Unveiling the Unique Mitogenome Structure of Phylloporus: Implications for Phylogeny and Evolution in Boletaceae
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Mitogenomes Assembly and Annotation
2.3. Sequence Analysis
2.4. Phylogenetic Analysis
2.5. Data Availability
3. Results
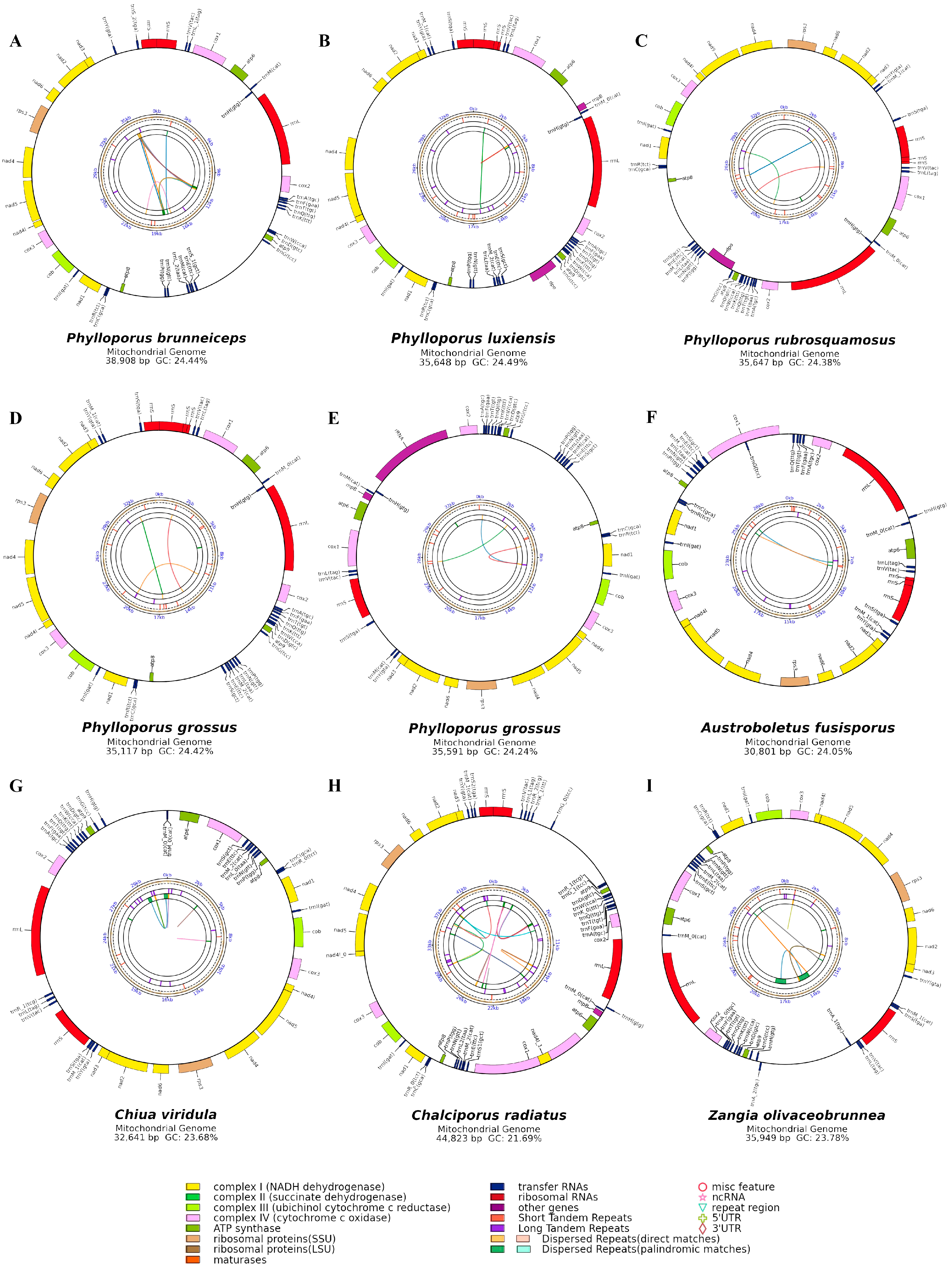
3.1. Characterization of Mitogenomes in Phylloporus
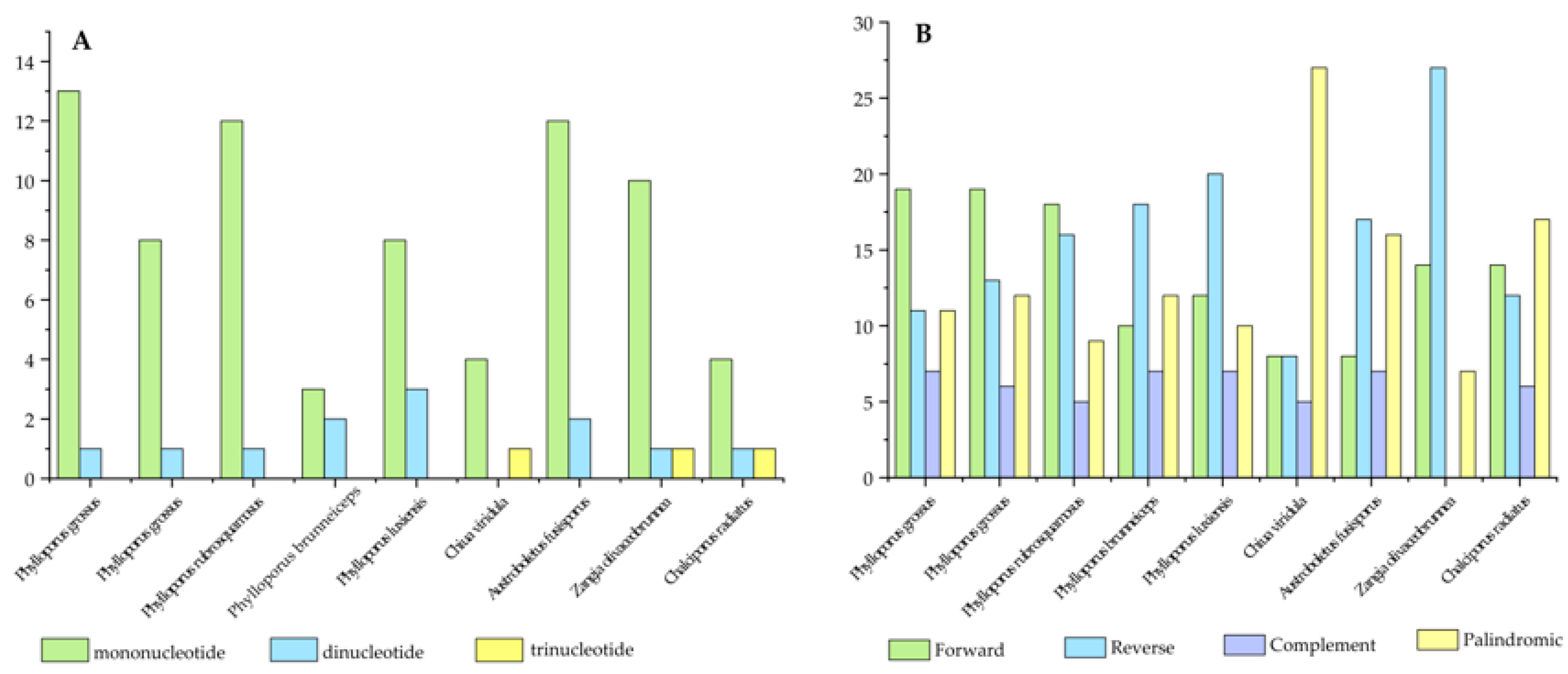
3.2. Analysis of Repetitive Sequences
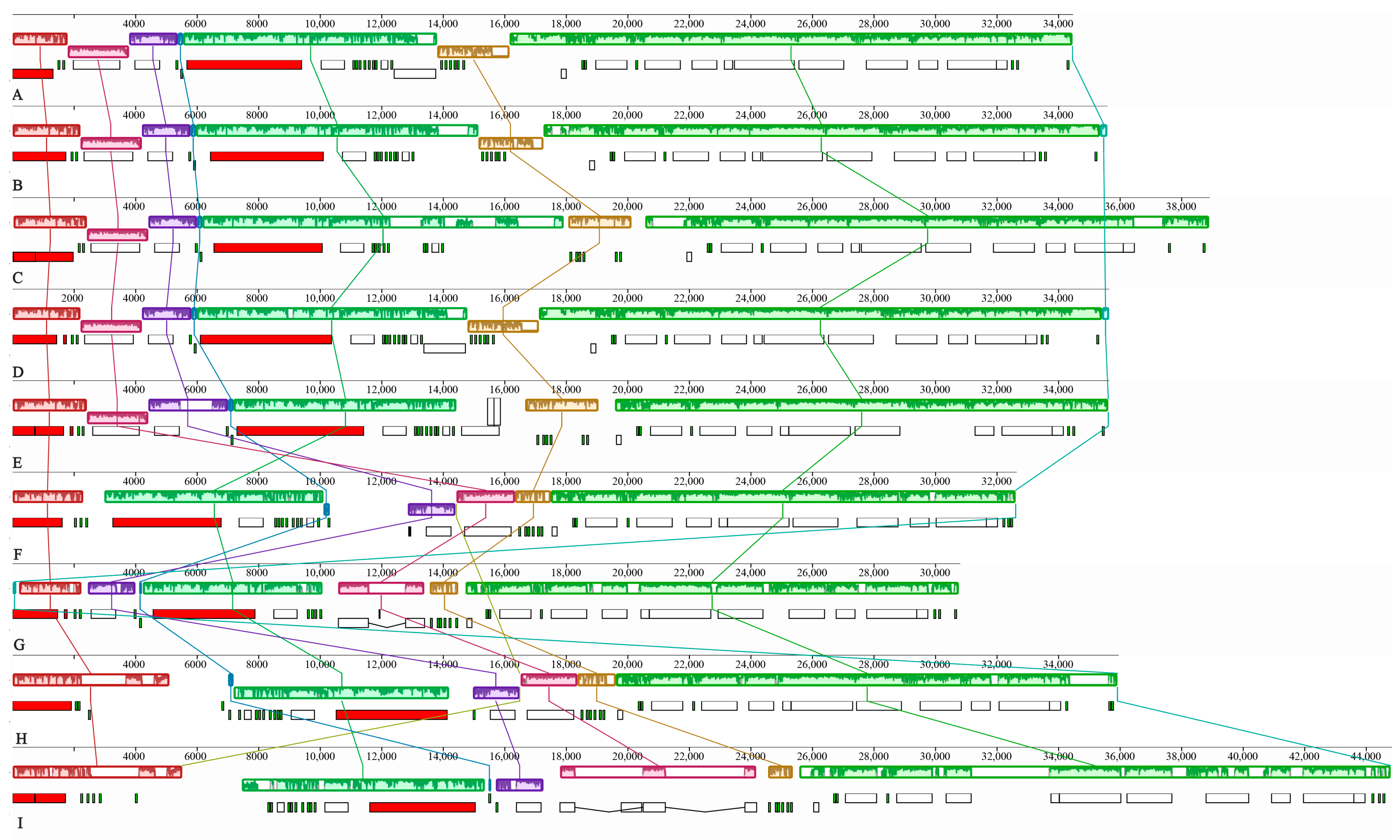
3.3. Genome Rearrangement Analysis
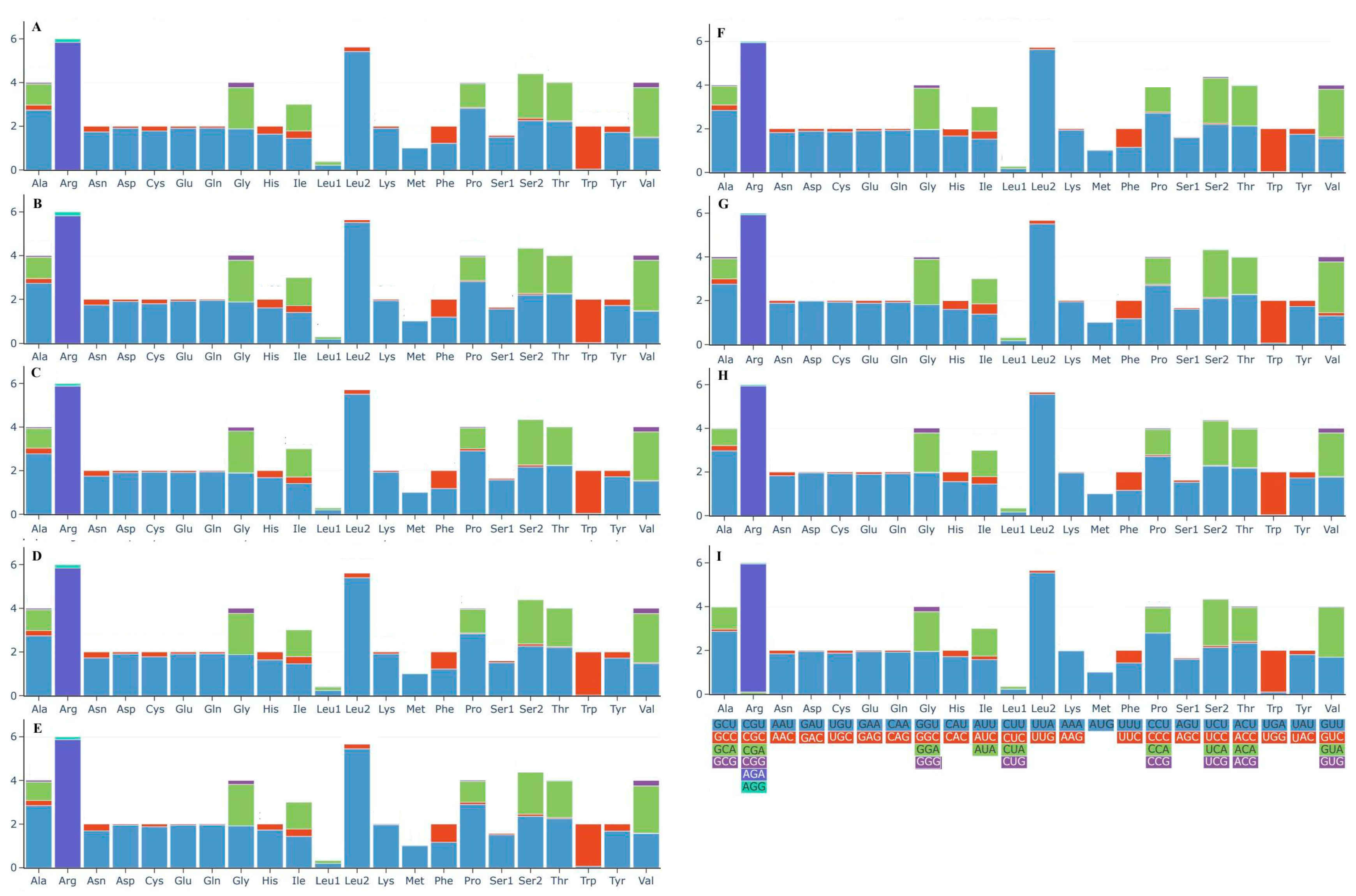
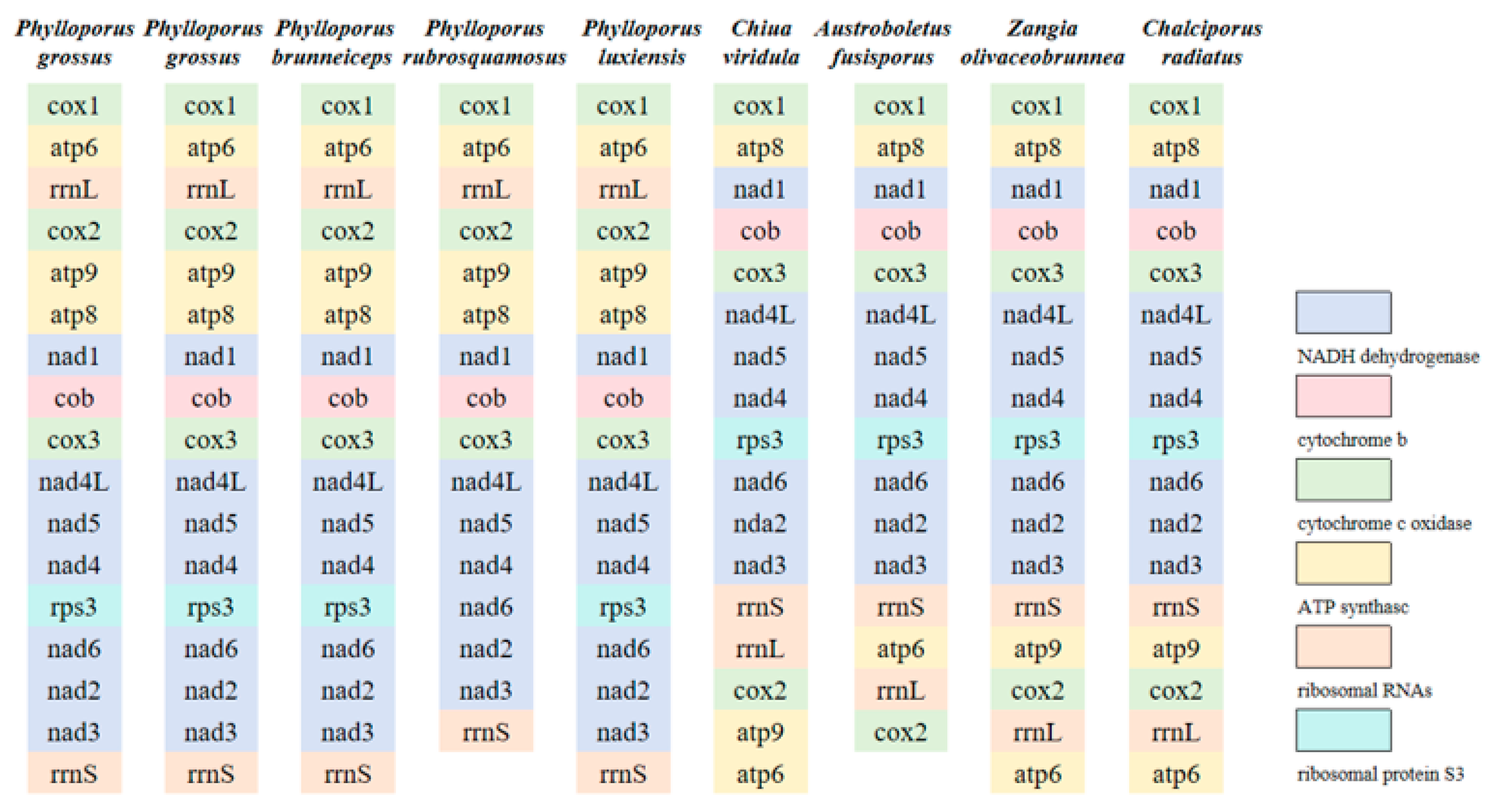
3.4. Protein-Coding Genes of the Nine Boletaceae Mitogenomes
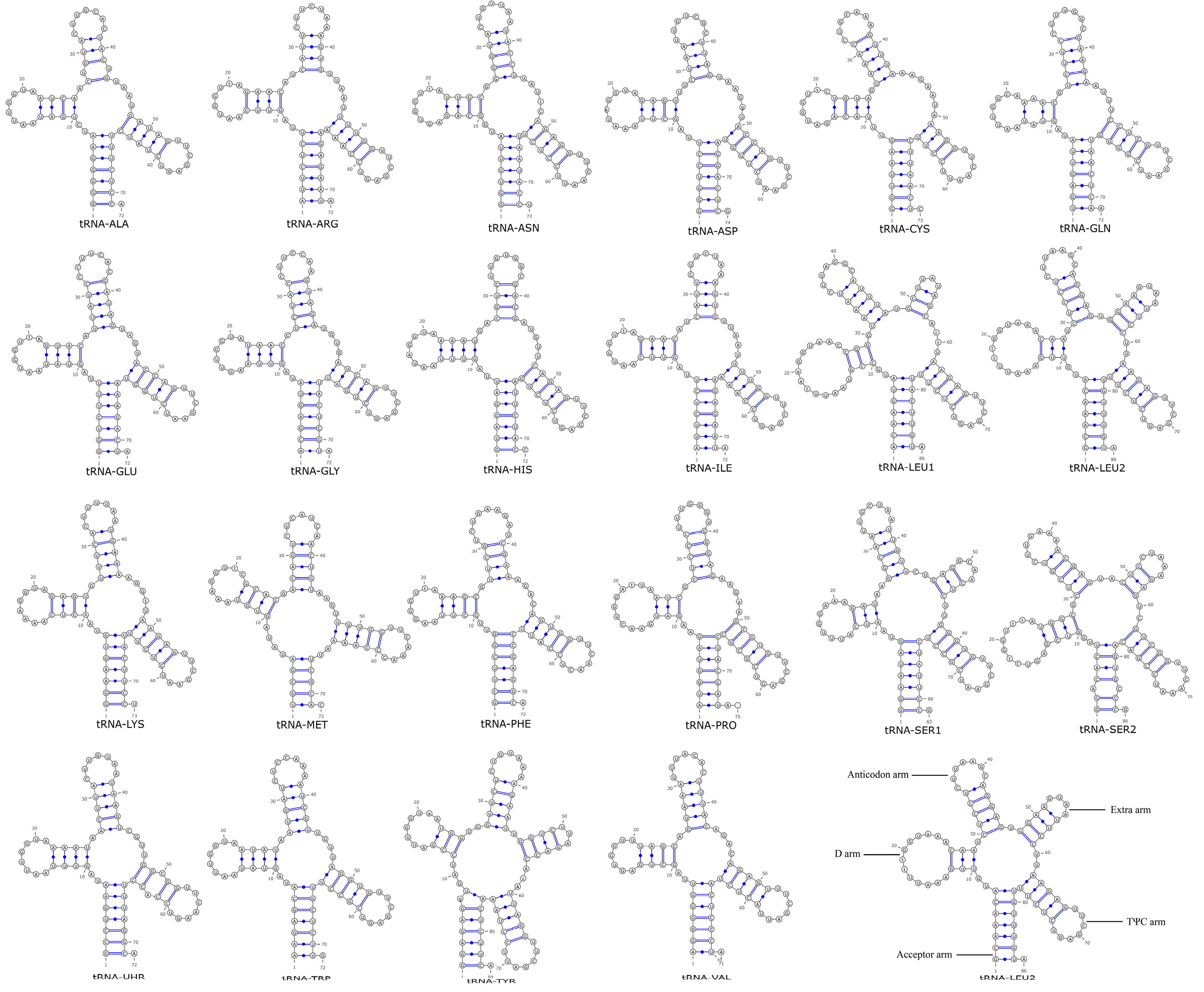
3.5. Analysis of the tRNA Secondary Structure in Phylloporus
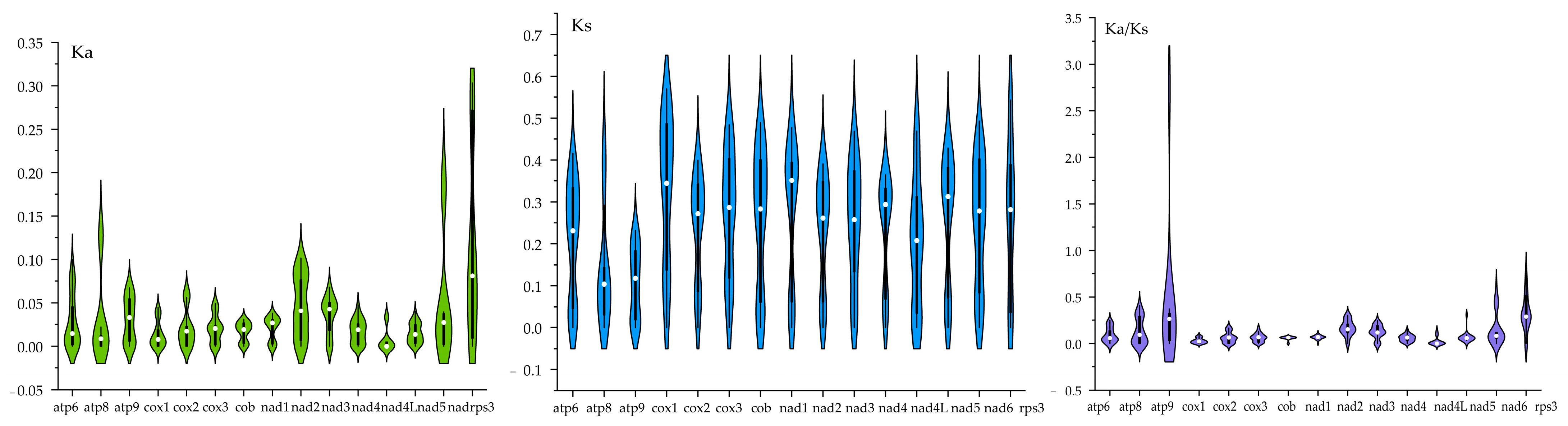
3.6. Evolutionary Rates of Core Genes in Boletaceae
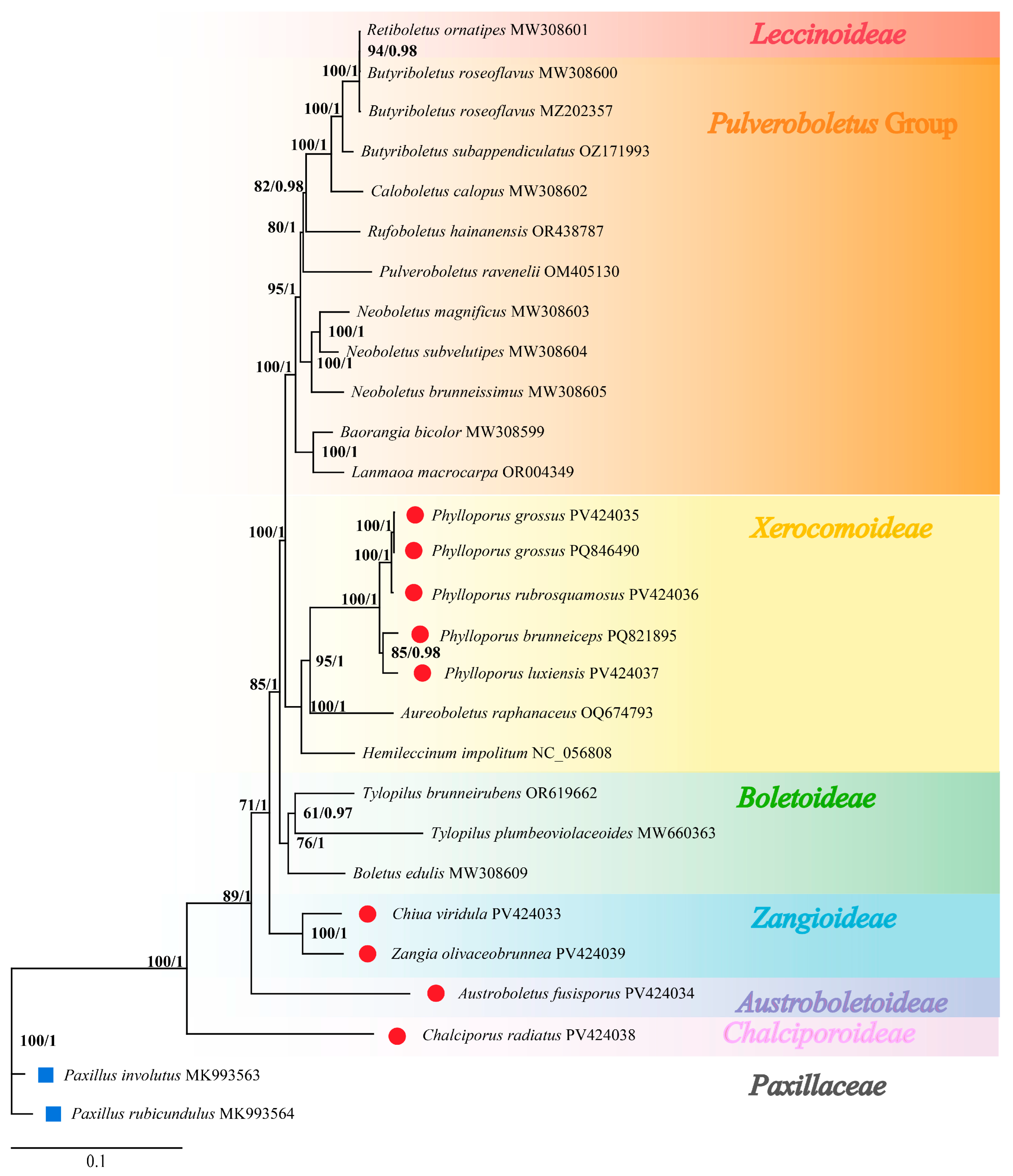
3.7. Phylogenetic Relationships
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zeng, N.-K.; Tang, L.-P.; Li, Y.-C.; Tolgor, B.; Zhu, X.-T.; Zhao, Q.; Yang, Z.L. The genus Phylloporus (Boletaceae, Boletales) from China: Morphological and multilocus DNA sequence analyses. Fungal Divers. 2012, 58, 73–101. [Google Scholar] [CrossRef]
- Ramos, A.; Leticia, M.; Bandala, V.M. Morphological and molecular characterization of ectomycorrhizas of Phylloporus (Boletales) and Quercus sapotifolia from tropical oak forest of eastern Mexico. Symbiosis 2023, 91, 45–54. [Google Scholar] [CrossRef]
- Courty, P.-E.; Buée, M.; Diedhiou, A.G.; Frey-Klett, P.; Le Tacon, F.; Rineau, F.; Turpault, M.-P.; Uroz, S.; Garbaye, J.J.S.B. The role of ectomycorrhizal communities in forest ecosystem processes: New perspectives and emerging concepts. Soil Biol. Biochem. 2010, 42, 679–698. [Google Scholar] [CrossRef]
- Wu, G.; Miyauchi, S.; Morin, E.; Kuo, A.; Drula, E.; Varga, T.; Kohler, A.; Feng, B.; Cao, Y.; Lipzen, A.; et al. Evolutionary innovations through gain and loss of genes in the ectomycorrhizal Boletales. New Phytol. 2022, 233, 1383–1400. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhong, X.; Ma, Z.; Tang, G.; Ding, L.; Sui, X.; Xu, J.; He, Y. Climate changes and their teleconnections with ENSO over the last 55 years, 1961–2015, in floods-dominated basin, Jiangxi province, China. Earth Space Sci. 2020, 7, e2019EA001047. [Google Scholar] [CrossRef]
