Abstract
Candida auris has emerged as a global public health threat due to its high mortality rates, multidrug resistance, and rapid transmission in healthcare settings. This study reports the first documented cases of C. auris candidemia in Portugal, comprising eight isolates from candidemia and colonised patients admitted to a major hospital in northern Portugal in 2023. Whole-genome sequencing (WGS) was performed to determine the phylogenetic relationships of the isolates, which were classified as belonging to Clade I. Genome sequencing also enabled the detection of missense mutations in antifungal resistance genes, which were correlated with antifungal susceptibility profiles determined according to EUCAST (European Committee on Antimicrobial Susceptibility Test) protocols and guidelines. All isolates exhibited resistance to fluconazole and amphotericin B according to the recently established EUCAST epidemiological cut-offs (ECOFFs). Most of the isolates showed a resistant phenotype to anidulafungin and micafungin. All isolates were resistant to caspofungin. Missense mutations identified included Y132F in ERG11, E709D in CDR1, A583S in TAC1b, K52N and E1464K in SNQ2, K74E in CIS2, M192I in ERG4, a novel mutation S237T in CRZ1, and variants in GCN5, a gene involved in chromatin remodelling and stress-response regulation. Identifying known and novel mutations highlights the evolution of antifungal resistance mechanisms in C. auris. These findings underscore the need for further research to understand C. auris resistance pathways and to guide effective clinical management strategies.
1. Introduction
Candida auris classification as a global public health threat is underscored by its epidemiological data, which reveals a widespread distribution across continents, affecting patients in over 60 countries [1,2]. This pathogen is associated with high mortality rates, and around 60% of patients infected may die from it, particularly in vulnerable populations in intensive care units (ICU) [2,3]. The rapid emergence and dissemination of C. auris highlight a urgent need for enhanced global surveillance and improved diagnostic capabilities. In 2022, the World Health Organisation classified this fungus as a priority pathogen due to its rapid transmission in healthcare settings and high mortality rate [4,5].
C. auris belongs to the Candida auris-Candida haemuli clade (CAH) from the family Metschnikowiaceae [6]. However, recent phylogenomic and comparative genomics analyses have proposed a reassignment to the genus Candidozyma [7]. The phylogenetic placement of this group has been challenging because members of the CAH clade share high sequence similarity in ribosomal markers, display convergent phenotypic traits, and appear to have undergone a relatively recent diversification. As a result, traditional phylogenetic markers often lack resolution, and only genome-scale analyses have provided sufficient phylogenetic signal to support their reassignment to Candidozyma [7].
To date, six geographically distinct C. auris clades have been described, including Clade I (South Asian), Clade II (East Asian), Clade III (South African), Clade IV (South American), Clade V (Iranian) and Clade VI (Singapore and Bangladesh) [1,8,9,10]. Clade-specific characteristics provide essential insights into C. auris epidemiology and clinical behaviour. Clades I, III, and IV are closely associated with outbreaks of invasive and multidrug-resistant infections, highlighting their significant role in healthcare-associated transmission [11]. In contrast, Clade II strains often display susceptibility to all antifungal agents and are predominantly linked to ear infections rather than invasive diseases. Clade II exhibits unique genomic features, such as distinct karyotypes characterised by extensive subtelomeric deletions and chromosomal rearrangements. This may contribute to its reduced propensity for causing systemic infections [12]. The clinical relevance of Clades V and VI is not yet fully understood due to limited studies so far.
Antifungal resistance mechanisms in C. auris isolates are multifaceted and involve significant fitness trade-offs, often compensated by adaptive mechanisms through regulatory network adaptations and increased stress tolerance that enable persistence and pathogenicity [13]. These compensatory adaptations highlight C. auris remarkable ability to balance the costs of antifungal resistance with fitness requirements, contributing to its persistence in healthcare and environmental settings [14].
In Portugal, a single case of C. auris belonging to Clade III was identified in 2023 in a colonised patient transferred from Angola, admitted to an ICU [15]. Our study documents the first reported cases of C. auris candidemia identified in 2023, highlighting the need for urgent surveillance and infection control measures. The primary objective was to characterise eight epidemiologically linked C. auris isolates obtained from candidemia and colonised patients admitted to the largest hospital in northern Portugal, focusing on their genetic and antifungal resistance profiles.
