative PCR
2.7. Statistical Analysis
3. Results
3.1. Effect of AaBRV1 on Colony Color, Growth Rate, and Mycelial Biomass of Its Host Fungus
3.2. Effect of AaBRV1 on Virulence of Its Host Fungus
3.3. Effect of AaBRV1 on Sensitivity to Three Fungicides of Its Host Fungus
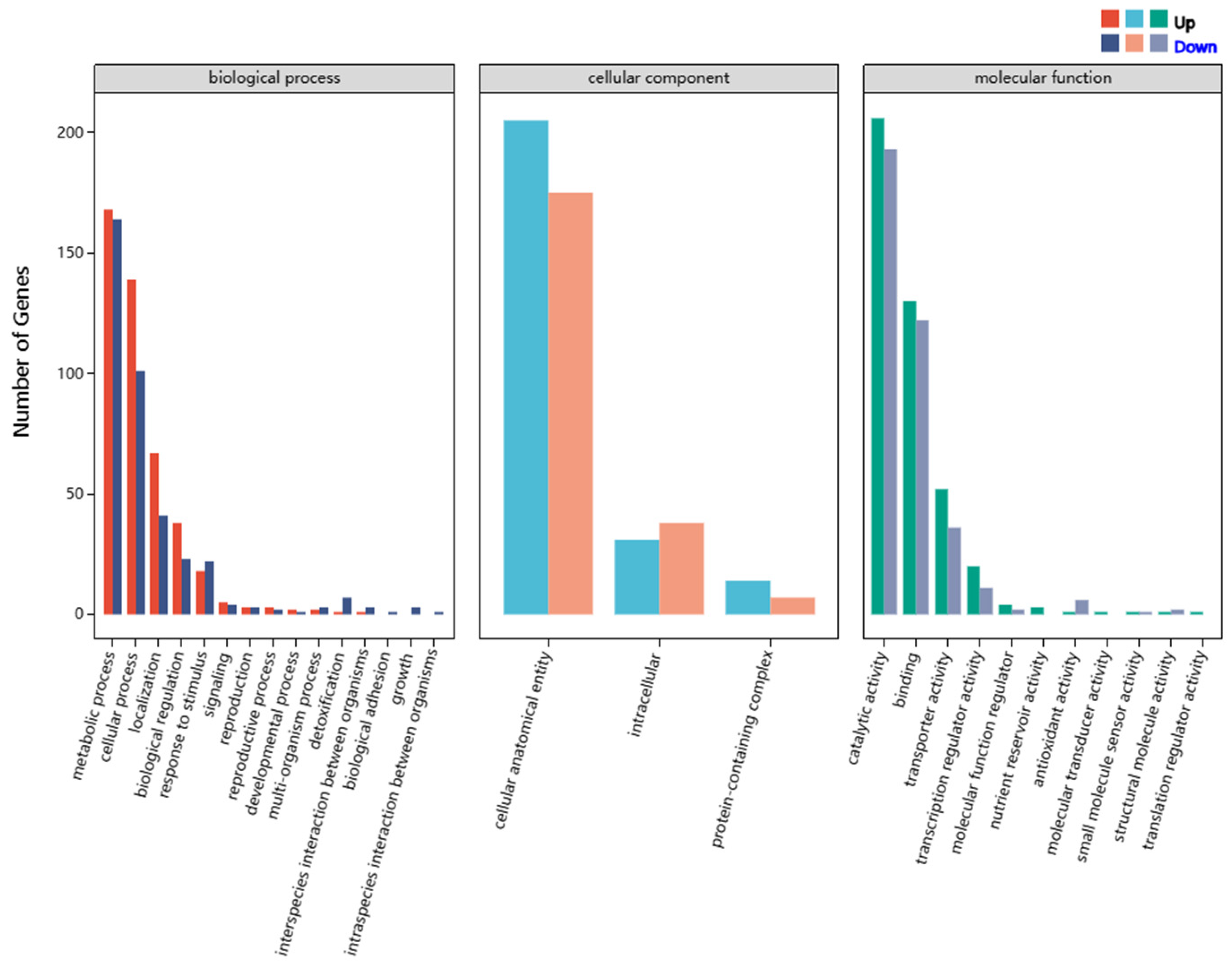
3.4. Overview of Differentially Expressed Genes (DEGs)
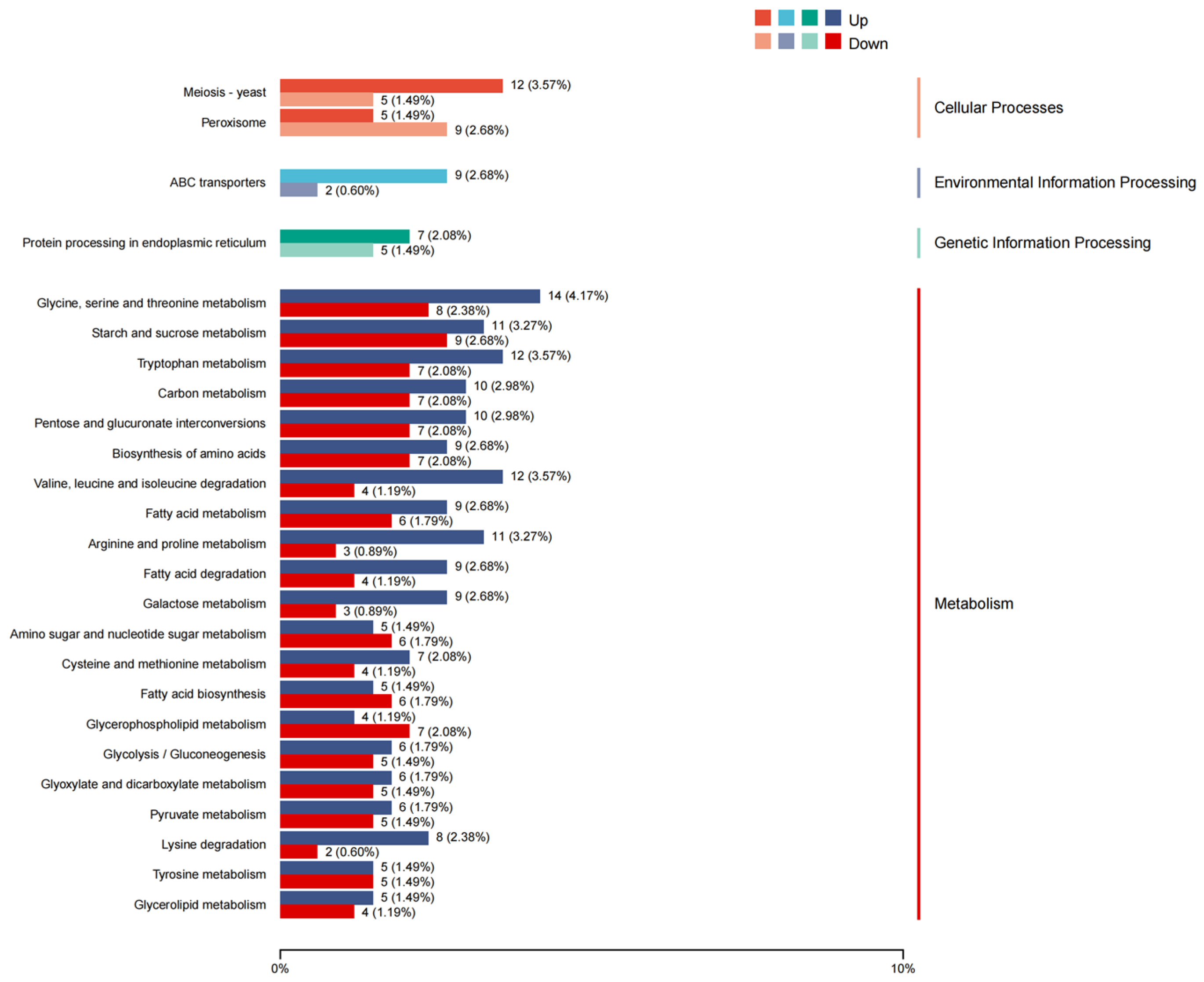
3.5. Influence of AaBRV1 on Its Host DEGs Related to MFS Multidrug Transporters and CYP450
3.6. Validation of Differentially Expressed Genes (DEGs)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Adriaenssens, E.M.; Lefkowitz, E.J.; Oksanen, H.M.; Siddell, S.G.; Zerbini, F.M.; Alfenas-Zerbini, P.; Aylward, F.O.; Dempsey, D.M.; Dutilh, B.E.; et al. Changes to virus taxonomy and the ICTV statutes ratified by the International Committee on Taxonomy of Viruses (2024). Arch. Virol. 2024, 169, 236. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Jin, F.; Zhang, J.; Yang, L.; Jiang, D.; Li, G. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J. Virol. 2012, 86, 6605–6619. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. Molecular characterization of a bipartite double-stranded RNA virus and its satellite-like RNA co-infecting the phytopathogenic fungus Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 406. [Google Scholar] [CrossRef] [PubMed]
- Ran, H.; Liu, L.; Li, B.; Cheng, J.; Fu, Y.; Jiang, D.; Xie, J. Co-infection of a hypovirulent isolate of Sclerotinia sclerotiorum with a new botybirnavirus and a strain of a mitovirus. Virol. J. 2016, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Marzano, S.L.; Domier, L.L. Novel mycoviruses discovered from metatranscriptomics survey of soybean phyllosphere phytobiomes. Virus Res. 2016, 213, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Fu, M.; Hong, N.; Zhai, L.; Xiao, F.; Wang, G. Characterization of a novel botybirnavirus isolated from a phytopathogenic Alternaria fungus. Arch. Virol. 2017, 162, 3907–3911. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Cai, L.; Fang, S.; Zheng, L.; Yan, F.; Zhang, S.; Liu, Y. The complete genomic sequence of a novel botybirnavirus isolated from a phytopathogenic Bipolaris maydis. Virus Genes 2018, 54, 733–736. [Google Scholar] [CrossRef]