- Zhang, J.; Song, H.; Hu, D. A Checklist of macrofungi in Jiangxi province. Biol. Hazard Sci. 2016, 39, 1–13. [Google Scholar] [CrossRef]
- Cao, K.Z.R. Jiangxi Jiuling Mountain Macrofungi Atlas; Jiangxi People’s Publishing House: Nanchang, China, 2022; ISBN 9787210143239. [Google Scholar]
- Zeb, U.; Aziz, T.; Azizullah, A.; Zan, X.Y.; Khan, A.A.; Bacha, S.A.S.; Cui, F.J. Complete mitochondrial genomes of edible mushrooms: Features, evolution, and phylogeny. Physiol. Plant. 2024, 176, e14363. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, T.-h.; Huang, O.; Zhu, K.-f.; Chen, S.-q.; Wang, Y.-a.; Wang, Y.; Wang, Y.-b.; Yu, H. Phylogenetic analysis of the mitochondrial genome of the Samsoniella hepiali holotype strain. Mycosystema 2022, 41, 1572–1584. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Z.; Yang, Z.; Sun, T.; Yang, S.; Yu, H. Morphology, phylogeny, and mitogenomics reveal a new entomopathogenic fungus, Blackwellomyces changningensis (Hypocreales, Clavicipitaceae), from southwestern China. Mycologia 2024, 117, 166–182. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, M.; Sun, Y.; Li, Q.; Liu, J.; Song, C.; Shang, X.; Tan, Q.; Zhang, L.; Yu, H. Whole-genome sequence of a high-temperature edible mushroom Pleurotus giganteus (zhudugu). Front. Microbiol. 2022, 13, 941889. [Google Scholar] [CrossRef]
- Liu, J.; Li, T.; Song, L.; Kong, J.; Niu, Q.; Wang, Y.; Wu, C.; Deng, B.; Wang, H.; Gai, Y. Comparative transcriptome of isonuclear alloplasmic strain revealed the important role of mitochondrial genome in regulating Flammulina filiformis. Agronomy 2023, 13, 998. [Google Scholar] [CrossRef]
