DNA Sequence Changes Resulting from Codon Optimization Affect Gene Expression in Pichia pastoris by Altering Chromatin Accessibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Plasmid Construction and Strain Generation
2.3. RNA Extraction and RT-qPCR
2.4. Chromatin Immuno-Precipitation (ChIP) Assay and qPCR
2.5. Chromatin Accessibility Assay
2.6. Data Analysis and Statistics
3. Results
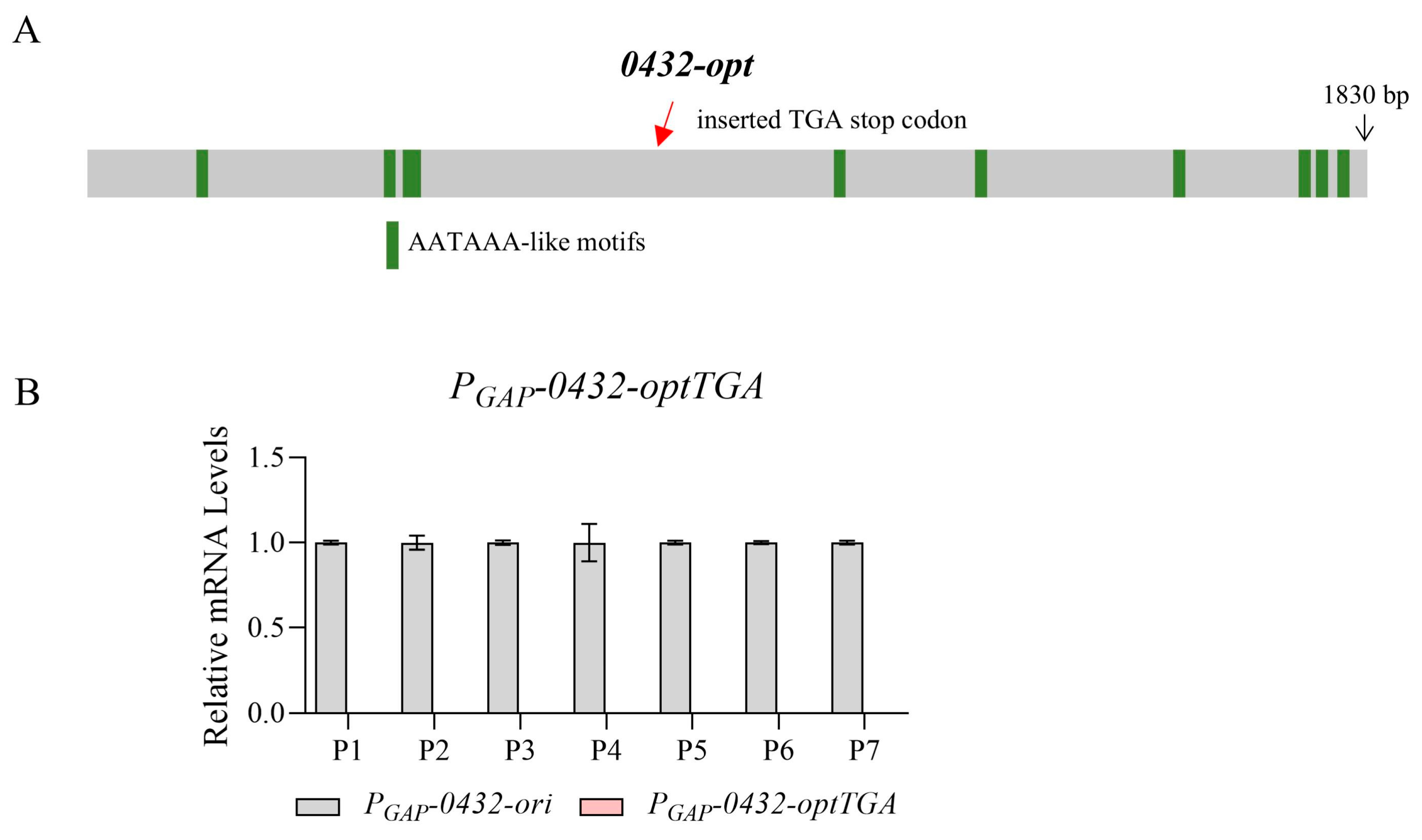
3.1. Codon Optimization Severely Abolished Mature mRNAs of 0432 and Fluc
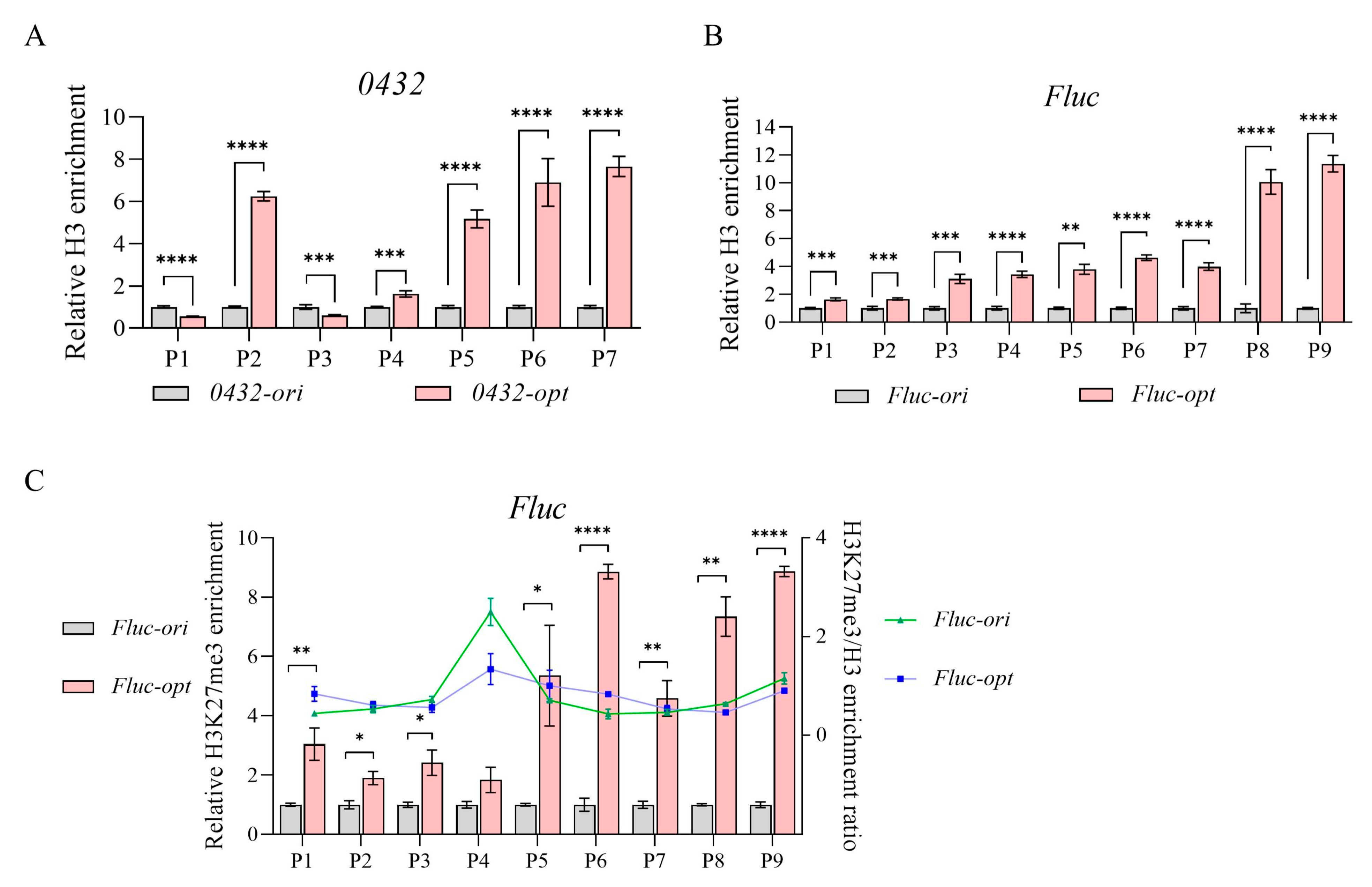
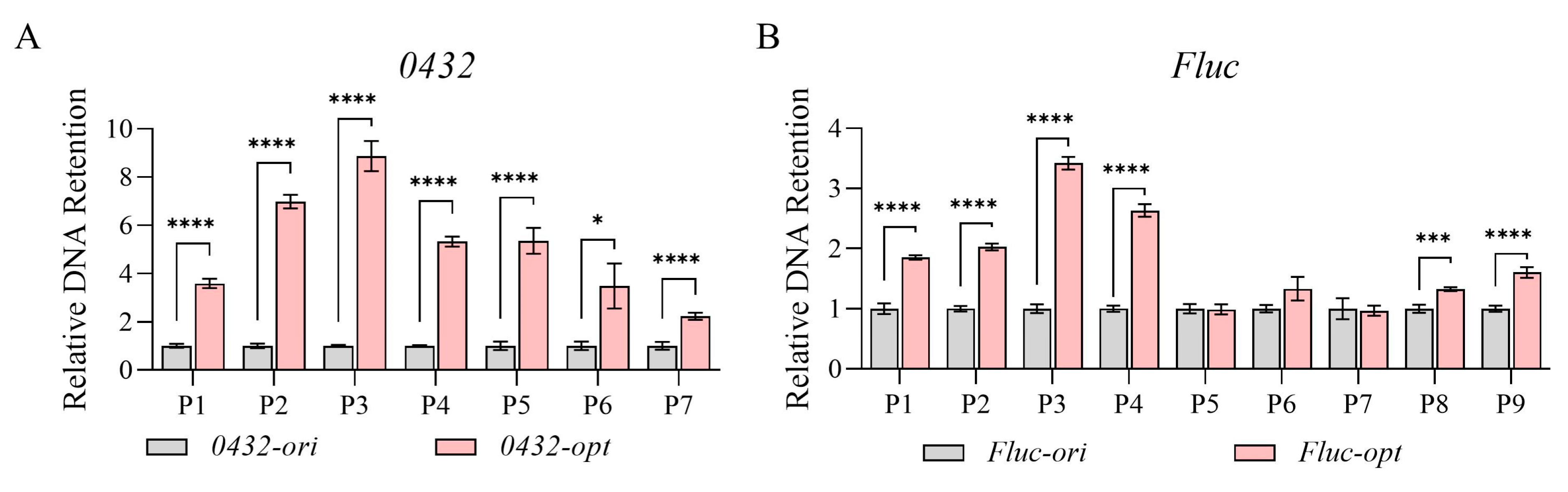
3.2. Codon Optimization Altered Nucleosome Occupancy and Chromatin Accessibility
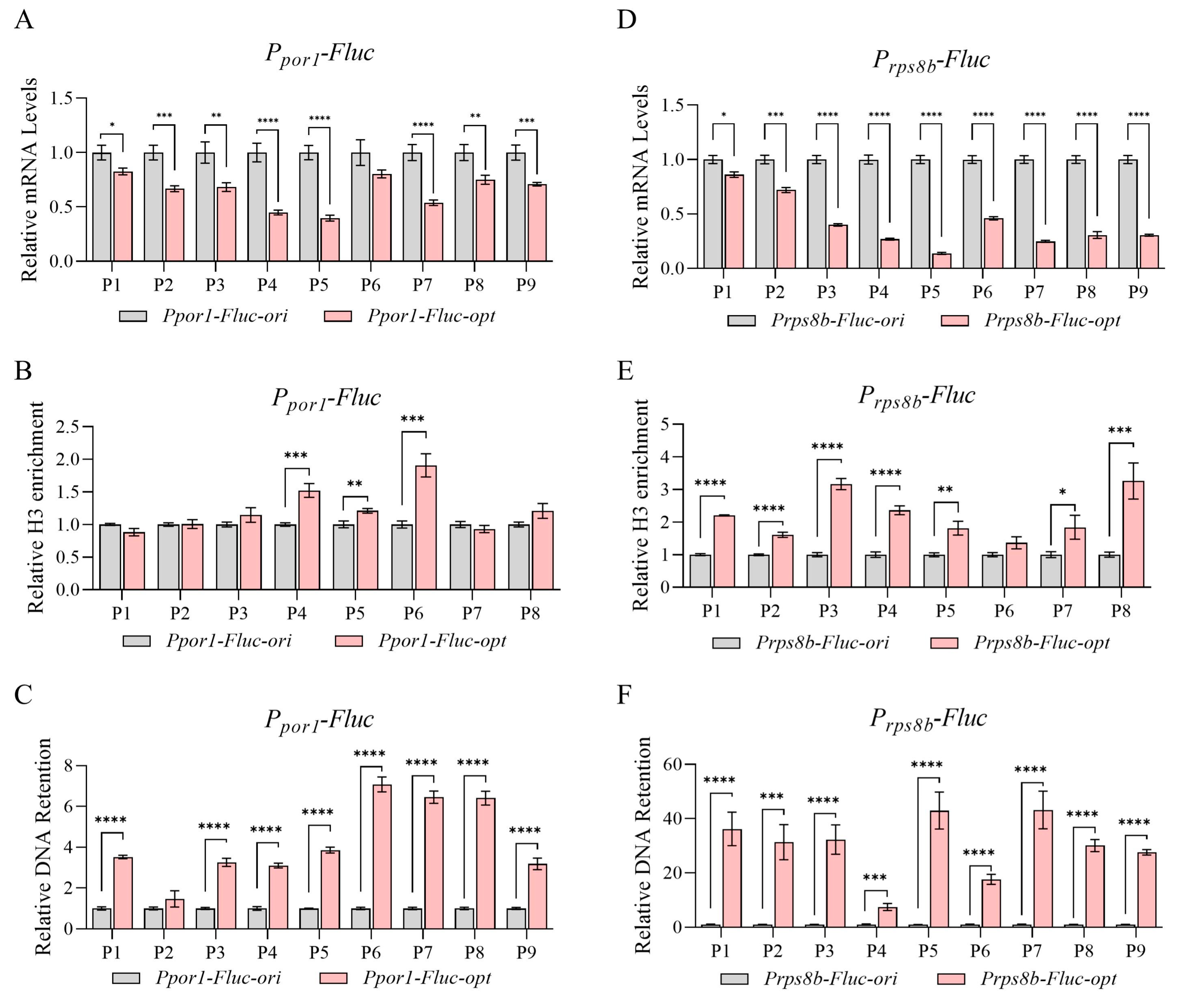
3.3. Transcriptional Repression and Reduced Chromatin Accessibility Caused by Codon Optimization Was Not Promoter-Specific
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brandis, G.; Hughes, D. The Selective Advantage of Synonymous Codon Usage Bias in Salmonella. PLoS Genet. 2016, 12, e1005926. [Google Scholar] [CrossRef] [PubMed]
- Szövényi, P.; Ullrich, K.K.; Rensing, S.A.; Lang, D.; Van Gessel, N.; Stenøien, H.K.; Conti, E.; Reski, R. Selfing in Haploid Plants and Efficacy of Selection: Codon Usage Bias in the Model Moss Physcomitrella Patens. Genome Biol. Evol. 2017, 9, 1528–1546. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ding, Y.; He, Y.; Chu, Y.; Zhao, P.; Ma, L.; Wang, X.; Li, X.; Liu, Y. The Effect of Multiple Evolutionary Selections on Synonymous Codon Usage of Genes in the Mycoplasma Bovis Genome. PLoS ONE 2014, 9, e108949. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, P.; Shah, P.; Rokas, A.; Liu, Y.; Johnson, C.H. Non-Optimal Codon Usage Is a Mechanism to Achieve Circadian Clock Conditionality. Nature 2013, 495, 116–120. [Google Scholar] [CrossRef]
