Multiomic Analysis Provided Insights into the Responses of Carbon Sources by Wood-Rotting Fungi Daldinia carpinicola
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture
2.2. Genome Sequencing
2.3. Genome Assembling
2.4. Prediction of the Protein-Coding Genes and Non-Coding RNAs
2.5. Gene Prediction and Gene Cluster Prediction
2.6. Comparative Genomic Analysis
2.7. Functional Genomic Annotation
2.8. Protein Subcellular Localization Analysis
2.9. Transcriptomic Sequencing and Analysis
2.10. Metabolomic Sequencing and Analysis
2.11. The Methods of Combined Transcriptomic and Metabolomic Analysis
3. Results
3.1. Genomic Assembly of D. carpinicola
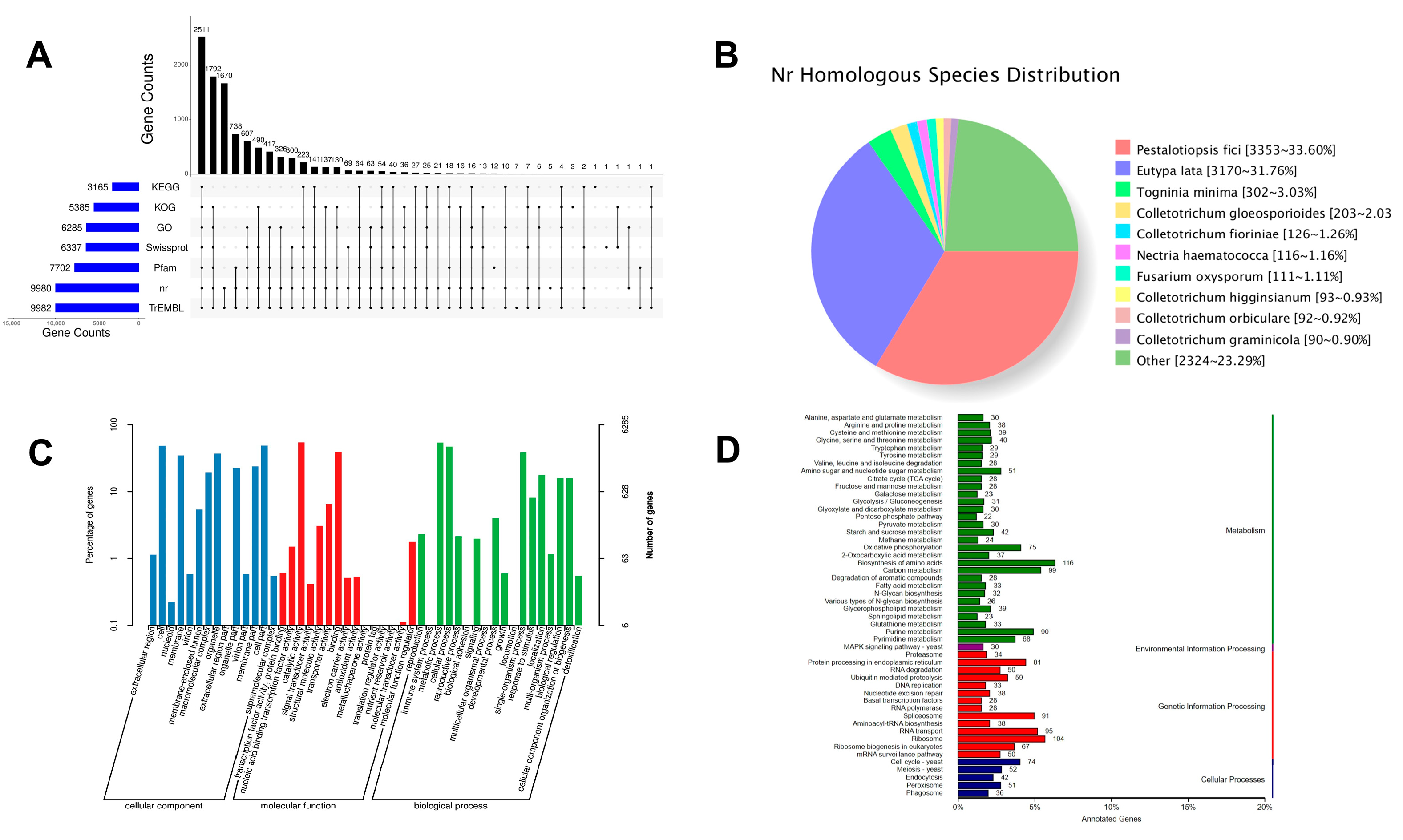
3.2. D. carpinicola Gene Prediction
3.3. D. carpinicola Genome Annotation
3.4. Genome Evolution Analyze
3.5. Comparative Analysis and Average Nucleotide Identity (ANI) Value Analysis
3.6. Gene Cluster Analysis
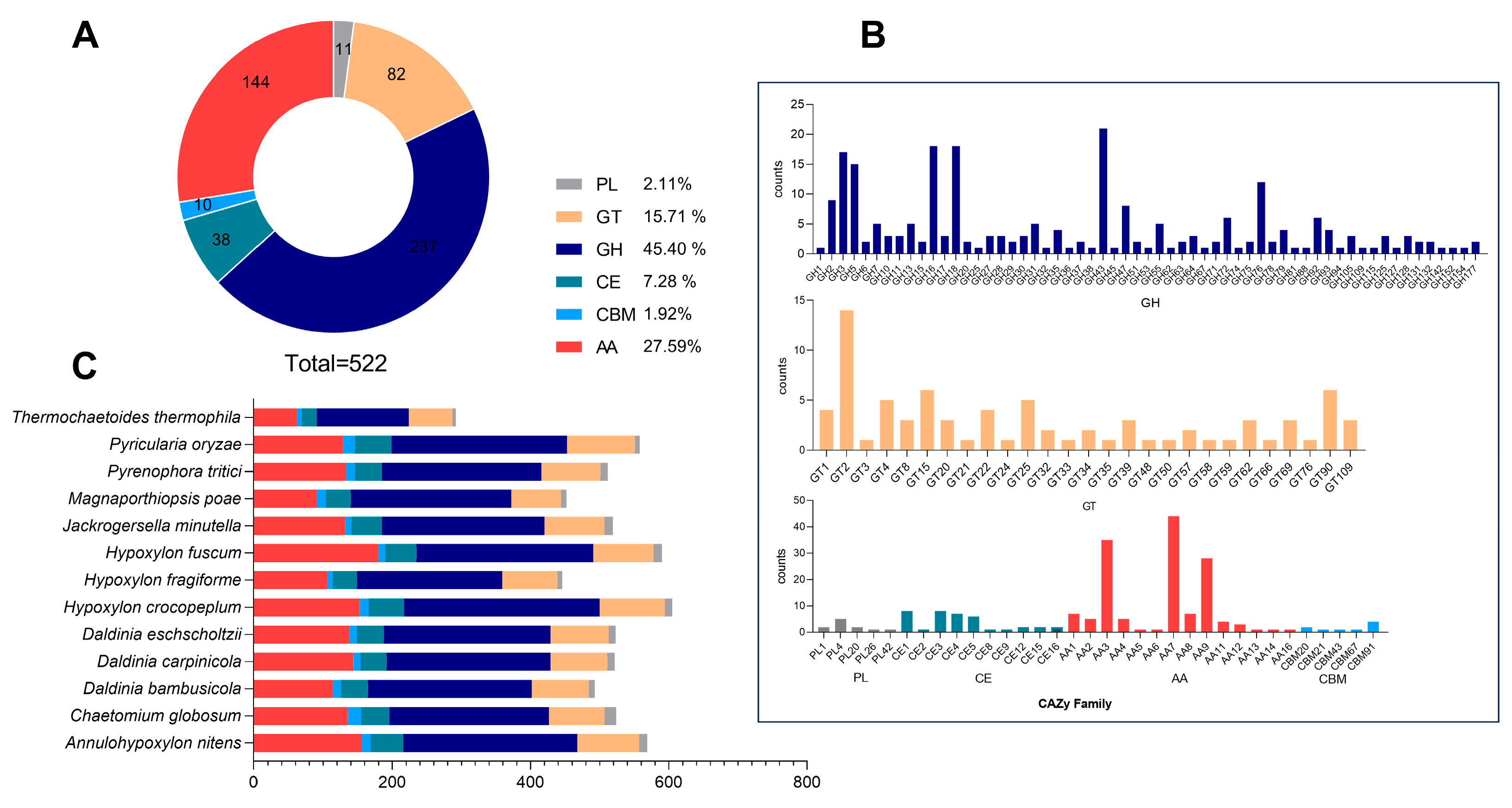
3.7. The CAZymes Analyze in the D. carpinicola Genome
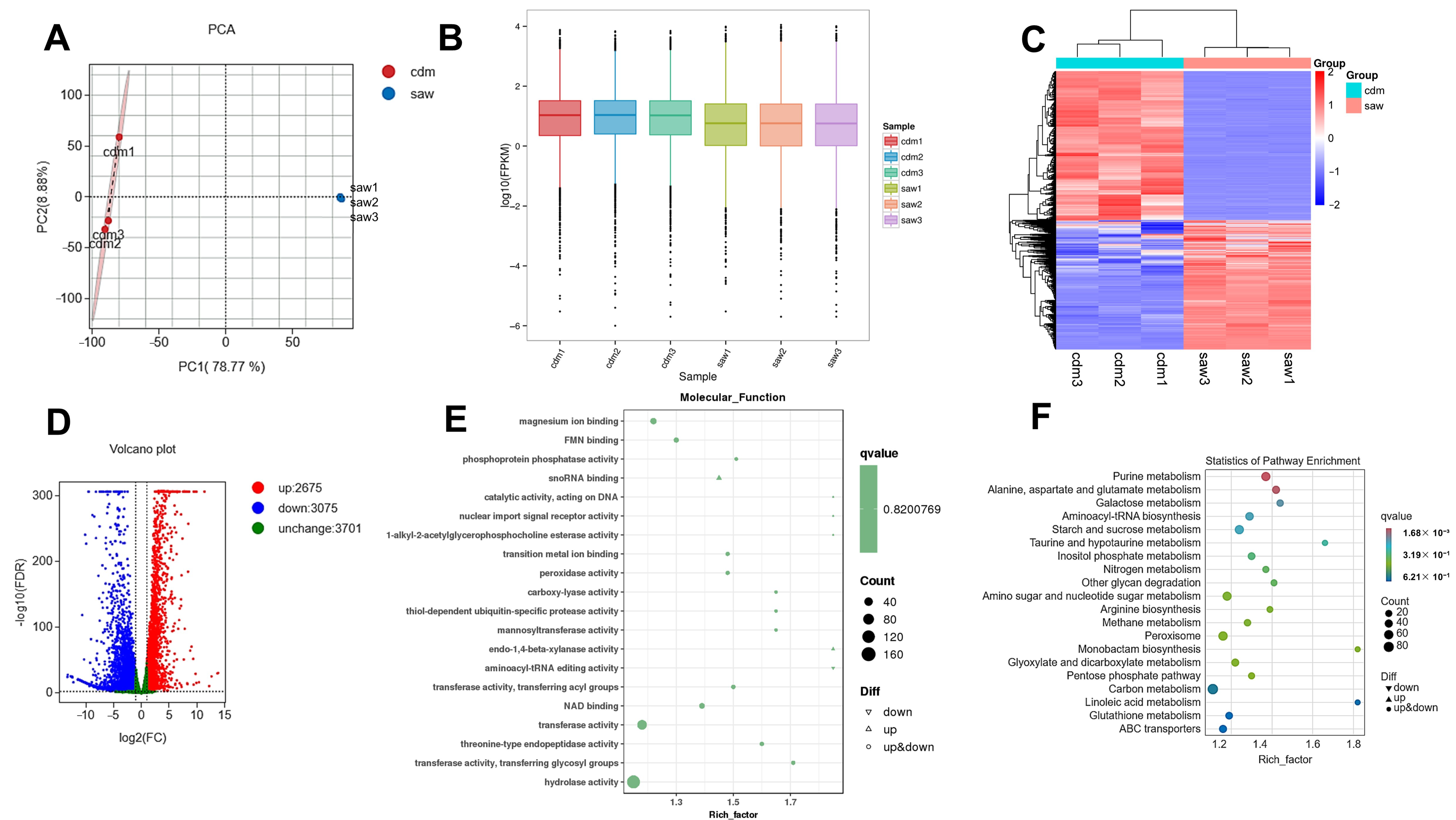
3.8. Transcriptome Analysis
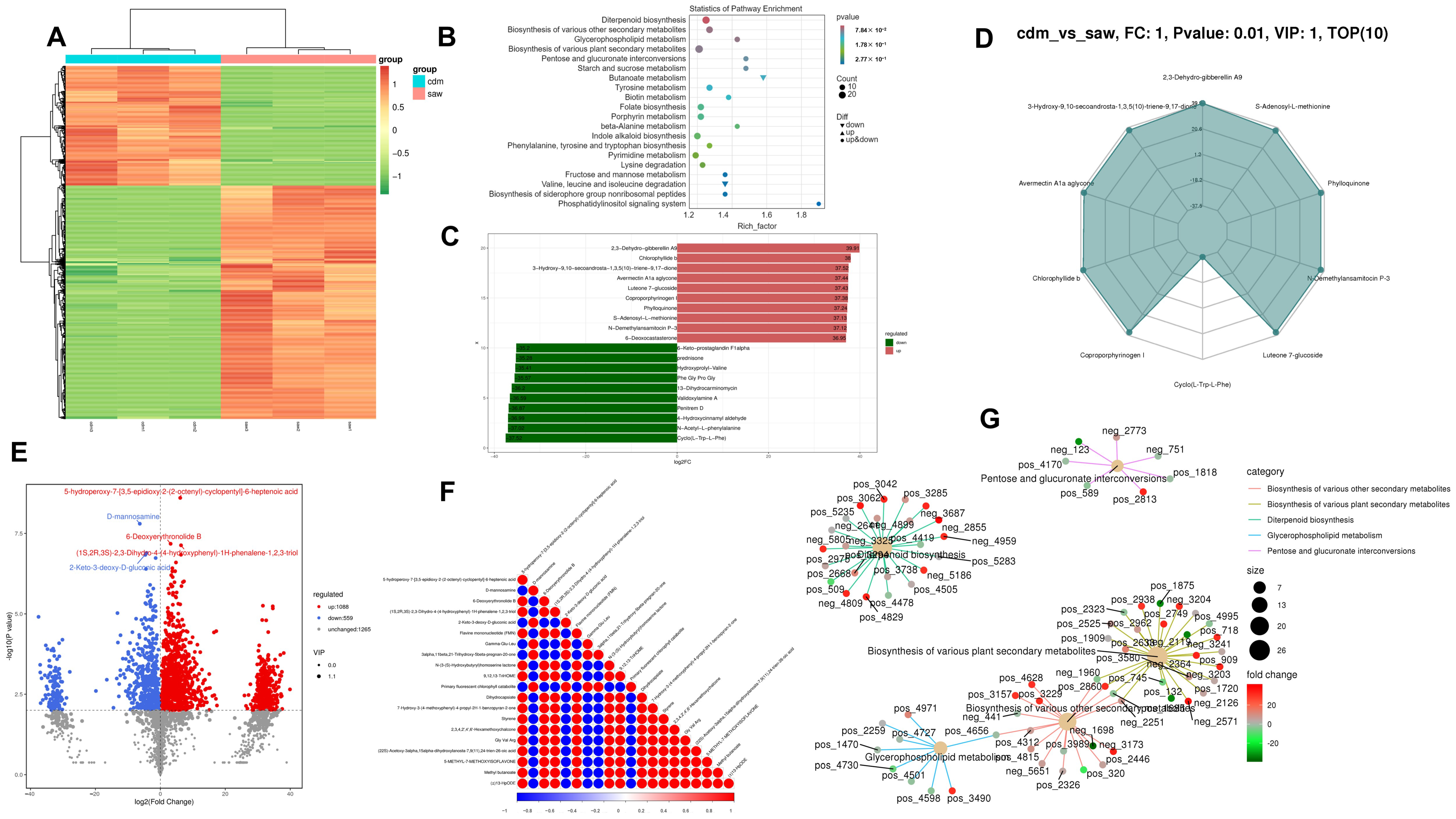
3.9. Metabolomics Analysis
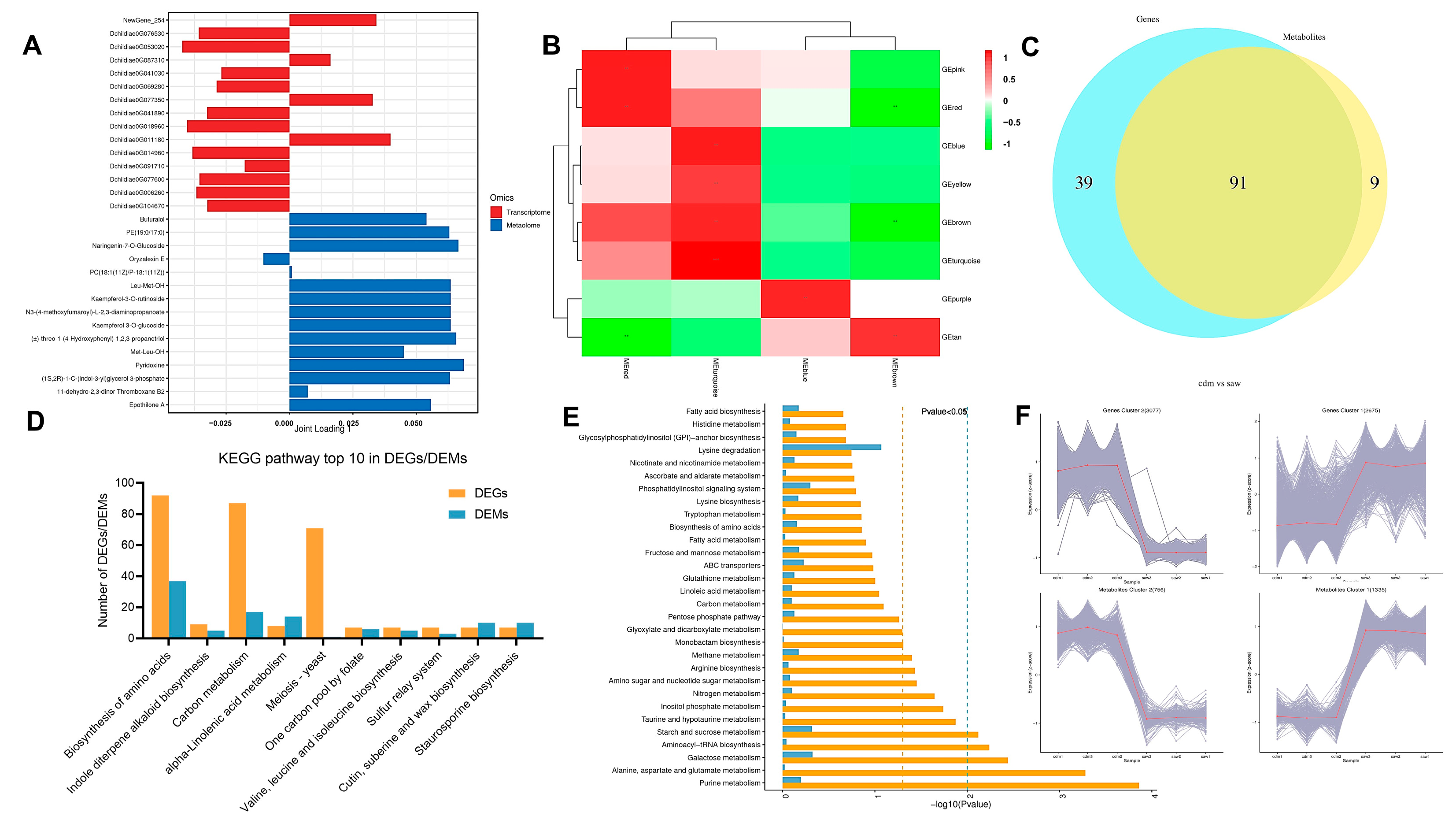
3.10. Combined Transcriptomic and Metabolomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alshammari, N.; Ameen, F.; AlKahtani, M.D.F.; Stephenson, S. Characterizing the Assemblage of Wood-Decay Fungi in the Forests of Northwest Arkansas. J. Fungi 2021, 7, 309. [Google Scholar] [CrossRef] [PubMed]
- Lasota, J.; Błońska, E.; Piaszczyk, W.; Wiecheć, M. How the deadwood of different tree species in various stages of decomposition affected nutrient dynamics? J. Soils Sediments 2017, 18, 2759–2769. [Google Scholar] [CrossRef]
- Li, T.; Cui, L.; Song, X.; Cui, X.; Wei, Y.; Tang, L.; Mu, Y.; Xu, Z. Wood decay fungi: An analysis of worldwide research. J. Soils Sediments 2022, 22, 1688–1702. [Google Scholar] [CrossRef]
- Haq, I.U.; Hillmann, B.; Moran, M.; Willard, S.; Knights, D.; Fixen, K.X.; Schilling, J.S. Bacterial communities associated with wood rot fungi that use distinct decomposition mechanisms. ISME Commun. 2022, 2, 26. [Google Scholar] [CrossRef]