- Li, Q.; Bao, Z.; Tang, K.; Feng, H.; Tu, W.; Li, L.; Han, Y.; Cao, M.; Zhao, C. First two mitochondrial genomes for the order Filobasidiales reveal novel gene rearrangements and intron dynamics of Tremellomycetes. IMA Fungus 2022, 13, 7. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Donath, A.; Juhling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.F.; Laforest, M.J.; Burger, G. Mitochondrial introns: A critical view. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Miyauchi, S.; Kiss, E.; Kuo, A.; Drula, E.; Kohler, A.; Sanchez-Garcia, M.; Morin, E.; Andreopoulos, B.; Barry, K.W.; Bonito, G.; et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat. Commun. 2020, 11, 5125. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ren, Y.; Xiang, D.; Shi, X.; Zhao, J.; Peng, L.; Zhao, G. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus 2020, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, P.; Li, L.; Feng, H.; Tu, W.; Bao, Z.; Xiong, C.; Gui, M.; Huang, W. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int. J. Biol. Macromol. 2021, 172, 560–572. [Google Scholar] [CrossRef]
- Shi, W.; Song, W.; Peng, Y.; Wang, S.; Yang, G.; Shi, C. The complete mitochondrial genome sequence and annot-ation of Tylopilus plumbeoviolaceoides (Boletaceae, Boletoideae). Mitochondrial DNA Part B Resour. 2022, 7, 999–1000. [Google Scholar] [CrossRef]
- Zheng, Y.t.; Chen, L.-l.; Zhao, K. Complete mitochondrial genome sequence of Lanmaoa macrocarpa (Boletales, Basidiomycota). Mitochondrial DNA Part B Resour. 2023, 8, 1067–1070. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Mu, X.H.; Liang, X.X.; Zheng, Y.T.; Zhao, K. Complete mitochondrial genome sequence of Aureoboletus raphanaceus (Boletales, Basidiomycota). Mitochondrial DNA Part B Resour. 2024, 9, 20–23. [Google Scholar] [CrossRef]
- Cho, S.E.; Kwag, Y.N.; Han, S.K.; Lee, D.H.; Kim, C.S. Complete mitochondrial genome sequence of Pulveroboletus ravenelii (Boletales, Basidiomycota). Mitochondrial DNA Part B Resour. 2022, 7, 1581–1582. [Google Scholar] [CrossRef]
- Zeng, Y.P.; Huang, J.Y.; Tu, L.; Zhao, K. Complete mitochondrial genome sequence of Butyriboletus hainanensis (Boletales, Basidiomycota). Mitochondrial DNA Part B Resour. 2024, 9, 46–49. [Google Scholar] [CrossRef]
- Huang, J.Y.; Tu, L.; Lv, Y.; Zhao, K. Complete mitochondrial genome sequence and phylogenetic analysis of Tylo-pilus brunneirubens (Boletales, Basidiomycota). Mitochondrial DNA Part B Resour. 2024, 9, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Torresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef]