- Zhou, M.; Guo, J.; Cha, J.; Chae, M.; Chen, S.; Barral, J.M.; Sachs, M.S.; Liu, Y. Non-Optimal Codon Usage Affects Expression, Structure and Function of Clock Protein FRQ. Nature 2013, 495, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Hense, W.; Anderson, N.; Hutter, S.; Stephan, W.; Parsch, J.; Carlini, D.B. Experimentally Increased Codon Bias in the Drosophila Adh Gene Leads to an Increase in Larval, but Not Adult, Alcohol Dehydrogenase Activity. Genetics 2010, 184, 547–555. [Google Scholar] [CrossRef]
- Zhou, Z.; Dang, Y.; Zhou, M.; Li, L.; Yu, C.-H.; Fu, J.; Chen, S.; Liu, Y. Codon Usage Is an Important Determinant of Gene Expression Levels Largely through Its Effects on Transcription. Proc. Natl. Acad. Sci. USA 2016, 113, E6117–E6125. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A Code within the Genetic Code: Codon Usage Regulates Co-Translational Protein Folding. Cell Commun. Signal 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yu, C.; Liu, Y. Codon Usage Regulates Protein Structure and Function by Affecting Translation Elongation Speed in Drosophila Cells. Nucleic Acids Res. 2017, 45, 8484–8492. [Google Scholar] [CrossRef]
- Saunders, R.; Deane, C.M. Synonymous Codon Usage Influences the Local Protein Structure Observed. Nucleic Acids Res. 2010, 38, 6719–6728. [Google Scholar] [CrossRef]
- Jeacock, L.; Faria, J.; Horn, D. Codon Usage Bias Controls mRNA and Protein Abundance in Trypanosomatids. Elife 2018, 7, e32496. [Google Scholar] [CrossRef] [PubMed]
