Advances in CRISPR/Cas9-Based Gene Editing in Filamentous Fungi
Abstract
1. Introduction
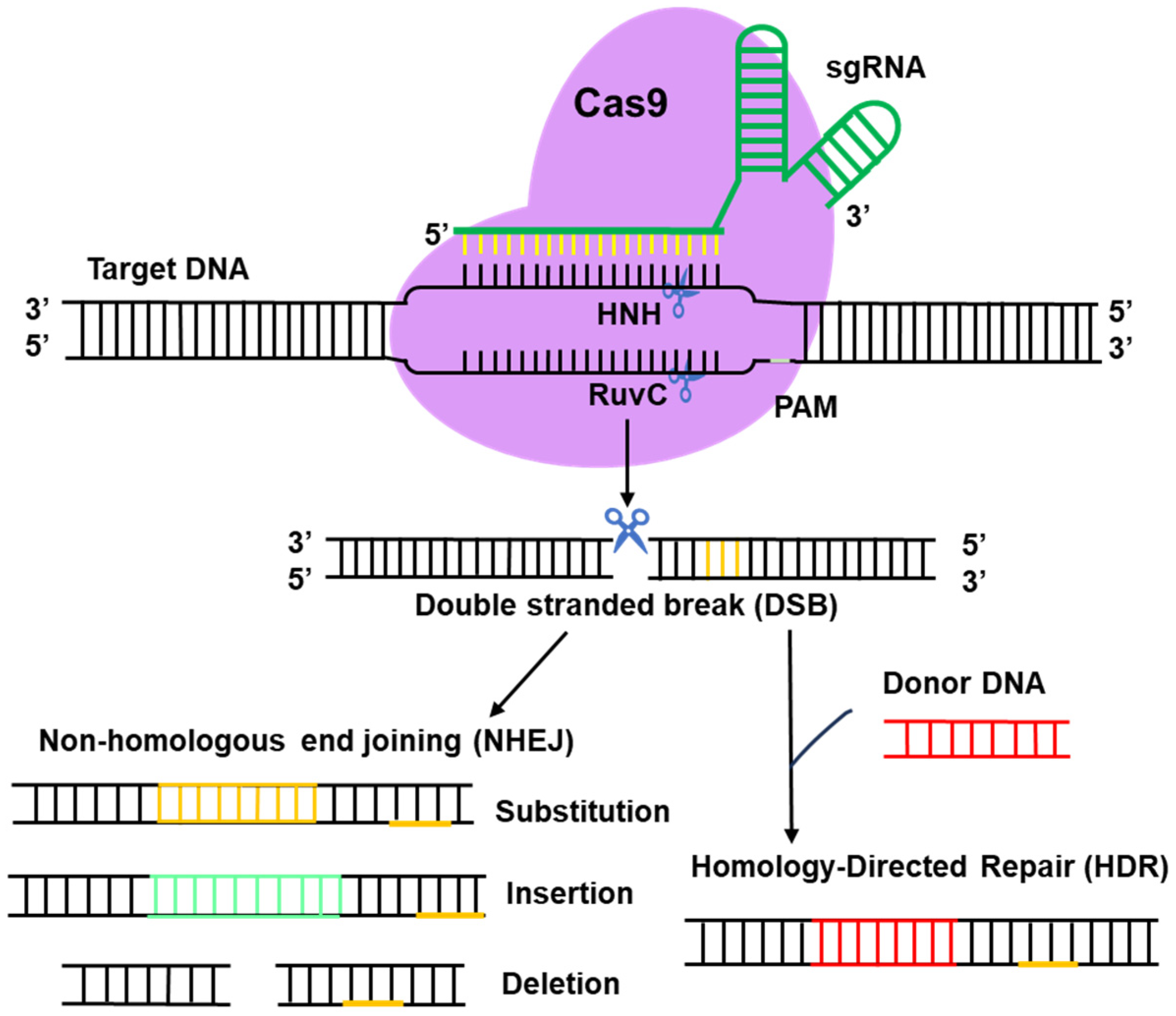
2. Brief About CRISPR/Cas9
3. Effects of Gene Editing on Filamentous Fungi
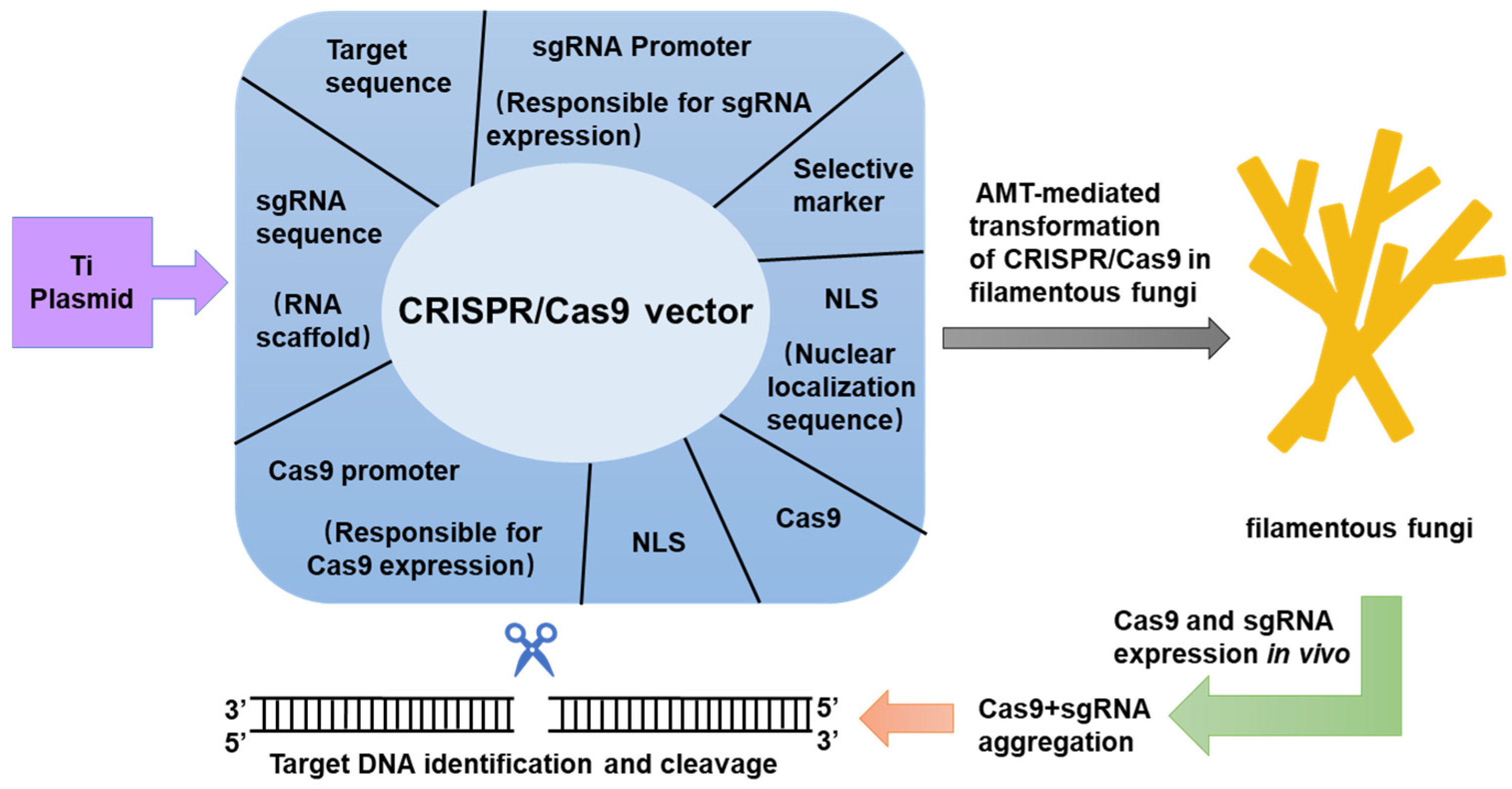
4. CRISPR/Cas9 Expression in Filamentous Fungi
4.1. Cas9 Protein Expression
4.2. Expression of sgRNA
5. Transformation of Cas9 Protein and sgRNA in Filamentous Fungi
6. Regulation of the HR/NHEJ Pathway in Filamentous Fungi
7. Application of CRISPR/Cas9 Technology in Filamentous Fungi
7.1. Application of CRISPR/Cas9 in Engineered Strains of Filamentous Fungi
7.2. Application of CRISPR/Cas9 Technology to Editing Filamentous Fungi in Agriculture
7.3. CRISPR/Cas9 Applied to Industrial Strains of Filamentous Fungi
7.4. CRISPR/Cas9 Applied to Filamentous Fungi Strains for the Pharmaceutical Industry
7.5. CRISPR/Cas9 Applied to Other Strains of Filamentous Fungi
8. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2101–2137. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, K.; Kapoor, N.; Kaur, H.; Abu-Seer, E.A.; Tariq, M.; Siddiqui, S.; Yadav, V.K.; Niazi, P.; Kumar, P.; Alghamdi, S. A Comprehensive Review of the Diversity of Fungal Secondary Metabolites and Their Emerging Applications in Healthcare and Environment. Mycobiology 2024, 52, 335–387. [Google Scholar] [CrossRef]
- Hyde, K.D. The numbers of fungi. Fungal Divers. 2022, 114, 1. [Google Scholar] [CrossRef]
- Troiano, D.; Orsat, V.; Dumont, M.J. Status of filamentous fungi in integrated biorefineries. Renew. Sustain. Energy Rev. 2020, 117, 109472. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Troein, C.; Siregar, S.; Gentile, L.; Abbondanza, G.; Peterson, C.; Persson, P.; Tunlid, A. Regulation of fungal decomposition at single-cell level. ISME J. 2020, 14, 896–905. [Google Scholar] [CrossRef]
- Sharma, A.; Sooch, B.S. Chapter 4—Fungi as sources of industrial enzymes: Sources, production, properties, structure and applications. In Fungal Biotechnology; Singh, R.S., Bhari, R., Eds.; Academic Press: Cambridge, MA, USA, 2025; pp. 69–95. [Google Scholar]
- Liu, D.; Garrigues, S.; de Vries, R.P. Heterologous protein production in filamentous fungi. Appl. Microbiol. Biotechnol. 2023, 107, 5019–5033. [Google Scholar] [CrossRef]
- Ullah, M.; Xia, L.; Xie, S.; Sun, S. CRISPR/Cas9-based genome engineering: A new breakthrough in the genetic manipulation of filamentous fungi. Biotechnol. Appl. Biochem. 2020, 67, 835–851. [Google Scholar] [CrossRef]
- Arazoe, T. Genome Editing Using CRISPR/Cas9 System in the Rice Blast Fungus. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2021; Volume 2356, pp. 149–160. [Google Scholar]