- Dang, Z.; Huang, L.; Jia, Y.; Lockhart, P.J.; Fong, Y.; Tian, Y. Identification of genic SSRs provide a perspective for studying environmental adaptation in the endemic shrub Tetraena mongolica. Genes 2020, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Maharachchikumbura, S.S.; Larignon, P.; Hyde, K.D.; Al-Sadi, A.M.; Liu, Z. Characterization of Neopestalotiopsis, Pestalotiopsis and Truncatella species associated with grapevine trunk diseases in France. Phytopathol. Mediterr. 2016, 55, 380–390. [Google Scholar] [CrossRef]
- Wu, G.; Feng, B.; Xu, J.; Zhu, X.-T.; Li, Y.-C.; Zeng, N.-K.; Hosen, M.I.; Yang, Z.L. Molecular phylogenetic analyses redefine seven major clades and reveal 22 new generic clades in the fungal family Boletaceae. Fungal Divers. 2014, 69, 93–115. [Google Scholar] [CrossRef]
- Wu, G.; Li, Y.; Zhu, X.; Zhao, K.; Han, L.; Cui, Y.; Li, F.; Xu, J.; Yang, Z. One hundred noteworthy boletes from China. Fungal Divers. 2016, 81, 25–188. [Google Scholar] [CrossRef]
- Chuankid, B.; Vadthanarat, S.; Hyde, K.D.; Thongklang, N.; Zhao, R.; Lumyong, S.; Raspé, O. Three new Phylloporus species from tropical China and Thailand. Mycol. Prog. 2019, 18, 603–614. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, R.; Su, M.; Wu, L.; Zeng, N. Phylloporus rubiginosus, a noteworthy lamellar bolete from tropical Asia. Guizhou Sci. 2019, 37, 1–5. [Google Scholar] [CrossRef]
- Wu, L.-L.; Liang, Z.-Q.; Su, M.-S.; Fan, Y.-G.; Zhang, P.; Jiang, S.; Chen, Y.-L.; Hao, Y.-J.; Zeng, N.-K. Updated taxonomy of Chinese Phylloporus (Boletaceae, Boletales): Six new taxa and four redescribed species. Mycol. Prog. 2021, 20, 1243–1273. [Google Scholar] [CrossRef]
- Kai, Z.N.; Ying, D.Q. Poisonous fungi of the order Boletomycetes in China. Mycol. Res. 2024, 22, 322–332. [Google Scholar] [CrossRef]
- Dentinger, B.T.; Ammirati, J.F.; Both, E.E.; Desjardin, D.E.; Halling, R.E.; Henkel, T.W.; Moreau, P.A.; Nagasawa, E.; Soytong, K.; Taylor, A.F.; et al. Molecular phylogenetics of porcini mushrooms (Boletus section Boletus). Mol. Phylogenet. Evol. 2010, 57, 1276–1292. [Google Scholar] [CrossRef]