- Gooch, V.D.; Mehra, A.; Larrondo, L.F.; Fox, J.; Touroutoutoudis, M.; Loros, J.J.; Dunlap, J.C. Fully Codon-Optimized Luciferase Uncovers Novel Temperature Characteristics of the Neurospora Clock. Eukaryot. Cell 2008, 7, 28–37. [Google Scholar] [CrossRef]
- Huang, M.; Gao, Y.; Zhou, X.; Zhang, Y.; Cai, M. Regulating Unfolded Protein Response Activator HAC1p for Production of Thermostable Raw-Starch Hydrolyzing α-Amylase in Pichia Pastoris. Bioproc. Biosyst. Eng. 2017, 40, 341–350. [Google Scholar] [CrossRef]
- Huang, H.; Yang, P.; Luo, H.; Tang, H.; Shao, N.; Yuan, T.; Wang, Y.; Bai, Y.; Yao, B. High-Level Expression of a Truncated 1,3-1,4-Beta-D-Glucanase from Fibrobacter Succinogenes in Pichia Pastoris by Optimization of Codons and Fermentation. Appl. Microbiol. Biotechnol. 2008, 78, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.Y.; Xu, T.T.; Zhao, Q.J.; Li, C.L. Codon Optimization Enhances the Expression of Porcine β-Defensin-2 in Escherichia Coli. Genet. Mol. Res. 2015, 14, 4978–4988. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, K.; Han, Y.; Xing, Y.; Zhang, Y.; Yang, Q.; Zhou, M. Codon Usage Bias Regulates Gene Expression and Protein Conformation in Yeast Expression System P. Pastoris. Microb. Cell Fact. 2021, 20, 91. [Google Scholar] [CrossRef] [PubMed]
- Buhr, F.; Jha, S.; Thommen, M.; Mittelstaet, J.; Kutz, F.; Schwalbe, H.; Rodnina, M.V.; Komar, A.A. Synonymous Codons Direct Cotranslational Folding toward Different Protein Conformations. Mol. Cell 2016, 61, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cai, G.; Wu, D.; Lu, J. Comparison of Two Codon Optimization Strategies Enhancing Recombinant Sus Scrofa Lysozyme Production in Pichia Pastoris. Cell. Mol. Biol. 2015, 61, 43–49. [Google Scholar]