- Liers, C.; Arnstadt, T.; Ullrich, R.; Hofrichter, M. Patterns of lignin degradation and oxidative enzyme secretion by different wood- and litter-colonizing basidiomycetes and ascomycetes grown on beech-wood. FEMS Microbiol. Ecol. 2011, 78, 91–102. [Google Scholar] [CrossRef]
- Hatakka, A.; Hammel, K.E. Fungal Biodegradation of Lignocellulose. In Industrial Applications; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 319–340. [Google Scholar] [CrossRef]
- Cheng, F.; Sheng, J.; Cai, T.; Jin, J.; Liu, W.; Lin, Y.; Du, Y.; Zhang, M.; Shen, L. A Protease-Insensitive Feruloyl Esterase from China Holstein Cow Rumen Metagenomic Library: Expression, Characterization, and Utilization in Ferulic Acid Release from Wheat Straw. J. Agric. Food Chem. 2012, 60, 2546–2553. [Google Scholar] [CrossRef]
- Cheng, F.; Sheng, J.; Dong, R.; Men, Y.; Gan, L.; Shen, L. Novel Xylanase from a Holstein Cattle Rumen Metagenomic Library and Its Application in Xylooligosaccharide and Ferulic Acid Production from Wheat Straw. J. Agric. Food Chem. 2012, 60, 12516–12524. [Google Scholar] [CrossRef]
- Jiang, W.; Zhu, D.; Zhao, L.; Liu, Y.; Wang, C.; Farid, M.S.; Gu, Y.; Li, J.; Li, T.; Sun, Y.; et al. l-Cysteine Treatment Delayed the Quality Deterioration of Fresh-Cut Button Mushrooms by Regulating Oxygen Metabolism, Inhibiting Water Loss, and Stimulating Endogenous H2S Production. J. Agric. Food Chem. 2022, 71, 974–984. [Google Scholar] [CrossRef]
- Muszyńska, B.; Grzywacz-Kisielewska, A.; Kała, K.; Gdula-Argasińska, J. Anti-inflammatory properties of edible mushrooms: A review. Food Chem. 2018, 243, 373–381. [Google Scholar] [CrossRef]
- Moiseenko, K.; Glazunova, O.; Shakhova, N.; Savinova, O.; Vasina, D.; Tyazhelova, T.; Psurtseva, N.; Fedorova, T. Data on the genome analysis of the wood-rotting fungus Steccherinum ochraceum LE-BIN 3174. Data Brief 2020, 29, 105169. [Google Scholar] [CrossRef]