- Li, H.; Wu, S.; Ma, X.; Chen, W.; Zhang, J.; Duan, S.; Gao, Y.; Kui, L.; Huang, W.; Wu, P.; et al. The Genome Sequences of 90 Mushrooms. Sci. Rep. 2018, 8, 9982. [Google Scholar] [CrossRef]
- Malik, M.; Malik, F.; Fatma, T.; Qasim Hayat, M.; Jamal, A.; Gul, A.; Faraz Bhatti, M. The complete mitochondrial genome of Penicillium expansum: Insights into the fungal evolution and phylogeny. Gene 2024, 910, 148315. [Google Scholar] [CrossRef]
- Li, Y.; Yang, T.; Qiao, J.; Liang, J.; Li, Z.; Sa, W.; Shang, Q. Whole-genome sequencing and evolutionary analysis of the wild edible mushroom, morchella eohespera. Front. Microbiol. 2023, 14, 1309703. [Google Scholar] [CrossRef]
- Yu, Y.; Li, H.T.; Wu, Y.H.; Li, D.Z. Correlation analysis reveals an important role of GC content in accumulation of deletion mutations in the coding region of angiosperm plastomes. J. Mol. Evol. 2021, 89, 73–80. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, J.B.; Ma, W.Z.; Pressel, S.; Liu, H.M.; Wu, Y.H.; Schneider, H. Chloroplast phylogenomics of liverworts: A reappraisal of the backbone phylogeny of liverworts with emphasis on Ptilidiales. Cladistics 2020, 36, 184–193. [Google Scholar] [CrossRef]
- Tao, J.; Wang, X.; Long, Y.; Gao, Z.; Zhang, G.; Guo, Z.; Wang, G.; Xu, G.; Wang, Y.; Liu, H. Determining gene order patterns in the Suillus and Boletales through comparative analysis of their mitogenomes. Int. J. Mol. Sci. 2024, 25, 9597. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Yan, Z.F.; Kook, M.; Li, C.T.; Yi, T.H. Genetic and chemical diversity of edible mushroom Pleurotus species. Biomed. Res. Int. 2022, 2022, 6068185. [Google Scholar] [CrossRef]
- Boiko, S.M. The trends in the spread of simple sequence repeats in the genomes of Schizophyllum commune. Mycologia 2023, 115, 288–298. [Google Scholar] [CrossRef]
- Gong, F.; Geng, Y.; Zhang, P.; Zhang, F.; Fan, X.; Liu, Y. Genetic diversity and structure of a core collection of Huangqi (Astragalus ssp.) developed using genomic simple sequence repeat markers. Genet. Resour. Crop Evol. 2022, 70, 571–585. [Google Scholar] [CrossRef]