- Behera, B.C. Citric acid from Aspergillus niger: A comprehensive overview. Crit. Rev. Microbiol. 2020, 46, 727–749. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Qi, C.; Shen, X.; Li, B.; Liu, C.; Huang, L.; Lan, H.; Chen, D.; Jiang, Y.; Wang, D. PAMPHLET: PAM Prediction HomoLogous-Enhancement Toolkit for precise PAM prediction in CRISPR-Cas systems. J. Genet. Genom. 2025, 52, 258–268. [Google Scholar] [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.-f.; Yu, T. CRISPR/Cas9 therapeutics: Progress and prospects. Signal Transduct. Target. Ther. 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Westermann, L.; Neubauer, B.; Köttgen, M. Nobel Prize 2020 in Chemistry honors CRISPR: A tool for rewriting the code of life. Pflug. Arch. Eur. J. Physiol. 2021, 473, 1–2. [Google Scholar] [CrossRef]
- Mani, I.; Arazoe, T.; Singh, V. Chapter Two—CRISPR-Cas systems for genome editing of mammalian cells. In Progress in Molecular Biology and Translational Science; Singh, V., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 181, pp. 15–30. [Google Scholar]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, K.-F.; Wang, W.-J.; Ma, Y.-R.; Shi, T.-Q.; Huang, H.; Ji, X.-J. Increasing the homologous recombination efficiency of eukaryotic microorganisms for enhanced genome engineering. Appl. Microbiol. Biotechnol. 2019, 103, 4313–4324. [Google Scholar] [CrossRef]
- McCarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Li, Y.; Xia, H.; Mao, Q. Generation of a novel HEK293 luciferase reporter cell line by CRISPR/Cas9-mediated site-specific integration in the genome to explore the transcriptional regulation of the PGRN gene. Bioengineered 2019, 10, 98–107. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Laughery, M.F.; Wyrick, J.J. Simple CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2019, 129, e110. [Google Scholar] [CrossRef]
- Modaffari, D.; Finlayson, A.; Miao, Y.; Wallace, E.; Sawin, K. Improved gene editing and fluorescent-protein tagging in Aspergillus nidulans using a Golden Gate-based CRISPR-Cas9 plasmid system. Wellcome Open Res. 2024, 9, 602. [Google Scholar] [CrossRef]
- Ali Agha, A.S.A.; Al-Samydai, A.; Aburjai, T. New Frontiers in CRISPR: Addressing Antimicrobial Resistance with Cas9, Cas12, Cas13, and Cas14. Heliyon 2025, 11, e42013. [Google Scholar] [CrossRef]
- Lee, D.; Muir, P.; Lundberg, S.; Lundholm, A.; Sandegren, L.; Koskiniemi, S. A CRISPR-Cas9 system protecting E. coli against acquisition of antibiotic resistance genes. Sci. Rep. 2025, 15, 1545. [Google Scholar] [CrossRef]
- Strzyz, P. CRISPR–Cas9 wins Nobel. Nat. Rev. Mol. Cell Biol. 2020, 21, 714. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 2018, 3, 135–149. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; El-Tanani, M.; Tambuwala, M.M. Principles of CRISPR-Cas9 technology: Advancements in genome editing and emerging trends in drug delivery. J. Drug Deliv. Sci. Technol. 2024, 92, 105338. [Google Scholar] [CrossRef]