- Achimón, F.; Krapacher, C.R.; Jacquat, A.G.; Pizzolitto, R.P.; Zygadlo, J.A. Carbon sources to enhance the biosynthesis of useful secondary metabolites in Fusarium verticillioides submerged cultures. World J. Microbiol. Biotechnol. 2021, 37, 78. [Google Scholar] [CrossRef] [PubMed]
- Ng, I.-S.; Chen, P.T.; Ju, Y.-M.; Tsai, S.-W. Novel Cellulase Screening and Optimal Production from the Wood Decaying Xylariaceae: Daldinia Species. Appl. Biochem. Biotechnol. 2010, 196, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wongkanoun, S.; Becker, K.; Boonmee, K.; Srikitikulchai, P.; Boonyuen, N.; Chainuwong, B.; Luangsa-Ard, J.; Stadler, M. Three novel species and a new record of Daldinia (Hypoxylaceae) from Thailand. Mycol. Prog. 2020, 19, 1113–1132. [Google Scholar] [CrossRef]
- Lu, S.-M.; Liao, C.-J.; Xu, Y.; Gui, T.-Y.; Sui, H.-L.; Zhou, M. Two new ketene derivatives from the endophytic fungus Daldinia eschscholtzii J11. Phytochem. Lett. 2023, 58, 81–85. [Google Scholar] [CrossRef]
- Ruiz-Villafán, B.; Cruz-Bautista, R.; Manzo-Ruiz, M.; Passari, A.K.; Villarreal-Gómez, K.; Rodríguez-Sanoja, R.; Sánchez, S. Carbon catabolite regulation of secondary metabolite formation, an old but not well-established regulatory system. Microb. Biotechnol. 2021, 15, 1058–1072. [Google Scholar] [CrossRef]
- Cepeda-García, C.; Domínguez-Santos, R.; García-Rico, R.O.; García-Estrada, C.; Cajiao, A.; Fierro, F.; Martín, J.F. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2014, 98, 7113–7124. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Keilwagen, J.; Wenk, M.; Erickson, J.L.; Schattat, M.H.; Grau, J.; Hartung, F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016, 44, e89. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2015, 210, 743–761. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Shen, X.; Liu, J.; Zhao, D.; Sun, Y.; Wang, L.; Liu, Y.; Gong, X.; Liu, Y.; et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 2016, 12, 116. [Google Scholar] [CrossRef]