- Ye, K.; Qin, J.; Yonghong, H. Decoding the complete mitochondrial genome of Hydrangea chinensis maxim.: Insights into genomic recombination, gene transfer, and RNA editing dynamics. BMC Plant Biol. 2025, 25, 1078. [Google Scholar] [CrossRef] [PubMed]
- Martin, W. Gene transfer from organelles to the nucleus: Frequent and in big chunks. Proc. Natl. Acad. Sci. USA 2003, 100, 8612–8614. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Beaudet, D.; Nadimi, M.; Iffis, B.; Hijri, M. Rapid mitochondrial genome evolution through invasion of mobile elements in two closely related species of arbuscular mycorrhizal fungi. PLoS ONE 2013, 8, e60768. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, M.M.; Almotairy, H.M. Analysis of heat shock proteins based on amino acids for the tomato genome. Genes 2022, 13, 2014. [Google Scholar] [CrossRef] [PubMed]
- Cvijovic, I.; Good, B.H.; Desai, M.M. The effect of strong purifying selection on genetic diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef] [PubMed]
| Family | Species | Genbank Number | Size (bp) | References | |
|---|---|---|---|---|---|
| Boletaceae | Aureoboletus raphanaceus | NC079662 | 42,157 | [36] | |
| Baorangia bicolor | MW308599 | 38,082 | [28] | ||
| Boletus edulis | MW308609 | 34,763 | [28] | ||
| Butyriboletus subappendiculatus | OZ171993 | 35,023 | Unpublished | ||
| Butyriboletus roseoflavus | MW308600 | 36,622 | [28] | ||
| Butyriboletus roseoflavus | MZ202357 | 36,551 | Unpublished | ||
| Caloboletus calopus | MW308602 | 32,883 | [28] | ||
| Hemileccinum impolitum | NC056808 | 39,362 | Unpublished | ||
| Lanmaoa macrocarpa | NC080885 | 38,139 | [30] | ||
| Neoboletus brunneissimus | MW308605 | 42,147 | [28] | ||
| Neoboletus magnificus | MW308603 | 39,449 | [28] | ||
| Neoboletus obscureumbrinus | MW308607 | 39,929 | [28] | ||
| Pulveroboletus ravenelii | NC061666 | 43,528 | [37] | ||
| Rufoboletus hainanensis | OR438787 | 36,592 | [38] | ||
| Retiboletus ornatipes | MW308601 | 36,785 | [28] | ||
| Tylopilus brunneirubens | NC084291 | 32,389 | [39] | ||