- Lin, J.; Feng, M.; Zhang, H.; She, Q. Characterization of a novel type III CRISPR-Cas effector provides new insights into the allosteric activation and suppression of the Cas10 DNase. Cell Discov. 2020, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Bhatia, S.; Yadav, S.K. CRISPR-Cas for genome editing: Classification, mechanism, designing and applications. Int. J. Biol. Macromol. 2023, 238, 124054. [Google Scholar] [CrossRef]
- Fang, S.; Song, X.; Cui, L.; Hu, L.; Wang, M.; Ai, L.; Wang, S. Application of the Streptococcus pyogenes CRISPR/Cas9 system in Lacticaseibacillus paracasei CGMCC4691. J. Future Foods 2025, 5, 520–527. [Google Scholar] [CrossRef]
- Maruyama, J.I. Genome Editing Technology and Its Application Potentials in the Industrial Filamentous Fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef]
- Yang, L.-Z.; Min, Y.-H.; Liu, Y.-X.; Gao, B.-Q.; Liu, X.-Q.; Huang, Y.; Wang, H.; Yang, L.; Liu, Z.J.; Chen, L.-L. CRISPR-array-mediated imaging of non-repetitive and multiplex genomic loci in living cells. Nat. Methods 2024, 21, 1646–1657. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Ceremuga, M.; Krzowski, L.; Saluk-Bijak, J.; Bijak, M. Various Aspects of a Gene Editing System-CRISPR-Cas9. Int. J. Mol. Sci. 2020, 21, 9604. [Google Scholar] [CrossRef]
- Song, R.; Zhai, Q.; Sun, L.; Huang, E.; Zhang, Y.; Zhu, Y.; Guo, Q.; Tian, Y.; Zhao, B.; Lu, H. CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 2019, 103, 6919–6932. [Google Scholar] [CrossRef]
- Jensen, E.D.; Ferreira, R.; Jakočiūnas, T.; Arsovska, D.; Zhang, J.; Ding, L.; Smith, J.D.; David, F.; Nielsen, J.; Jensen, M.K.; et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb. Cell Fact. 2017, 16, 46. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Xu, X.; Wang, Y.; Chen, W.; Wang, Y.; Wu, Z.; Tang, N.; Wang, Y.; Zhao, S.; et al. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat. Catal. 2020, 3, 813–823. [Google Scholar] [CrossRef]
- Nayfach, S.; Bhatnagar, A.; Novichkov, A.; Estevam, G.O.; Kim, N.; Hill, E.; Ruffolo, J.A.; Silverstein, R.; Gallagher, J.; Kleinstiver, B.; et al. Engineering of CRISPR-Cas PAM recognition using deep learning of vast evolutionary data. bioRxiv 2025. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Du, Y.; Liu, Y.; Hu, J.; Peng, X.; Liu, Z. CRISPR/Cas9 systems: Delivery technologies and biomedical applications. Asian J. Pharm. Sci. 2023, 18, 100854. [Google Scholar] [CrossRef]
- Chen, K.; Maimaitirexiati, G.; Zhang, Q.; Li, Y.; Liu, X.; Tang, H.; Gao, X.; Wang, B.; Yu, T.; Guo, S. CRISPR-Cas9-based one-step multiplexed genome editing through optimizing guide RNA processing strategies in Pichia pastoris. Synth. Syst. Biotechnol. 2025, 10, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Ju, S.; Jung, W.J.; Jeong, T.Y.; Yoon, D.E.; Lee, J.H.; Yang, J.; Lee, H.; Choi, J.; Kim, H.S.; et al. Robust genome editing activity and the applications of enhanced miniature CRISPR-Cas12f1. Nat. Commun. 2025, 16, 677. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Kaur, E.; Raghavan, S.C.; Sengupta, S. Regulation of pathway choice in DNA repair after double-strand breaks. Curr. Opin. Pharmacol. 2025, 80, 102496. [Google Scholar] [CrossRef]