2. Materials and Methods
2.1. Patients, Strain Identification and Clinical Data
Between June and October 2023, clinical samples from eight patients admitted to São João Local Health Unit (ULSSJ), the biggest hospital in northern Portugal, were positive for C. auris. In five patients, C. auris was isolated in Sabouraud dextrose agar from routine surveillance screening swabs (axilla and groin) and urine. In three patients, C. auris was isolated from positive blood cultures. All isolates were identified by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (Vitek MS V3®) (bioMérieux, Craponne, France). The clinical data of the patients at the time of the first C. auris sample collection are summarised in Table 1. The Ethical competent authority approved this study—CES (Comissão de Ética para a Saúde, ULSSJ) and the Administration Council and Epidemiology Unit of ULSSJ—project code 67-21.
2.2. DNA Extraction
Genomic DNA was extracted following the instructions in the manual of the NZY Plant/Fungi gDNA Isolation kit, with some modifications. Yeast cells were disrupted in tubes containing zirconium beads and lysis buffer after ten cycles of 30 s each, at maximum velocity, using the Minilys homogeniser (Bertin Technologies, Montigny-le-Bretonneux, France). The Nanodrop (Thermo Fisher Scientific, Wilmington, NC, USA) and agarose electrophoresis assessed genomic DNA quantity and quality.
2.3. Genome Sequencing and Assembly
Yeast strains were sequenced from 1200 ng of genomic DNA. Library construction was performed using the Home-made Whole-Genome Sequencing prep kit (based on the Kapa HyperPrep kit). The integrity of the library fragment size was checked with the 4200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA), and its concentration was measured using the Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). The generated DNA fragments were sequenced in the Illumina Novaseq platfor (m, using 150 bp paired-end sequencing reads. After trimming the low-quality reads from output reads, the raw data quality was checked with the FastQC software (version 0.12.1) [16]. The trimmed sequences were assembled using CLC Genomics Workbench v.12.0.3 (https://www.qiagenbioinformatics.com/, accessed on 27 August 2025).
The high-quality sequencing reads were mapped against the reference genome of C. auris (Clade I) strain B8441, isolated from a patient’s blood in Pakistan. The trimmed sequence reads were used to perform a de novo assembly approach using an algorithm based on de Bruijn graphs. After the initial contig creation, the reads were mapped back to the contigs for assembly correction. The genome assembly quality was evaluated with QUAST v.5.1.0 [17].
2.4. InDel and Structural Variants Detection
After the mapping, a variant calling algorithm was applied to detect the variants that satisfy the requirements specified by the following filters: direction filtering = remove variants that do not have a statistically similar distribution in the forwards and reverse reads; minimum frequency = 20%; minimum quality = 25%; and minimum coverage = 50%. InDel and Structural Variants were detected to obtain the list of insertions and deletions in the samples based on the following criteria: minimum number of reads = 30; and p-value threshold = 0.05.
2.5. Phylogenomic Analysis
Phylogenomic analysis of clinical strains of C. auris was performed for accurate species identification. C. auris strains of the six prevalent clades (clades I–VI) were selected for phylogenetic reconstruction (Supplementary Materials Table S1). The phylogenetic inference was carried out with the Reference sequence Alignment-based Phylogeny builder (REALPHY v.1.13) (https://realphy.unibas.ch/realphy/, accessed on 27 August 2025) [18], which uses input raw short sequence read data with read length parameter 150 (RL = 150). The tree was constructed based on the maximum-likelihood principle, PhyML, optimised for speed in handling long sequence alignments. The general time-reversible (GTR) and gamma-distributed rate was used as a model of nucleotide evolution, as set by default. Constructed phylogeny was visualised and edited using the Interactive Tree of Life website (iTOL v.6) (https://itol.embl.de/, accessed on 27 August 2025) [19].