| Tylopilus plumbeoviolaceoides | NC056835 | 37,242 | [29] | ||
| Paxillaceae | Paxillus involutus | NC045203 | 39,109 | [27] | |
| Paxillus rubicundulus | NC045204 | 41,061 | [27] | ||
| Species Name | Genbank Number | Length(bp) | GCRate(%) | AT-Skew | GC-Skew | tRNAs | Introns | Number of Repeats |
|---|---|---|---|---|---|---|---|---|
| Austroboletus fusisporus | PV424034 | 30,801 | 24.05 | −0.03 | 0.01 | 20 | 1 | 5 |
| Chalciporus radiatus | PV424038 | 44,823 | 21.69 | −0.02 | −0.02 | 24 | 0 | 27 |
| Chiua viridula | PV424033 | 32,641 | 23.68 | 0.06 | −0.02 | 25 | 0 | 15 |
| Phylloporus brunneiceps | PQ821895 | 38,908 | 24.44 | 0.04 | −0.04 | 28 | 0 | 6 |
| Phylloporus grossus | PQ846490 | 35,591 | 24.24 | 0.06 | −0.03 | 24 | 0 | 3 |
| Phylloporus grossus | PV424035 | 35,117 | 24.42 | 0.05 | −0.03 | 24 | 0 | 3 |
| Phylloporus luxiensis | PV424037 | 35,648 | 24.49 | 0.06 | −0.03 | 24 | 1 | 16 |
| Phylloporus rubrosquamosus | PV424036 | 35,647 | 24.38 | 0.05 | −0.03 | 24 | 1 | 4 |
| Zangia olivaceobrunnea | PV424039 | 35,949 | 23.78 | −0.02 | −0.03 | 26 | 0 | 7 |
| Species Name | Genbank Number | Length (bp) | GC Rate (%) | AT-Skew | GC-Skew | Number of Repeats |
|---|---|---|---|---|---|---|
| Austroboletus fusisporus | PV424034 | 30,801 | 24.05 | −0.03 | 0.01 | 5 |
| Chalciporus radiatus | PV424038 | 44,823 | 21.69 | −0.02 | −0.02 | 27 |
| Chiua viridula | PV424033 | 32,641 | 23.68 | 0.06 | −0.02 | 15 |
| Phylloporus brunneiceps | PQ821895 | 38,908 | 24.44 | 0.04 | −0.04 | 6 |
| Phylloporus grossus | PQ846490 | 35,591 | 24.24 | 0.06 | −0.03 | 3 |
| Phylloporus grossus | PV424035 | 35,117 | 24.42 | 0.05 | −0.03 | 3 |
| Phylloporus luxiensis | PV424037 | 35,648 | 24.49 | 0.06 | −0.03 | 16 |
| Phylloporus rubrosquamosus | PV424036 | 35,647 | 24.38 | 0.05 | −0.03 | 4 |
| Zangia olivaceobrunnea | PV424039 | 35,949 | 23.78 | −0.02 | −0.03 | 7 |
| Aureoboletus raphanaceus | NC079662 | 42,157 | 22.73 | 0.03 | 0.03 | 8 |
| Baorangia bicolor | MW308599 | 38,082 | 23.61 | 0.02 | 0.01 | 12 |
| Boletus edulis | MW308609 | 34,763 | 23.83 | 0.02 | −0.06 | 12 |
| Butyriboletus subappendiculatus | OZ171993 | 35,023 | 22.82 | 0.03 | 0.02 | 13 |