- Liao, H.; Wu, J.; VanDusen, N.J.; Li, Y.; Zheng, Y. CRISPR-Cas9-mediated homology-directed repair for precise gene editing. Mol. Ther.—Nucleic Acids 2024, 35, 102344. [Google Scholar] [CrossRef] [PubMed]
- Torres-Ruiz, R.; Rodriguez-Perales, S. CRISPR-Cas9 technology: Applications and human disease modelling. Brief. Funct. Genom. 2017, 16, 4–12. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, M.; Lin, S.; Huang, M.; Chen, S.; Chen, S. The application of CRISPR/Cas9–based genome-wide screening to disease research. Mol. Cell. Probes 2025, 79, 102004. [Google Scholar] [CrossRef]
- Deng, H.; Gao, R.; Liao, X.; Cai, Y. CRISPR system in filamentous fungi: Current achievements and future directions. Gene 2017, 627, 212–221. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Lightfoot, J.D.; Fuller, K.K. CRISPR/Cas9-Mediated Gene Replacement in the Fungal Keratitis Pathogen Fusarium solani var. petroliphilum. Microorganisms 2019, 7, 457. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, X.; Xiong, W.; Zhang, K.; Shen, W.; Zhang, Y.; Xu, X.; Zhong, C.; Zhang, Y.; Tian, T.; et al. Reducing CRISPR-Cas9 off-target effects by optically controlled chemical modifications of guide RNA. Cell Chem. Biol. 2024, 31, 1839–1851.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cobine, P.A.; Coleman, J.J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet. Biol. 2018, 117, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.Q.; Gao, J.; Wang, W.J.; Wang, K.F.; Xu, G.Q.; Huang, H.; Ji, X.J. CRISPR/Cas9-Based Genome Editing in the Filamentous Fungus Fusarium fujikuroi and Its Application in Strain Engineering for Gibberellic Acid Production. ACS Synth. Biol. 2019, 8, 445–454. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi. Comput. Struct. Biotechnol. J. 2019, 17, 761–769. [Google Scholar] [CrossRef]
- Schuster, M.; Schweizer, G.; Kahmann, R. Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Genet. Biol. 2018, 112, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Han, Y.; Wang, C.; Jiang, C.; Xu, J.R. Targeted Deletion of the USTA and UvSLT2 Genes Efficiently in Ustilaginoidea virens with the CRISPR-Cas9 System. Front. Plant Sci. 2018, 9, 699. [Google Scholar] [CrossRef]
- Xia, X.; Li, S.; Wang, N.; Cheng, P.; Zhu, B.; Zhang, P.; Yang, D.; Lin, H.; Niu, L. Convenient, high-efficiency multiplex genome editing in autotetraploid alfalfa using endogenous U6 promoters and visual reporters. aBIOTECH 2025, 6, 81–90. [Google Scholar] [CrossRef]
- Zhang, Y.; Ouyang, L.; Nan, Y.; Chu, J. Efficient gene deletion and replacement in Aspergillus niger by modified in vivo CRISPR/Cas9 systems. Bioresour. Bioprocess. 2019, 6, 4. [Google Scholar] [CrossRef]
- Zheng, X.; Cairns, T.; Zheng, P.; Meyer, V.; Sun, J. Protocol for gene characterization in Aspergillus niger using 5S rRNA-CRISPR-Cas9-mediated Tet-on inducible promoter exchange. STAR Protoc. 2022, 3, 101838. [Google Scholar] [CrossRef]