- Zhai, L.; Yang, M.; Zhang, M.; Hong, N.; Wang, G. Characterization of a botybirnavirus conferring hypovirulence in the phytopathogenic fungus Botryosphaeria dothidea. Viruses 2019, 11, 266. [Google Scholar] [CrossRef]
- Shamsi, W.; Sato, Y.; Jamal, A.; Shahi, S.; Kondo, H.; Suzuki, N.; Bhatti, M.F. Molecular and biological characterization of a novel botybirnavirus identified from a Pakistani isolate of Alternaria alternata. Virus Res. 2019, 263, 119–128. [Google Scholar] [CrossRef]
- Cottet, L.; Potgieter, C.A.; Castro, M.E.; Castillo, A. Molecular characterization of a new botybirnavirus that infects Botrytis cinerea. Arch. Virol. 2019, 164, 1479–1483. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, S.; Xiao, X.; Cheng, J.; Fu, Y.; Chen, T.; Jiang, D.; Xie, J. Discovery of two mycoviruses by high-throughput sequencing and assembly of mycovirus-derived small silencing RNAs from a hypovirulent strain of Sclerotinia sclerotiorum. Front. Microbiol. 2019, 10, 1415. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhou, K.; Wu, M.; Zhang, J.; Yang, L.; Chen, W.; Li, G. Viral cross-class transmission results in disease of a phytopathogenic fungus. ISME J. 2022, 16, 2763–2774. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, W.; Kondo, H.; Ulrich, S.; Rigling, D.; Prospero, S. Novel RNA viruses from the native range of Hymenoscyphus fraxineus, the causal fungal agent of ash dieback. Virus Res. 2022, 320, 198901. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Shi, X.; He, Y.; Chen, J.; Xu, Q.; Shafik, K.; Fu, L.; Yin, Y.; Kotta-Loizou, I.; Xu, W. A novel botybirnavirus with a unique satellite dsRNA causes latent infection in Didymella theifolia isolated from tea plants. Microbiol. Spectr. 2023, 11, e0003323. [Google Scholar] [CrossRef]
- Pan, X.; Xue, C.Q.; Liu, C.J.; Yan, S.W.; Lv, T.T.; Dai, J.L.; Zhang, X.T.; Li, H.L.; Li, J.H.; Gao, F. Molecular characterization of a new botybirnavirus that infects Alternaria sp. from tobacco. Arch. Virol. 2024, 169, 149. [Google Scholar]
- Ma, G.; Liang, Z.; Hua, H.; Zhou, T.; Wu, X. Complete genome sequence of a new botybirnavirus isolated from a phytopathogenic Alternaria alternata in China. Arch. Virol. 2019, 164, 1225–1228. [Google Scholar] [CrossRef]
- Liang, Z.; Hua, H.; Wu, C.; Zhou, T.; Wu, X. A botybirnavirus isolated from Alternaria tenuissima confers hypervirulence and decreased sensitivity of its host fungus to difenoconazole. Viruses 2022, 14, 2093. [Google Scholar] [CrossRef]