| Butyriboletus roseoflavus | MW308600 | 36,622 | 23.13 | 0.03 | 0.02 | 14 |
| Butyriboletus roseoflavus | MZ202357 | 36,551 | 23.13 | 0.03 | 0.02 | 12 |
| Caloboletus calopus | MW308602 | 32,883 | 23.21 | 0.03 | 0.02 | 17 |
| Hemileccinum impolitum | NC056808 | 39,362 | 23.98 | 0.02 | 0.01 | 5 |
| Lanmaoa macrocarpa | NC080885 | 38,139 | 23.5 | 0.03 | 0.03 | 14 |
| Neoboletus brunneissimus | MW308605 | 42,147 | 22.96 | 0.03 | 0.02 | 14 |
| Neoboletus magnificus | MW308603 | 39,449 | 22.93 | 0.03 | 0.02 | 11 |
| Neoboletus obscureumbrinus | MW308607 | 39,929 | 23.1 | 0.03 | 0.04 | 9 |
| Pulveroboletus ravenelii | NC061666 | 43,528 | 23.35 | −0.02 | 0.05 | 12 |
| Rufoboletus hainanensis | OR438787 | 36,592 | 23.95 | −0.04 | −0.02 | 5 |
| Retiboletus ornatipes | MW308601 | 36,785 | 23.19 | 0.03 | 0.01 | 13 |
| Tylopilus brunneirubens | NC084291 | 32,389 | 23.8 | −0.02 | 0.05 | 9 |
| Tylopilus plumbeoviolaceoides | NC056835 | 37,242 | 23.02 | 0.02 | 0.01 | 9 |
| Paxillus involutus | NC045203 | 39,109 | 21.75 | −0.09 | 0.06 | 53 |
| Paxillus rubicundulus | NC045204 | 41,061 | 21.03 | −0.09 | 0.05 | 47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.-Y.; Zhang, Z.; Mao, M.-W.; Zhao, K.; Yang, S. Unveiling the Unique Mitogenome Structure of Phylloporus: Implications for Phylogeny and Evolution in Boletaceae. J. Fungi 2025, 11, 831. https://doi.org/10.3390/jof11120831
Huang J-Y, Zhang Z, Mao M-W, Zhao K, Yang S. Unveiling the Unique Mitogenome Structure of Phylloporus: Implications for Phylogeny and Evolution in Boletaceae. Journal of Fungi. 2025; 11(12):831. https://doi.org/10.3390/jof11120831
Chicago/Turabian StyleHuang, Jie-Yu, Zhen Zhang, Ming-Wei Mao, Kuan Zhao, and Shan Yang. 2025. "Unveiling the Unique Mitogenome Structure of Phylloporus: Implications for Phylogeny and Evolution in Boletaceae" Journal of Fungi 11, no. 12: 831. https://doi.org/10.3390/jof11120831
APA StyleHuang, J.-Y., Zhang, Z., Mao, M.-W., Zhao, K., & Yang, S. (2025). Unveiling the Unique Mitogenome Structure of Phylloporus: Implications for Phylogeny and Evolution in Boletaceae. Journal of Fungi, 11(12), 831. https://doi.org/10.3390/jof11120831