- Escudero-Leyva, E.; Quirós-Guerrero, L.; Vásquez-Chaves, V.; Pereira-Reyes, R.; Chaverri, P.; Tamayo-Castillo, G. Differential Volatile Organic Compound Expression in the Interaction of Daldinia eschscholtzii and Mycena citricolor. ACS Omega 2023, 8, 31373–31388. [Google Scholar] [CrossRef]
- Pandey, A.; Banerjee, D. Daldinia bambusicola Ch4/11 an Endophytic Fungus Producing Volatile Organic Compounds Having Antimicrobial and Olio Chemical Potential. J. Adv. Microbiol. 2015, 1, 330–337. [Google Scholar] [CrossRef]
- Victor, C.; Moses, O.; Eze, A.; Festus, O.; Charles, E. Isolation, identification, and evaluation of biological activities of Daldinia eschscholtzii, an endophytic fungus isolated from the leaves of Musa paradisiaca. GSC Biol. Pharm. Sci. 2020, 12, 216–228. [Google Scholar] [CrossRef]
- Chen, J.-X.; Yang, X.-Q.; Wang, X.-Y.; Han, H.-L.; Cai, Z.-J.; Xu, H.; Yang, Y.-B.; Ding, Z.-T. Antifeedant, Antifungal Cryptic Polyketides with Six Structural Frameworks from Tea Endophyte Daldinia eschscholtzii Propelled by the Antagonistic Coculture with Phytopathogen Colletotrichum pseudomajus and Different Culture Methods. J. Agric. Food Chem. 2023, 72, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Gao, X.; Zhang, M.; Hu, C.; Yang, W.; Guo, L.; Yang, S.; Yu, H.; Yu, H. Whole Genome Sequence of an Edible Mushroom Oudemansiella raphanipes (Changgengu). J. Fungi 2023, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Bao, H.; Bau, T. Analyses of transcriptomes and the first complete genome of Leucocalocybe mongolica provide new insights into phylogenetic relationships and conservation. Sci. Rep. 2021, 11, 2930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Deng, W.; Yan, W.; Li, T. Whole Genome Sequence of an Edible and Potential Medicinal Fungus, Cordyceps guangdongensis. G3 Genes Genomes Genet. 2018, 8, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wu, F.; Si, J.; Zhao, Y.-F.; Dai, Y.-C. Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2017, 111, 50–58. [Google Scholar] [CrossRef]