- Yurchenko, E.; Karpova, D.; Burovinskaya, M.; Vinogradova, S. Leaf spot caused by Alternaria spp. is a new disease of grapevine. Plants 2024, 13, 3335. [Google Scholar] [CrossRef]
- Wang, F.; Saito, S.; Michailides, T.J.; Xiao, C.L. Fungicide resistance in Alternaria alternata from blueberry in California and its impact on control of Alternaria rot. Plant Dis. 2022, 106, 1446–1453. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Ma, L.; An, Y.; Wang, H.; Wu, W. Laboratory screening of control agents against isolated fungal pathogens causing postharvest diseases of pitaya in Guizhou, China. Front. Chem. 2022, 10, 942185. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, X.; Hua, H.; Zhou, T.; Wu, X. Molecular and biological characterization of a novel strain of Alternaria alternata chrysovirus 1 identified from the pathogen Alternaria tenuissima causing watermelon leaf blight. Virus Res. 2020, 280, 197904. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.; Zhang, X.; Xia, J.; Wu, X. A novel strain of Fusarium oxysporum virus 1 isolated from Fusarium oxysporum f. sp. niveum strain X-GS16 influences phenotypes of F. oxysporum strain HB-TS-YT-1hyg. J. Fungi 2024, 10, 252. [Google Scholar] [CrossRef]
- Hua, H.; Zhang, X.; Liu, L.; Wu, X. A novel strain of Fusarium oxysporum alternavirus 1 isolated from Fusarium oxysporum f. sp. melonis strain T-BJ17 confers hypovirulence and increases the sensitivity of its host fungus to difenoconazole and pydiflumetofen. Viruses 2024, 16, 901. [Google Scholar] [CrossRef]
- Higuchi, A.; Tojo, M.; Mochizuki, T. Sensitivity of Globisporangium ultimum to the fungicide metalaxyl is enhanced by the infection with a toti-like mycovirus. Microbiol. Res. 2024, 285, 127742. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, X.; Liang, X.; Chen, D.; Xie, C.; Kang, Z.; Zheng, L. A novel hexa-segmented dsRNA mycovirus confers hypovirulence in the phytopathogenic fungus Diaporthe pseudophoenicicola. Environ. Microbiol. 2022, 24, 4274–4284. [Google Scholar] [CrossRef]
- de Waard, M.A.; Andrade, A.C.; Hayashi, K.; Schoonbeek, H.J.; Stergiopoulos, I.; Zwiers, L.H. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 2006, 62, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier Jr, M.H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef]