- Yuan, G.; Deng, S.; Czajka, J.J.; Dai, Z.; Hofstad, B.A.; Kim, J.; Pomraning, K.R. CRISPR-Cas9/Cas12a systems for efficient genome editing and large genomic fragment deletions in Aspergillus niger. Front. Bioeng. Biotechnol. 2024, 12, 1452496. [Google Scholar] [CrossRef]
- van Leeuwe, T.M.; Arentshorst, M.; Ernst, T.; Alazi, E.; Punt, P.J.; Ram, A.F.J. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 2019, 6, 13. [Google Scholar] [CrossRef]
- Yoshioka, I.; Kirimura, K. Rapid and marker-free gene replacement in citric acid-producing Aspergillus tubingensis (A. niger) WU-2223L by the CRISPR/Cas9 system-based genome editing technique using DNA fragments encoding sgRNAs. J. Biosci. Bioeng. 2021, 131, 579–588. [Google Scholar] [CrossRef]
- Chey, Y.C.J.; Gierus, L.; Lushington, C.; Arudkumar, J.C.; Geiger, A.B.; Staker, L.G.; Robertson, L.J.; Pfitzner, C.; Kennedy, J.G.; Lee, R.H.B.; et al. Optimal SpCas9- and SaCas9-mediated gene editing by enhancing gRNA transcript levels through scaffold poly-T tract reduction. BMC Genom. 2025, 26, 138. [Google Scholar] [CrossRef] [PubMed]
- Riesenberg, S.; Helmbrecht, N.; Kanis, P.; Maricic, T.; Pääbo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 2022, 13, 489. [Google Scholar] [CrossRef]
- Zhao, X.; Song, H.; Liu, J.; Feng, K.; Wu, Q.; Arif, T.; Cao, Y.; Zhang, L. Efficient Protoplast Isolation and PEG-mediated Transformation protocols for blueberry Vaccinium corymbosum. Sci. Hortic. 2025, 340, 113916. [Google Scholar] [CrossRef]
- Wang, S.; Wang, G.; Li, H.; Li, F.; Wang, J. Agrobacterium tumefaciens-mediated transformation of embryogenic callus and CRISPR/Cas9-mediated genome editing in ‘Feizixiao’ litchi. Hortic. Plant J. 2023, 9, 947–957. [Google Scholar] [CrossRef]
- Park, S.-J.; Park, S.J.; Kwon, Y.W.; Choi, E.-H. Synergistic combination of RAD51-SCR7 improves CRISPR-Cas9 genome editing efficiency by preventing R-loop accumulation. Mol. Ther.—Nucleic Acids 2024, 35, 102274. [Google Scholar] [CrossRef] [PubMed]
- Almotiri, A.; Abogosh, A.; Abdelfattah, A.; Alowaisy, D.; Rodrigues, N.P. Treating genetic blood disorders in the era of CRISPR-mediated genome editing. Mol. Ther. 2025. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoon, S.; Kim, K.P. Boosting CRISPR-Cas9 efficiency through enhanced homologous recombination. Mol. Ther.—Nucleic Acids 2024, 35, 102329. [Google Scholar] [CrossRef]
- Zheng, C.; Zhang, G.; Dean, L.J.; Sontheimer, E.J.; Xue, W. The reverse transcriptase domain of prime editors contributes to DNA repair in mammalian cells. Nat. Biotechnol. 2025, 1–8. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Ray, U.; Raghavan, S.C. Modulation of DNA double-strand break repair as a strategy to improve precise genome editing. Oncogene 2020, 39, 6393–6405. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, S.; Liu, Q.; Chen, F.; Shao, Y. CRISPR/Cas9 system is a suitable gene targeting editing tool to filamentous fungus Monascus pilosus. Appl. Microbiol. Biotechnol. 2024, 108, 154. [Google Scholar] [CrossRef]