2.6. Antifungal Susceptibility Profile
Minimal inhibitory concentrations (MIC) for fluconazole (FLC), voriconazole (VRC), Posaconazole (PSC), isavuconazole (ISC), amphotericin B (AmB) and echinocandins, caspofungin (CSF), anidulafungin (ANF) and micafungin (MCF) were determined by microdilution assay according to the European Committee on Antimicrobial Susceptibility Testing E.Def.7.4 (EUCAST) and according to the M27-A3 protocol of the Clinical laboratory Standard Institute (CLSI) [20,21,22]. Since there are currently no established C. auris-specific susceptibility breakpoints, the strains were classified by using tentative breakpoints (TBP), according to CDC guidelines (https://www.cdc.gov/candida-auris/hcp/laboratories/antifungal-susceptibility-testing.html?CDC_AAref_Val=https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html, accessed on 27 August 2025), epidemiological cutoff values (ECVs) defined by M27-M57S protocol [23] and the recently defined epidemiological cutoffs (ECOFF) determined by EUCAST [20].
2.7. Molecular Detection of Antifungal Resistance Gene Mutations
Several genes involved in antifungal resistance in C. auris were selected based on their associated mechanisms in drug resistance or tolerance (Table 2). The sequences of all genes analysed were extracted from the genome of C. auris B8441 (clade I) available in the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 27 August 2025). Reference genes were aligned against the genome assemblies generated in this study using BLAST+ v.2.11.0 [24] to identify the corresponding gene regions. Variant calling for single-nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) was performed using FreeBayes v1.3.10 with default parameters. Variants were annotated and translated into amino acid substitutions using SnpEff v5.2 [25]. All mutations were further confirmed by visually inspecting the alignment of the reads in Integrative Genomics Viewer v2.1.2 [26]. Moreover, antifungal resistance point mutations in the genes were assessed and confirmed using the Solu online platform (Solu Healthcare Inc., Helsinki, Finland, https://platform.solu.bio/, accessed on 27 August 2025). Alignments of genes and corresponding amino acid sequences were generated with Clustal Omega v1.2.4 (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 27 August 2025).

Table 1.
Demographic and clinical characteristics of patients with C. auris positive cultures.
Table 1.
Demographic and clinical characteristics of patients with C. auris positive cultures.
| Candidemia | Colonisation | |||||||
|---|---|---|---|---|---|---|---|---|
| SCO 267 | SCO 276 | SCO 279 | SCO248 | SCO 275 | SCO 240 | SCO242 | SCO266 | |
| Sex/age | Male/66 | Male/67 | Female/58 | Female/64 | Male/51 | Male/59 | Male/77 | Male/63 |
| Sample | blood culture | blood culture | blood culture | CVC | CVC | rectal swab | urine | axillary and groin swab |
| Hospital ward | ICU | surgery | medicine | infectiology | orthopaedics | surgery | surgery | surgery |
| Days of hospitalisation until the first isolation | 91 | 106 | 110 | 22 | 132 | 34 | 15 | 79 |
| ICU before isolation | yes | yes | no | yes | no | yes | no | no |
| Antifungal treatment | micafungin | Caspofungin + amphotericin B | caspofungin | no | no | no | no | no |
| Underlying disease | Oesophageal neoplasia, dyslipidemia | Gastric adenocarcinoma, peritonitis, HTN, dyslipidemia | Severe gonarthrosis, infection after total knee arthroplasty, DM2, HTN, dyslipidemia | hepatic abscess by Clostridium difficile, DM2, HTN, dyslipidemia | chronic osteomyelitis | Colon neoplasia, faecal peritonitis | Pancreatic adenocarcinoma DM2, HTN | Biliary tract neoplasia, acute cholangitis with hepatic abscess |
| Outcome | deceased | deceased | alive | alive | alive | alive | alive | deceased |
DM2: diabetes mellitus type 2; HTN: hypertension; CVC: central venous catheter.

Table 2.
Genes analysed for antifungal resistance in Candida auris, grouped by functional category. The table lists the gene categories, individual genes, and their associated mechanisms or functions in drug resistance or tolerance.
Table 2.