- Lee, K.; Poudel, Y.B.; Glinkerman, C.M.; Boger, D.L. Total synthesis of dihydrolysergic acid and dihydrolysergol: Development of a divergent synthetic strategy applicable to rapid assembly of D-ring analogs. Tetrahedron 2015, 71, 5897–5905. [Google Scholar] [CrossRef]
- Wagger, J.; Požes, A.; Požgan, F. Synthesis of European pharmacopoeial impurities A, B, C, and D of cabergoline. RSC Adv. 2013, 3, 23146–23156. [Google Scholar] [CrossRef][Green Version]
- Arnold, S.L.; Panaccione, D.G. Biosynthesis of the Pharmaceutically Important Fungal Ergot Alkaloid Dihydrolysergic Acid Requires a Specialized Allele of cloA. Appl. Environ. Microbiol. 2017, 83, e00805-17. [Google Scholar] [CrossRef]
- Awakawa, T.; Yang, X.; Wakimoto, T.; Abe, I. Pyranonigrin E: A PKS-NRPS Hybrid Metabolite from Aspergillus niger Identified by Genome Mining. ChemBioChem 2013, 14, 2095–2099. [Google Scholar] [CrossRef]
- Jalal, M.A.F.; Love, S.K.; van der Helm, D. Nα-Dimethylcoprogens Three novel trihydroxamate siderophores from pathogenic fungi. Biol. Met. 1988, 1, 4–8. [Google Scholar] [CrossRef]
- Maguvu, T.E.; Travadon, R.; Cantu, D.; Trouillas, F.P. Whole genome sequencing and analysis of multiple isolates of Ceratocystis destructans, the causal agent of Ceratocystis canker of almond in California. Sci. Rep. 2023, 13, 14873. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.R.; Salazar, M.P.; Schaap, P.J.; van de Vondervoort, P.J.; Culley, D.; Thykaer, J.; Frisvad, J.C.; Nielsen, K.F.; Albang, R.; Albermann, K.; et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011, 21, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Xie, B.; Yang, Z.; Chen, Z.; Chen, B.; Deng, Y.; Jiang, Y.; van Peer, A.F. Identification and expression analysis of a new glycoside hydrolase family 55 exo-β-1,3-glucanase-encoding gene in Volvariella volvacea suggests a role in fruiting body development. Gene 2013, 527, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the Large Glycoside Hydrolase Family 43 into Subfamilies: A Motivation for Detailed Enzyme Characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Kawai, R.; Igarashi, K.; Yoshida, M.; Kitaoka, M.; Samejima, M. Hydrolysis of β-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-β-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 2005, 71, 898–906. [Google Scholar] [CrossRef]