- Lv, D.; Zhang, W.; Meng, X.; Liu, W. Single Mutation in Transcriptional Activator Xyr1 Enhances Cellulase and Xylanase Production in Trichoderma reesei on Glucose. J. Agric. Food Chem. 2023, 71, 11993–12003. [Google Scholar] [CrossRef]
- Jiménez, A.; Muñoz-Fernández, G.; Ledesma-Amaro, R.; Buey, R.M.; Revuelta, J.L. One-vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii. Microb. Biotechnol. 2019, 12, 1293–1301. [Google Scholar] [CrossRef]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X. Mutations in ORP1 Conferring Oxathiapiprolin Resistance Confirmed by Genome Editing using CRISPR/Cas9 in Phytophthora capsici and P. sojae. Phytopathology 2018, 108, 1412–1419. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Zhang, J.; Li, K.; Sun, Y.; Yao, C.; Liu, W.; Liu, H.; Zhong, Y. An efficient CRISPR/Cas9 genome editing system based on a multiple sgRNA processing platform in Trichoderma reesei for strain improvement and enzyme production. Biotechnol. Biofuels Bioprod. 2024, 17, 22. [Google Scholar] [CrossRef]
- Mathis, H.; Naquin, D.; Margeot, A.; Bidard, F. Enhanced heterologous gene expression in Trichoderma reesei by promoting multicopy integration. Appl. Microbiol. Biotechnol. 2024, 108, 470. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Duan, C.; Pan, Y.; Liu, G. Improvement of the CRISPR-Cas9 mediated gene disruption and large DNA fragment deletion based on a chimeric promoter in Acremonium chrysogenum. Fungal Genet. Biol. 2020, 134, 103279. [Google Scholar] [CrossRef] [PubMed]
- Todokoro, T.; Hata, Y.; Ishida, H. CRISPR/Cas9 improves targeted knock-in efficiency in Aspergillus oryzae. Biotechnol. Notes 2024, 5, 58–63. [Google Scholar] [CrossRef]
- Watts, A.; Sankaranarayanan, S.; Watts, A.; Raipuria, R.K. Optimizing protein expression in heterologous system: Strategies and tools. Meta Gene 2021, 29, 100899. [Google Scholar] [CrossRef]
- Grüttner, S.; Kempken, F. A user-friendly CRISPR/Cas9 system for mutagenesis of Neurospora crassa. Sci. Rep. 2024, 14, 20469. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Cerezo, S.; Kun, R.S.; de Vries, R.P.; Garrigues, S. CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer. Enzym. Microb. Technol. 2020, 133, 109463. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, P.; Tu, W.; Gao, Q.; Wang, C.; Tan, L.; Zhao, L.; Han, H.; Ma, L.; Otsuki, K.; et al. Highly anticipated natural diterpenoids as an important source of new drugs in 2013–2023. Chin. Chem. Lett. 2025, 36, 109955. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Abdel-Ghany, S.E.; Ali, G.S. Genome editing approaches: Manipulating of lovastatin and taxol synthesis of filamentous fungi by CRISPR/Cas9 system. Appl. Microbiol. Biotechnol. 2017, 101, 3953–3976. [Google Scholar] [CrossRef]