Genes analysed for antifungal resistance in Candida auris, grouped by functional category. The table lists the gene categories, individual genes, and their associated mechanisms or functions in drug resistance or tolerance.
| Category | Genes | Mechanism/Function | References |
|---|---|---|---|
| Drug targets and ergosterol biosynthesis (Azoles/Polyenes) | ERG11 | Azole target; mutations reduce drug binding | [27,28] |
| ERG2, ERG3, ERG4, ERG5, ERG6, ERG10, ERG25 | Enzymes in ergosterol pathway; alterations affect membrane composition and polyene/azole susceptibility | ||
| Efflux pumps (ABC/MFS transporters) | CDR1, CDR2, CDR4, MDR1, SNQ2 | Drug transporters that expel antifungals; overexpression or mutations increase azole resistance | [29,30] |
| Transcription factors | TAC1B, UPC2, MRR1, CRZ1 | Regulate expression of efflux pumps or ergosterol pathway; CRZ1 modulates stress response and tolerance | [29,31] |
| Epigenetic regulators | GCN5, SET1, SET2, RTT109, DOT1 | chromatin remodelling and antifungal stress response gene expression | [32] |
| Echinocandin target/1,3-β-glucan synthase | FKS1, FKS2 | Subunits of 1,3-β-glucan synthase; hotspot mutations confer echinocandin resistance | [33] |
| Stress response and antifungal tolerance | HSP90, CNA1 | Chaperone and calcineurin subunit; modulate stress response and tolerance to echinocandins/azoles | [34] |
| Other tolerance/stress and metabolism | CIS2 | Cystathionine γ-lyase; involved in sulfur metabolism, redox homeostasis, and stress response | [35] |
| Pheromone/export and signalling | STE6 | ABC transporter for a-factor pheromone export; may influence mating and signalling | [36] |
3. Results
3.1. Case Patient Description and Timeline
Clinical characteristics of the three candidemia and five asymptomatic carriage patients with C. auris are detailed in Table 1.
Three colonisation sites were identified: in the skin (2 patients, isolates SCO240 and SCO266), in the central venous catheter (2 patients, isolates SCO248 and SCO275), and in urine (1 patient, SCO242). The patient median age was 63.1 (range: 51–77); two were females, and six were males. The patients were hospitalised between May and October 2023, with four patients simultaneously admitted to the same ward (surgery, isolates SCO276, SCO240, SCO242, SCO266) (Figure 1).
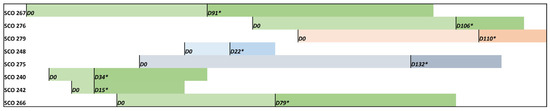
Figure 1.
Timeline representing hospitalisation days (D0—admission) and the first C. auris positive detection (*) from colonised and candidemia patients. Green colour refers to patients admitted to the same hospital ward (surgery), grey to orthopaedics, light blue to infectiology and salmon to medicine.
SCO240 and SCO242 isolates were isolated from patients sharing the same room, and SCO266 and SCO276 were recovered from patients in close rooms. The median time from admission to the first C. auris isolation was 106 and 34 days for candidemia and colonised patients, respectively; however, isolation times vary from 15 to 132 days. Three patients developed C. auris candidemia (isolates SCO267, SCO276 and SCO279); however, no information regarding colonisation sites or early detection was obtained before the onset of the invasive infection. Only candidemia patients received antifungal therapy, mainly with echinocandins. Three candidemia patients died (infected with SCO266, SCO267 and SCO276 strains), although death was not exclusively attributed to C. auris infection. All the patients had no recent travel history, nor were they transferred from other hospitals or healthcare facilities. No environmental samples were obtained, nor were contacts outside the hospital, such as family members, swabbed.
3.2. Genome Statistics and Phylogenomic Analysis
The genome assemblies of the eight C. auris strains showed an average length of 12.2 Mb, a contig count of 7, GC content of 45.3%, a scaffold N50 value of 2.3 bp, and a coverage depth for quality-trimmed reads ranging from 149-fold to 247-fold (Table 3). The accession numbers for the assemblies generated in this study are presented in Table 3. C. auris clade I, commonly called the South Asian, comprises isolates from cases in India, Pakistan, the United Arab Emirates, Lebanon, Italy, China, and Brazil. Phylogenetic analysis using whole-genome sequencing revealed that all resistant C. auris strains analysed in this study (SCO strains) belong to Clade I (Figure 2).

Table 3.
Genomic features of eight clinical strains of Candida auris from this study.