- Couturier, M.; Roussel, A.; Rosengren, A.; Leone, P.; Stålbrand, H.; Berrin, J.-G. Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 β-(1,4)-Mannanases from Podospora anserina Reveal Differences upon Manno-oligosaccharide Catalysis. J. Biol. Chem. 2013, 288, 14624–14635. [Google Scholar] [CrossRef]
- Dodd, D.; Kiyonari, S.; Mackie, R.I.; Cann, I.K.O. Functional Diversity of Four Glycoside Hydrolase Family 3 Enzymes from the Rumen Bacterium Prevotella bryantii B1 4. J. Bacteriol. 2010, 192, 2335–2345. [Google Scholar] [CrossRef]
- Wu, W.; Davis, R.W.; Tran-Gyamfi, M.B.; Kuo, A.; LaButti, K.; Mihaltcheva, S.; Hundley, H.; Chovatia, M.; Lindquist, E.; Barry, K.; et al. Characterization of four endophytic fungi as potential consolidated bioprocessing hosts for conversion of lignocellulose into advanced biofuels. Appl. Microbiol. Biotechnol. 2017, 101, 2603–2618. [Google Scholar] [CrossRef]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Poulsen, J.-C.N.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of Lignocellulosic Biomass Hydrolysis by Proteins of Glycoside Hydrolase Family 61: Structure and Function of a Large, Enigmatic Family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef]
- Chan, C.L.; Yew, S.M.; Ngeow, Y.F.; Na, S.L.; Lee, K.W.; Hoh, C.-C.; Yee, W.-Y.; Ng, K.P. Genome analysis of Daldinia eschscholtzii strains UM 1400 and UM 1020, wood-decaying fungi isolated from human hosts. BMC Genom. 2015, 16, 966. [Google Scholar] [CrossRef]
- Masigol, H.; Grossart, H.-P.; Taheri, S.R.; Mostowfizadeh-Ghalamfarsa, R.; Pourmoghaddam, M.J.; Bouket, A.C.; Khodaparast, S.A. Utilization of Low Molecular Weight Carbon Sources by Fungi and Saprolegniales: Implications for Their Ecology and Taxonomy. Microorganisms 2023, 11, 782. [Google Scholar] [CrossRef] [PubMed]