- Qin, H.; Xiao, H.; Zou, G.; Zhou, Z.; Zhong, J.-J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 2017, 56, 57–61. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J.-I. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]
- Wei, T.-Y.; Wu, Y.-J.; Xie, Q.-P.; Tang, J.-W.; Yu, Z.-T.; Yang, S.-B.; Chen, S.-X. CRISPR/Cas9-Based Genome Editing in the Filamentous Fungus Glarea lozoyensis and Its Application in Manipulating gloF. ACS Synth. Biol. 2020, 9, 1968–1977. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Liu, D.; Liu, D.; Zhang, C.; Lin, L.; Zhu, Z.; Li, H.; Dai, Y.; Wang, X.; et al. MtTRC-1, a Novel Transcription Factor, Regulates Cellulase Production via Directly Modulating the Genes Expression of the Mthac-1 and Mtcbh-1 in Myceliophthora thermophila. Appl. Environ. Microbiol. 2022, 88, e0126322. [Google Scholar] [CrossRef] [PubMed]
- Benites-Pariente, J.S.; Samolski, I.; Ludeña, Y.; Villena, G.K. CRISPR/Cas9 mediated targeted knock-in of eglA gene to improve endoglucanase activity of Aspergillus fumigatus LMB-35Aa. Sci. Rep. 2024, 14, 19661. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, C.; Pei, L.; Qian, Q.; Lu, L. Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems. J. Fungi 2023, 9, 719. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, N.; Song, Y.; Gao, J.; Nian, L.; Zhou, J.; Zhang, B.; Liu, Z.; Zheng, Y. Development of a marker recyclable CRISPR/Cas9 system for scarless and multigene editing in Fusarium fujikuroi. Biotechnol. J. 2024, 19, e2400164. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Tang, J.; Dang, Y.; Sun, L.; Liu, B.; Li, H.; He, X.; Shuai, Q.; Peng, Z.; et al. Development of a quinic acid-induced CRISPR/Cas9 genome editing system and its application for the activation of ilicicolin H biosynthesis in Trichoderma reesei. Int. J. Biol. Macromol. 2024, 279 Pt. 4, 135339. [Google Scholar] [CrossRef]
- Zhao, S.; Yin, R.; Zhang, M.; Zhai, Z.; Shen, Z.; Mou, Y.; Xu, D.; Zhou, L.; Lai, D. Efficient gene editing in the slow-growing, non-sporulating, melanized, endophytic fungus Berkleasmium sp. Dzf12 using a CRISPR/Cas9 system. World J. Microbiol. Biotechnol. 2024, 40, 176. [Google Scholar] [CrossRef]
- Ree Yoon, H.; Han, S.; Chul Shin, S.; Cheong Yeom, S.; Jin Kim, H. Improved natural food colorant production in the filamentous fungus Monascus ruber using CRISPR-based engineering. Food Res. Int. 2023, 167, 112651. [Google Scholar] [CrossRef]
- Chen, B.X.; Xue, L.N.; Wei, T.; Wang, N.; Zhong, J.R.; Ye, Z.W.; Guo, L.Q.; Lin, J.F. Multiplex gene precise editing and large DNA fragment deletion by the CRISPR-Cas9-TRAMA system in edible mushroom Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2982–2991. [Google Scholar] [CrossRef]

| Strain Name | Experimental Strategy | Editing Efficiency | Major Findings | References |
|---|---|---|---|---|