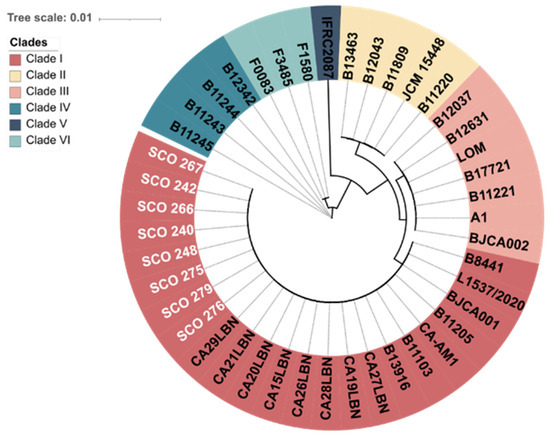
Figure 2.
Phylogenetic tree showing the genetic relationships among isolates from six distinct clades. Resistant Candida auris isolates from this study are highlighted in white. The scale bar indicates the evolutionary distance.
3.3. Genetic Variants Detection
The genetic variant analysis of C. auris strains revealed the presence of different mutation types, including single-nucleotide variants (SNVs), deletions, insertions, replacements, and multi-nucleotide variants (MNVs) (Supplementary Materials Table S2). The total number of SNVs was consistent across the strains, ranging from 60,619 (SCO 267) to 61,659 (SCO 275), varying by approximately 1000 variants. Regarding deletions, strain SCO 275 showed the highest count (2418), while strain SCO 276 had the lowest (2214). As for insertions, the numbers ranged from 3101 (SCO 276) to 3674 (SCO 275), with SCO 275 also showing the highest number of such variants. The strains displayed more minor variations for replacements, with SCO 275 having the highest number (337) and SCO 276 having the lowest (308). Finally, the MNVs, referring to variants involving multiple nucleotides, also exhibited similar numbers across strains, with SCO 275 showing the highest (3224) and SCO 267 being the lowest (3130) (Supplementary Materials Table S2).
3.4. Antifungal Susceptibility
Antifungal MIC values, determined according to EUCAST, susceptibility profiles, and phenotypes of the C. auris isolates from colonised patients and patients with candidemia are presented in Table 4. MIC values and susceptibility profiles determined according CLSI protocol are detailed in Supplementary Materials Table S3 [21,22]. All isolates showed high MIC values to fluconazole and amphotericin B, categorised as a resistant phenotype according to EUCAST ECOFFs, being one isolate (SCO242) classified as increased exposure (IE) (Table 4) [20,37]. Most of the isolates showed a resistant phenotype to anidulafungin and micafungin, and all isolates were resistant to caspofungin.

Table 4.
Antifungal minimal inhibitory concentration (MIC) and susceptibility phenotype of C. auris isolates according to EUCAST guidelines for antifungal susceptibility testing.
3.5. Analysis of Resistance-Associated Mutations
We identified several mutations in antifungal resistance–associated genes across our C. auris isolates (Figure 3, Table 5). In the ergosterol biosynthesis pathway, the ERG11 gene carried the missense mutation Y132F (c.396A>T), a well-characterised mutation conferring resistance to azoles [38,39]. In ERG4, we detected the substitution M192I (c.576G>T), which may affect sterol composition and modulate azole susceptibility. Among efflux-related genes, the CDR1 gene contained the substitution E709D (c.2126A>T), while its transcriptional regulator TAC1b harboured the A583S (c.861C>A) mutation, potentially impacting efflux activity. In the ABC transporter SNQ2, two additional variants were identified: K52N (c.72C>T) and E1464K (c.4306T>A), both previously associated with resistant isolates and possibly contributing to efflux-mediated azole tolerance [40].
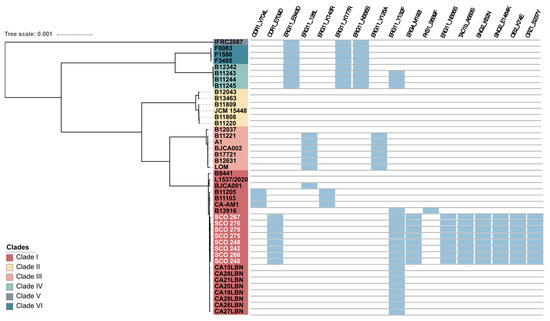
Figure 3.