- Sigoillot, J.C.; Berrin, J.G.; Bey, M.; Lesage-Meessen, L.; Levasseur, A.; Lomascolo, A.; Record, E.; Uzan-Boukhris, E. Fungal Strategies for Lignin Degradation. In Advances in Botanical Research; Elsevier Science & Technology: Oxford, UK, 2012; Volume 61, pp. 263–308. ISBN 0065-2296. [Google Scholar]
- Brunner, I.; Fischer, M.; Rüthi, J.; Stierli, B.; Frey, B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS ONE 2018, 13, e0202047. [Google Scholar] [CrossRef] [PubMed]
- Brailsford, F.L.; Glanville, H.C.; Marshall, M.R.; Yates, C.A.; Owen, A.T.; Golyshin, P.N.; Johnes, P.J.; Jones, D.L. Land cover and nutrient enrichment regulates low-molecular weight dissolved organic matter turnover in freshwater ecosystems. Limnol. Oceanogr. 2021, 66, 2979–2987. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, X.; Singh, D.; Yu, H.; Yang, X. Biological pretreatment of lignocellulosics: Potential, progress and challenges. Biofuels 2010, 1, 177–199. [Google Scholar] [CrossRef]
- Martin-Vicente, A.; Souza, A.C.O.; Guruceaga, X.; Thorn, H.I.; Xie, J.; Nywening, A.V.; Ge, W.; Fortwendel, J.R. A conserved fungal morphogenetic kinase regulates pathogenic growth in response to carbon source diversity. Nat. Commun. 2024, 15, 8945. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, L.; Tang, H.; Wang, J.; Ren, L.; Cui, G.; Zhao, Y.; Tang, J.; Su, P.; Wang, Y.; et al. Engineering the microenvironment of P450s to enhance the production of diterpenoids in Saccharomyces cerevisiae. Acta Pharm. Sin. B 2024, 14, 4608–4618. [Google Scholar] [CrossRef]
- Ma, D.; Du, G.; Fang, H.; Li, R.; Zhang, D. Advances and prospects in microbial production of biotin. Microb. Cell Factories 2024, 23, 1–14. [Google Scholar] [CrossRef]
- Li, J.; Lai, Z.; Zhang, W.; Zeng, L.; Cui, S. Modular assembly of indole alkaloids enabled by multicomponent reaction. Nat. Commun. 2023, 14, 4806. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Serrano-Amatriain, C.; Ledesma-Amaro, R.; Jiménez, A. Formation of folates by microorganisms: Towards the biotechnological production of this vitamin. Appl. Microbiol. Biotechnol. 2018, 102, 8613–8620. [Google Scholar] [CrossRef]