- Song, J.X.; Zhang, S.Z.; Lu, L. Fungal cytochrome P450 protein Cyp51: What we can learn from its evolution, regulons and Cyp51-based azole resistance. Fungal Biol. Rev. 2018, 32, 131–142. [Google Scholar] [CrossRef]
- Ma, G.; Bao, S.; Zhao, J.; Sui, Y.; Wu, X. Morphological and molecular characterization of Alternaria species causing leaf blight on watermelon in China. Plant Dis. 2021, 105, 60–70. [Google Scholar] [CrossRef]
- Hu, H.J.; Wang, J.R.; Cheng, X.H.; Liu, Y.; Zhang, X.Y. Preliminary studies on the effects of oyster mushroom spherical virus China strain on the mycelial growth and fruiting body yield of the edible mushroom Pleurotus ostreatus. Biology 2022, 11, 574. [Google Scholar] [CrossRef]
- Lalitha, S. Primer premier 5. Biotechnol. Softw. Int. Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, C.; Hua, H.; Cai, Q.; Wu, X. Characterization of the first alternavirus identified in Fusarium avenaceum, the causal agent of potato dry rot. Viruses 2023, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Botella, L.; Suzuki, N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu. Rev. Phytopathol. 2022, 60, 307–336. [Google Scholar] [CrossRef] [PubMed]
- Filippou, C.; Coutts, R.H.A.; Kotta-Loizou, I.; El-Kamand, S.; Papanicolaou, A. Transcriptomic analysis reveals molecular mechanisms underpinning mycovirus-mediated hypervirulence in Beauveria bassiana infecting Tenebrio molitor. J. Fungi 2025, 11, 63. [Google Scholar] [CrossRef]
- Zhu, J.Z.; Qiu, Z.L.; Gao, B.D.; Li, X.G.; Zhong, J. A novel partitivirus conferring hypovirulence by affecting vesicle transport in the fungus Colletotrichum. mBio 2024, 15, e0253023. [Google Scholar] [CrossRef]
- Sun, N.; Li, D.; Fonzi, W.; Li, X.; Zhang, L.; Calderone, R. Multidrug-resistant transporter mdr1p-mediated uptake of a novel antifungal compound. Antimicrob. Agents Chemother. 2013, 57, 5931–5939. [Google Scholar] [CrossRef]


| Samples | Clean Reads | ≥Q30 | Mapped Reads | Mapped Reads Ratio |
|---|---|---|---|---|
| SD-BZF-19 (OS-1) | 42,366,376 | 96.84% | 37,749,516 | 89.10% |
| SD-BZF-19 (OS-2) | 45,057,340 | 96.92% | 40,344,549 | 89.54% |
| SD-BZF-19 (OS-3) | 41,652,466 | 96.79% | 37,533,659 | 90.11% |
| SD-BZF-19-G14 (VF-1) | 40,630,300 | 97.55% | 36,727,212 | 90.39% |
| SD-BZF-19-G14 (VF-2) | 43,131,488 | 96.90% | 38,787,040 | 89.93% |
| SD-BZF-19-G14 (VF-3) | 40,859,104 | 97.39% | 36,183,361 | 88.56% |
| #ID | log2FC | Regulated | Pfam_Annotation |