Summary of the putative molecular mechanisms underlying antifungal resistance (blue boxes) found in C. auris isolates belonging to the six clades identified. The eight isolates from the present study are highlighted in white. The scale bar indicates the evolutionary distance.

Table 5.
List of variants reported in Candida auris isolates in Portugal. Newly reported variants are highlighted in bold.
Although its contribution to antifungal resistance in C. auris is negligible, a novel substitution in the transcription factor CRZ1 (S237Y, c.710C>A) was detected [32].
Concerning to recently epigenetic regulators involved in antifungal resistance [32], an in-frame deletion of six nucleotides in the GCN5 gene (c.439_444del) was detected in five isolates (SCO 240, SCO 248, SCO 266, SCO 275 and SCO 279) comparative to the B8441 reference genome, resulting in the deletion of two amino acids (p.(Glu134_Asn135del)) (Table 5). This variant was confirmed by inspection of the read alignments against the assemblies. The deletion lies within the N-terminal low-complexity region of Gcn5, whose functional significance is unknown. Other epigenetic regulators, STE1, SET2, RTT109 and DOT1 were also screened for genomic mutations, but no variants were found.
Similarly, no variants were detected in FKS1, FKS2, CDR2, CDR4, ERG2, ERG3, ERG5, ERG6, ERG 10, ERG 25, MDR1, MRR1, HSP90, CNA1, UPC2, and CIS2 genes in the set of C. auris clinical isolates, suggesting that it is unlikely that these genes play a role in the resistance phenotypes observed in these isolates. In the STE6 gene, a synonymous substitution (c.2157G>T) was identified, causing no alteration in the amino acid sequence and therefore, with no protein functional impact.
4. Discussion
The interaction between C. auris and the skin microbiota is complex, influencing both the persistence of the pathogen on the host and its potential for transmission [3]. Colonisation often precedes infection, with the skin microbiota serving as a critical reservoir for the yeast [41,42]. Understanding these dynamics is crucial for developing effective infection control and prevention strategies. Whole-genome sequencing allows, for instance, detailed characterisation of C. auris strains from colonised patients, providing insights into their genetic diversity, antifungal resistance profiles, and potential transmission pathways. Epidemiological studies have reported colonisation rates ranging from 5% to 70%, depending on the geographic location, patient population, and local infection control practices [41,42,43].
We report for the first time in Portugal, eight cases of candidemia and colonisation with C. auris in 2023, with potential epidemiological linkage. Most patients enrolled in this study had multiple health conditions, such as diabetes mellitus or neoplasia, yet only one patient had a prior ICU admission. The median from hospital admission to onset of candidemia was 106 days, longer than the time reported by other studies [44,45]. This is consistent with the notion that C. auris infections frequently occur several weeks after hospital admission, particularly among patients with prolonged hospital stays, multiple admissions, or those requiring intensive care [1,46]. These risk factors for C. auris infections are closely tied to healthcare settings and involve critical illness, invasive devices, antimicrobial use, and immunosuppression [1,47,48]. Effective infection control practices, early diagnosis, and antifungal stewardship are essential to mitigate risk.
Whole-genome sequencing revealed that all isolates belonged to Clade I, with single nucleotide variant (SNV counts ranging from 60,619 to 61,058. While crude SNP counts alone are insufficient to infer genetic relatedness accurately, it is plausible that all strains of C. auris, which belong to Clade I, were hospital-acquired, potentially originating from patients admitted simultaneously and sharing rooms. Two candidemia patients (SCO267 and SCO276) and one colonised patient (SCO266) died, most probably due to their complex health state and other concomitant pathologies. Future analyses using MLST or core genome phylogenomic approaches would be required to confirm these isolates’ genetic relationships and transmission pathways.