- Liu, K.; Ouyang, Y.; Lin, R.; Ge, C.; Zhou, M. Strong Negative Correlation between Codon Usage Bias and Protein Structural Disorder Impedes Protein Expression after Codon Optimization. J. Biotechnol. 2022, 343, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Tomari, Y. Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol. Cell 2016, 61, 874–885. [Google Scholar] [CrossRef]
- Igolkina, A.A.; Zinkevich, A.; Karandasheva, K.O.; Popov, A.A.; Selifanova, M.V.; Nikolaeva, D.; Tkachev, V.; Penzar, D.; Nikitin, D.M.; Buzdin, A. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks. Cells 2019, 8, 1034. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Dang, Y.; Zhou, M.; Yuan, H.; Liu, Y. Codon Usage Biases Co-Evolve with Transcription Termination Machinery to Suppress Premature Cleavage and Polyadenylation. Elife 2018, 7, e33569. [Google Scholar] [CrossRef]
- Xing, Y.; Gong, R.; Xu, Y.; Liu, K.; Zhou, M. Codon Usage Bias Affects α-Amylase mRNA Level by Altering RNA Stability and Cytosine Methylation Patterns in Escherichia Coli. Can. J. Microbiol. 2020, 66, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Mansisidor, A.R.; Risca, V.I.I. Chromatin Accessibility: Methods, Mechanisms, and Biological Insights. Nucleus 2022, 13, 236–276. [Google Scholar] [CrossRef]
- Misteli, T. Protein Dynamics: Implications for Nuclear Architecture and Gene Expression. Science 2001, 291, 843–847. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The Molecular Hallmarks of Epigenetic Control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L.; Safer, J.P.; Stanchfield, J.E. Structural Repeating Units in Chromatin. I. Evidence for Their General Occurrence. Exp. Cell Res. 1976, 97, 101–110. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin Accessibility and the Regulatory Epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.S.; Jia, S. Heterochromatin Revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef]