|---|---|---|---|
| gene12067 | −2.7290 | down | Major Facilitator Superfamily |
| gene3093 | −1.8004 | down | Major Facilitator Superfamily |
| gene6558 | −2.1461 | down | Major Facilitator Superfamily |
| gene1907 | 1.7180 | up | Major Facilitator Superfamily |
| gene3128 | −1.5539 | down | Major Facilitator Superfamily |
| gene12139 | 4.7976 | up | Major Facilitator Superfamily |
| gene449 | −1.8754 | down | Major Facilitator Superfamily |
| gene13064 | 2.3708 | up | Major Facilitator Superfamily |
| gene4157 | 2.1862 | up | Major Facilitator Superfamily |
| gene9346 | −2.9509 | down | Major Facilitator Superfamily |
| gene3911 | −1.2205 | down | Major Facilitator Superfamily |
| gene8132 | −2.2791 | down | Major Facilitator Superfamily |
| gene7671 | −4.0058 | down | Major Facilitator Superfamily |
| gene12402 | −2.9413 | down | Major Facilitator Superfamily |
| gene7749 | −2.1046 | down | Major Facilitator Superfamily |
| gene4382 | −1.7318 | down | Major Facilitator Superfamily |
| gene10728 | 3.4501 | up | Major Facilitator Superfamily |
| gene10144 | −1.4685 | down | Major Facilitator Superfamily |
| gene14 | −1.2037 | down | Major Facilitator Superfamily |
| gene20 | 3.2432 | up | Major Facilitator Superfamily |
| gene6734 | 4.0763 | up | Major Facilitator Superfamily |
| gene4149 | 2.5069 | up | Major Facilitator Superfamily |
| gene12318 | −1.7286 | down | Major Facilitator Superfamily |
| gene2867 | 5.8406 | up | Major Facilitator Superfamily |
| gene6803 | 3.5240 | up | Major Facilitator Superfamily |
| gene4338 | −1.4488 | down | Major Facilitator Superfamily |
| gene10118 | 1.3871 | up | Major Facilitator Superfamily |
| gene10927 | 1.2162 | up | Major Facilitator Superfamily |
| gene11344 | 2.3732 | up | Major Facilitator Superfamily |
| gene10023 | 2.3275 | up | Major Facilitator Superfamily |
| gene7724 | −1.9912 | down | Major Facilitator Superfamily |
| gene1004 | 1.6274 | up | Major Facilitator Superfamily |
| gene1015 | −1.5243 | down | Major Facilitator Superfamily |
| gene12533 | 2.8530 | up | Major Facilitator Superfamily |
| gene11267 | −1.3916 | down | Major Facilitator Superfamily |
| gene3700 | −1.0512 | down | Major Facilitator Superfamily |
| #ID | log2FC | Regulated | Pfam_Annotation |
|---|---|---|---|
| gene344 | 1.6649 | up | Cytochrome P450 |
| gene12733 | −3.5506 | down | Cytochrome P450 |