Antifungal susceptibility testing revealed a concerning multidrug-resistance profile. All isolates showed high MIC values to fluconazole and amphotericin B according to EUCAST ECOFFS [20,37], consistent with the high rates of azole resistance reported in Clade I isolates [1,28,35]. Resistance to fluconazole is a hallmark of Clade I isolates, with studies consistently reporting resistance rates exceeding 90% [47]. For instance, in a large-scale survey of C. auris isolates from India, more than 90% of Clade I isolates were resistant to fluconazole, while up to 5% were resistant to echinocandins [1,49]. In our study, voriconazole and posaconazole exhibited slightly better activity against C. auris isolates than fluconazole, although cross-resistance limits their therapeutic utility [1]. MIC values for isavuconazole ranged from 0.015 to 2 µg/mL. Although no ECVs or breakpoints are established for isavuconazole, MIC values vary significantly across studies and clades, including Clade I [1,50]. For instance, a global surveillance study of C. auris isolates from Clade I display MICs ≥4 µg/mL, indicating reduced susceptibility [50]. As frequently happens when using azole therapeutics, cross-resistance to isavuconazole and fluconazole is common in Clade I isolates due to similar target-binding sites in the lanosterol 14α-demethylase enzyme, limiting the utility of isavuconazole as a treatment option for fluconazole-resistant isolates [47,50,51]. Clinical use of isavuconazole for C. auris infections should be guided by antifungal susceptibility testing, and it is typically considered only when other triazoles and echinocandins are ineffective or contraindicated [51,52].
Resistance to azoles arises from the overexpression of efflux pumps, ATP-binding cassette (ABC) transporters, such as CDR1 and MDR1, and mutations in the ERG11 gene, the target of azoles [31]. Mutations in the ERG11 gene, such as Y132F and K143R, can reduce drug binding and are commonly suggested as responsible for the cross-resistance mechanisms shared between azole drugs [53,54]. The ERG11 mutation Y132F was detected in our cohort, consistent with the observed fluconazole resistance. The E709D mutation in the CDR1 gene was also identified. However, its role in antifungal resistance remains unclear and may act in combination with other resistance-associated mutations, such as A583S in the TAC1B gene [55]. Mutations in CDR1 and TAC1b may further contribute to antifungal resistance by enhancing efflux activity [56]. However, further research is necessary to elucidate the functional impact and specific roles of E709D and A583S mutations in mediating antifungal resistance in C. auris. Two variants in the SNQ2 ABC transporter (K52N, E1464K) were detected. While their functional impact is poorly characterised, upregulation of SNQ2 has been associated with multidrug resistance in clinical isolates, suggesting a potential role in modulating antifungal susceptibility. These findings highlight the contribution of efflux systems to antifungal tolerance in C. auris, demanding further functional validation.
Although less common, amphotericin B resistance involves alterations in the ergosterol biosynthesis pathway, which may impact membrane integrity; however, it can be balanced by lipid metabolism and membrane structure modifications [28]. Resistance to amphotericin B is variably reported in Clade I isolates, with rates ranging from 10% to 30% [47]. Loss-of-function mutations in the ERG6 and ERG3 genes, involved in ergosterol biosynthesis, have been associated with amphotericin B resistance [57,58]. In this study, all C. auris isolates displayed a resistance profile to amphotericin B, with one considered susceptible with increased exposure according to EUCAST [20,37]. However, no mutations were detected in ERG6 and ERG3. Instead, a missense substitution (M192I) was identified in ERG4, a downstream enzyme in the ergosterol pathway. Such changes may affect sterol compositions, potentially contributing to amphotericin B tolerance. In S. cerevisiae, ERG4 modifications have been shown to influence polyene sensitivity [59], thus corroborating that downstream sterol biosynthetic enzymes can modulate antifungal susceptibility. Nevertheless, no evidence links the ERG4 M192I variant to antifungal resistance in C. auris. Future studies are needed to determine whether the ERG4 M192I mutation modifies membrane sterol composition and consequently affects drug tolerance.
C. auris Clade I isolates are usually considered susceptible to echinocandins, including caspofungin, micafungin, and anidulafungin [55,60]. However, emerging reports have described the development of echinocandin resistance in Clade I strains [60]. In this study, most isolates exhibited a resistant phenotype for anidulafungin (7 out of 8) and micafungin (6 out of 8), and all isolates were resistant to caspofungin. However, when MIC values were determined and interpreted according to the CLSI protocol [21], a wild-type phenotype to anidulafin and micafungin was observed. Typically, echinocandin resistance is usually associated with mutations in the FKS1 and FKS2 genes, encoding the β-1,3-glucan synthase, often resulting in a fitness cost due to cell wall synthesis disruption [49,61]; nevertheless, no mutation in these genes was detected in our isolates.