- Hodges, C.; Bintu, L.; Lubkowska, L.; Kashlev, M.; Bustamante, C. Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science 2009, 325, 626–628. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Reversing Histone Methylation. Nature 2005, 436, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, J.; Lian, Y.; Fan, C.; Zhang, P.; Wu, Y.; Li, X.; Xiong, F.; Li, X.; Li, G.; et al. Linking Long Non-Coding RNAs and SWI/SNF Complexes to Chromatin Remodeling in Cancer. Mol. Cancer 2017, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Voss, T.C.; Hager, G.L. Dynamic Regulation of Transcriptional States by Chromatin and Transcription Factors. Nat. Rev. Genet. 2014, 15, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Dann, G.P.; Liszczak, G.P.; Bagert, J.D.; Müller, M.M.; Nguyen, U.T.T.; Wojcik, F.; Brown, Z.Z.; Bos, J.; Panchenko, T.; Pihl, R.; et al. ISWI Chromatin Remodellers Sense Nucleosome Modifications to Determine Substrate Preference. Nature 2017, 548, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lin, R.; Cui, L.; Jiang, T.; Shi, J.; Lu, C.; Li, P.; Zhou, M. Alter Codon Bias of the P. Pastoris Genome to Overcome a Bottleneck in Codon Optimization Strategy Development and Improve Protein Expression. Microbiol. Res. 2024, 282, 127629. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Latrasse, D.; Benhamed, M. Histone Modification ChIP-Seq on Arabidopsis Thaliana Plantlets. Bio. Protoc. 2021, 11, e4211. [Google Scholar] [CrossRef]