| Contig Length (bp) | Contigs | Scaffold N50 (bp) | Scaffold N90 (bp) | GC Content (%) | Gaps Number | Library (bp) | Mapped (%) | Properly Mapped (%) | Coverage (%) | Depth (X) |
|---|---|---|---|---|---|---|---|---|---|---|
| 35,934,728 | 46 | 1,323,482 | 550,949 | 44.81 | 0 | 350 | 81.71 | 78.97 | 99.97 | 74.29 |
| Content | Number/Length |
|---|---|
| Gene number | 10,596 |
| Gene length (bp) | 23,949,977 |
| Average gene length (bp) | 2260.28 |
| Exon length (bp) | 21,747,637 |
| Average exon length (bp) | 670.77 |
| Exon number | 32,422 |
| Average exon number | 3.06 |
| CDS length (bp) | 16,005,309 |
| Average CDS length (bp) | 509.17 |
| CDS number | 31,434 |
| Average CDS number | 2.97 |
| Intron length (bp) | 2,202,340 |
| Average Intron length (bp) | 100.9 |
| Intron number | 21,826 |
| Average Intron number per gene | 2.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Ma, X.; Zhang, Y.; Sun, Y.; Yu, H.; Han, J.; Ma, M.; Wan, L.; Cheng, F. Multiomic Analysis Provided Insights into the Responses of Carbon Sources by Wood-Rotting Fungi Daldinia carpinicola. J. Fungi 2025, 11, 115. https://doi.org/10.3390/jof11020115
Yang P, Ma X, Zhang Y, Sun Y, Yu H, Han J, Ma M, Wan L, Cheng F. Multiomic Analysis Provided Insights into the Responses of Carbon Sources by Wood-Rotting Fungi Daldinia carpinicola. Journal of Fungi. 2025; 11(2):115. https://doi.org/10.3390/jof11020115
Chicago/Turabian StyleYang, Peng, Xingchi Ma, Yu Zhang, Yanan Sun, Hao Yu, Jiandong Han, Meng Ma, Luzhang Wan, and Fansheng Cheng. 2025. "Multiomic Analysis Provided Insights into the Responses of Carbon Sources by Wood-Rotting Fungi Daldinia carpinicola" Journal of Fungi 11, no. 2: 115. https://doi.org/10.3390/jof11020115
APA StyleYang, P., Ma, X., Zhang, Y., Sun, Y., Yu, H., Han, J., Ma, M., Wan, L., & Cheng, F. (2025). Multiomic Analysis Provided Insights into the Responses of Carbon Sources by Wood-Rotting Fungi Daldinia carpinicola. Journal of Fungi, 11(2), 115. https://doi.org/10.3390/jof11020115