| gene8445 | −1.3397 | down | Cytochrome P450 |
| gene12537 | 1.8953 | up | Cytochrome P450 |
| gene9662 | −5.4342 | down | Cytochrome P450 |
| gene10926 | 1.5227 | up | Cytochrome P450 |
| gene2102 | −3.7723 | down | Cytochrome P450 |
| gene6912 | 3.1839 | up | Cytochrome P450 |
| gene7128 | −2.7300 | down | Cytochrome P450 |
| gene3654 | −4.2753 | down | Cytochrome P450 |
| gene2397 | −1.9457 | down | Cytochrome P450 |
| gene12168 | −9.0107 | down | Cytochrome P450 |
| gene13312 | 1.9832 | up | Cytochrome P450 |
| gene5352 | −5.5725 | down | Cytochrome P450 |
| gene9354 | −3.9415 | down | Cytochrome P450 |
| gene10880 | −1.1803 | down | Cytochrome P450 |
| gene5571 | −4.3013 | down | Cytochrome P450 |
| gene7176 | −1.1220 | down | Cytochrome P450 |
| gene8218 | −2.9576 | down | Cytochrome P450 |
| gene6073 | −3.5630 | down | Cytochrome P450 |
| gene10344 | −7.0703 | down | Cytochrome P450 |
| gene4550 | 1.5708 | up | Cytochrome P450 |
| gene11675 | 2.0081 | up | Cytochrome P450 |
| gene4276 | −2.8887 | down | Cytochrome P450 |
| gene11233 | 1.8129 | up | Cytochrome P450 |
| gene6652 | 2.6545 | up | Cytochrome P450 |
| gene8442 | 1.7749 | up | Cytochrome P450 |
| gene1667 | −1.8225 | down | Cytochrome P450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhu, Q.; Chen, Z.; Chen, J.; Liu, Z.; Wu, X. Alternaria alternata botybirnavirus 1 (AaBRV1) Infection Affects the Biological Characteristics of Its Host Fungus Alternaria alternata. J. Fungi 2025, 11, 376. https://doi.org/10.3390/jof11050376
Zhang X, Zhu Q, Chen Z, Chen J, Liu Z, Wu X. Alternaria alternata botybirnavirus 1 (AaBRV1) Infection Affects the Biological Characteristics of Its Host Fungus Alternaria alternata. Journal of Fungi. 2025; 11(5):376. https://doi.org/10.3390/jof11050376
Chicago/Turabian StyleZhang, Xinyi, Qiqi Zhu, Ziyuan Chen, Ju Chen, Zhijun Liu, and Xuehong Wu. 2025. "Alternaria alternata botybirnavirus 1 (AaBRV1) Infection Affects the Biological Characteristics of Its Host Fungus Alternaria alternata" Journal of Fungi 11, no. 5: 376. https://doi.org/10.3390/jof11050376
APA StyleZhang, X., Zhu, Q., Chen, Z., Chen, J., Liu, Z., & Wu, X. (2025). Alternaria alternata botybirnavirus 1 (AaBRV1) Infection Affects the Biological Characteristics of Its Host Fungus Alternaria alternata. Journal of Fungi, 11(5), 376. https://doi.org/10.3390/jof11050376