| A. oryzae | Cas9 (amyB promoter), sgRNA (U6 promoter), NHEJ repair | 10–20% | CRISPR/Cas9-mediated genome editing was achieved for the first time in A. oryzae, demonstrating the feasibility of the system. | [92] |
| Glarea lozoyensis | Cas9 (trpC promoter), sgRNA (U6 promoter), NHEJ repair | Approximately 80% | CRISPR/Cas9-based gene editing tool is efficient for manipulating genes in G. lozoyensis | [93] |
| Myceliophthora thermophila | Cas9 (Ptef1 promoter), sgRNA (U6 promoter), HR repair | Approximately 100% | Characterized a novel regulator MtTRC-1 in M. thermophila, which regulated cellulase production through direct transcriptional regulation of the Mthac-1 and Mtcbh-1 genes. | [94] |
| A. fumigatus | Cas9 (tef1 promoter), sgRNA (gpdA promoter), HR repair | Approximately 10% | A CRISPR/Cas9-mediated gene-editing strategy for improving the endoglucanase activity of A. fumigatus LMB-35Aa strain was successfully used, which constitutes the first report of heterologous cellulase production in filamentous fungi using this technology. | [95] |
| A. nidulans | Cas9 (PgpdA promoter), sgRNA (T7 promoter), MMEJ repair | Approximately 100% | The production of L-malic acid was enhanced by approximately 9.6 times. | [96] |
| F. fujikuroi | Cas9 (Ptef1 promoter), sgRNA (5S rRNA promoter), HR repair | dual: 25% 75% triple: 12.5~37.5% | The production of GA3 was enhanced by approximately 50.19%. | [97] |
| T. reesei | Cas9 (qai5 promoter), sgRNA (T7 promoter), HR repair | 46.7% | The production yield of ilicicolin H reached 4.8 mg/L. | [98] |
| Berkleasmium sp. Dzf12 | Cas9 (Ptef1 promoter), sgRNA (U6 snRNA promoter), HR repair | 16.6%~50% | DHN and spirobisnaphthalenes were found to have a biosynthetic relationship. | [99] |
| M. ruber | Cas9 (PgpdA promoter), sgRNA (T7 promoter), NHEJ repair | 18.2% | MpigI and MpigI’ were directly related in the production of Monascus pigments. | [100] |
| Cordyceps militaris | Cas9 (Pcmgpd promoter), sgRNA (Ptrpc promoter), NHEJ repair | 17.9% | Protein modification and promoter strength evaluation were performed, along with the deletion of 10 kb biosynthetic clusters. | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, B.; Li, Y.; Wang, T.; Li, D.; Jia, S. Advances in CRISPR/Cas9-Based Gene Editing in Filamentous Fungi. J. Fungi 2025, 11, 350. https://doi.org/10.3390/jof11050350
Ma B, Li Y, Wang T, Li D, Jia S. Advances in CRISPR/Cas9-Based Gene Editing in Filamentous Fungi. Journal of Fungi. 2025; 11(5):350. https://doi.org/10.3390/jof11050350
Chicago/Turabian StyleMa, Bin, Yimiao Li, Tinghui Wang, Dongming Li, and Shuang Jia. 2025. "Advances in CRISPR/Cas9-Based Gene Editing in Filamentous Fungi" Journal of Fungi 11, no. 5: 350. https://doi.org/10.3390/jof11050350
APA StyleMa, B., Li, Y., Wang, T., Li, D., & Jia, S. (2025). Advances in CRISPR/Cas9-Based Gene Editing in Filamentous Fungi. Journal of Fungi, 11(5), 350. https://doi.org/10.3390/jof11050350