- Nepon-Sixt, B.S.; Alexandrow, M.G. DNase I Chromatin Accessibility Analysis. Bio. Protoc. 2019, 9, e3444. [Google Scholar] [CrossRef]
- Stein, C.B.; Field, A.R.; Mimoso, C.A.; Zhao, C.; Huang, K.-L.; Wagner, E.J.; Adelman, K. Integrator Endonuclease Drives Promoter-Proximal Termination at All RNA Polymerase II-Transcribed Loci. Mol. Cell 2022, 82, 4232–4245.e11. [Google Scholar] [CrossRef]
- Rodríguez-Molina, J.B.; West, S.; Passmore, L.A. Knowing When to Stop: Transcription Termination on Protein-Coding Genes by Eukaryotic RNAPII. Mol. Cell 2023, 83, 404–415. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhang, H.-W.; Wu, X.-X.; Zhang, Y. Structural Basis of Exoribonuclease-Mediated mRNA Transcription Termination. Nature 2024, 628, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Klauer, A.A.; van Hoof, A. Degradation of mRNAs That Lack a Stop Codon: A Decade of Nonstop Progress. Wiley Interdiscip. Rev. RNA 2012, 3, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Crawford, G.E.; Davis, S.; Scacheri, P.C.; Renaud, G.; Halawi, M.J.; Erdos, M.R.; Green, R.; Meltzer, P.S.; Wolfsberg, T.G.; Collins, F.S. DNase-Chip: A High-Resolution Method to Identify DNase I Hypersensitive Sites Using Tiled Microarrays. Nat. Methods 2006, 3, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhu, J.; Zhu, Q.; Xing, Y.; Cai, M.; Jiang, T.; Zhou, M.; Zhang, Y. Identification and Characterization of Novel Promoters for Recombinant Protein Production in Yeast Pichia Pastoris. Yeast 2018, 35, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L. XSTREME: Comprehensive Motif Analysis of Biological Sequence Datasets. BioRxiv 2021. [Google Scholar] [CrossRef]
- Du, K.; Luo, Q.; Yin, L.; Wu, J.; Liu, Y.; Gan, J.; Dong, A.; Shen, W.-H. OsChz1 Acts as a Histone Chaperone in Modulating Chromatin Organization and Genome Function in Rice. Nat. Commun. 2020, 11, 5717. [Google Scholar] [CrossRef]
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase Titration Reveals Differences between Nucleosome Occupancy and Chromatin Accessibility. Nat. Commun. 2016, 7, 11485. [Google Scholar] [CrossRef]
- Rahaman, S.; Faravelli, S.; Voegeli, S.; Becskei, A. Polysome Propensity and Tunable Thresholds in Coding Sequence Length Enable Differential mRNA Stability. Sci. Adv. 2023, 9, eadh9545. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Guo, L.; Fang, B.; Shi, J.; Zhou, M. DNA Sequence Changes Resulting from Codon Optimization Affect Gene Expression in Pichia pastoris by Altering Chromatin Accessibility. J. Fungi 2025, 11, 282. https://doi.org/10.3390/jof11040282
Lu C, Guo L, Fang B, Shi J, Zhou M. DNA Sequence Changes Resulting from Codon Optimization Affect Gene Expression in Pichia pastoris by Altering Chromatin Accessibility. Journal of Fungi. 2025; 11(4):282. https://doi.org/10.3390/jof11040282
Chicago/Turabian StyleLu, Chaoyu, Linna Guo, Bohao Fang, Jiacheng Shi, and Mian Zhou. 2025. "DNA Sequence Changes Resulting from Codon Optimization Affect Gene Expression in Pichia pastoris by Altering Chromatin Accessibility" Journal of Fungi 11, no. 4: 282. https://doi.org/10.3390/jof11040282
APA StyleLu, C., Guo, L., Fang, B., Shi, J., & Zhou, M. (2025). DNA Sequence Changes Resulting from Codon Optimization Affect Gene Expression in Pichia pastoris by Altering Chromatin Accessibility. Journal of Fungi, 11(4), 282. https://doi.org/10.3390/jof11040282