Recently, the role of epigenetic regulators in C. auris antifungal resistance was comprehensively described [36]. Notably, the gene encoding the acetyltransferase catalytic subunit GCN5, responsible for histone H3 acetylation, was shown to have a crucial role in the transcription of genes involved in ergosterol biosynthesis, drug efflux, cell wall integrity and echinocandin resistance through the calcineurin signalling pathway. Therefore, Gcn5 seems to be a master regulator for C. auris drug resistance, associated with azole and polyene tolerance, and echinocandin resistance [32]. Interestingly, in the set of C. auris isolates, five of them displayed a variant of the GCN5 gene (c.439_444del), resulting in the in-frame deletion of two amino acids (Glu134_Asn135del), whose functional impact is unknown. Although the deletion we report is partial and located outside the annotated catalytic HAT domain, its recurrent presence in multiple clinical isolates raises the possibility of a modulatory effect on Gcn5 function. Definitive conclusions will require targeted functional assays, and we report this variant as an intriguing candidate for further investigation of GCN5-mediated pathways in antifungal tolerance. Besides GCN5, other genes such as SET1, SET2, RTT109 and DOT1 were recently associated with azole and echinocandin tolerance [36]. In the set of C. auris isolates, no mutation was detected in these genes.
We also identify a novel missense mutation in the calcineurin pathway target CRZ1 (S237Y). CRZ1 is a transcription factor involved in stress response and cell wall integrity, whose role in drug resistance is still controversial among Candida species [34,62,63]. In C. auris, growth under caspofungin exposure increases the expression of the CRZ1 gene in a Gcn5-dependent manner. Furthermore, the crz1Δ mutants displayed the same sensitivity to caspofungin as the wild-type strain, dissociating its role in echinocandin resistance [32].
In addition to the canonical resistance-associated genes, we investigated CIS2 and STE6, which have been implicated in experimental or clinical studies. Recently, a substitution in CIS2 (A27T), encoding a γ-glutamyltranspeptidase involved in xenobiotic detoxification, was linked to increased echinocandin MICs in experimentally evolved C. auris strains [35]. Another variant (K74E) was reported in susceptible and resistant isolates, suggesting some CIS2 polymorphisms may not be under antifungal selective pressure. In our isolates, no nonsynonymous variants were detected in CIS2. Regarding STE6, an a-pheromone ABC transporter reported to modulate antimycotic responses [36], we identified only a synonymous substitution (c.2157G>T), which is unlikely to impact protein function. Nevertheless, given the potential involvement of these genes in antifungal tolerance and adaptation, continued surveillance and functional studies are warranted.
This study provides several insights regarding C. auris infections. However, several limitations warrant consideration. First, the small sample size restricts the strength of the evidence for predicting risk factors associated with C. auris candidemia. Second, the lack of comprehensive surveillance cultures, including samples from various body sites, the hospital environment, and healthcare staff, limits the understanding of transmission patterns among patients. Also, the study’s retrospective nature precluded the collection of a complete exposure history. Despite the limitations, our findings provide important insights into the genetic determinants of antifungal resistance and potential transmission pathways of C. auris in Portuguese healthcare settings. Understanding these mechanisms is crucial to developing novel therapeutic approaches and targeting infection control strategies to guide the formulation of more robust hygiene practices, environmental cleaning protocols, and patient management strategies to reduce the spread of C. auris. This will guide future research into the molecular basis of resistance and persistence in this emerging pathogen.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof11100716/s1. Table S1: List of Candida auris strains used in this study; Table S2: Genetic variants detected in Candida auris isolates; Table S3: Antifungal minimal inhibitory concentration (MIC) and susceptibility phenotype of C. auris isolates according to CLSI guidelines for antifungal susceptibility testing. Strains were classified according CLSI defined ECVs and CDC.
Author Contributions
Conceptualization, I.M.M., M.F.M.G. and S.C.d.O.; Methodology, I.M.M. and M.F.M.G.; Software, S.H.; Validation, J.A.P. and J.T.G.; Formal analysis, M.F.M.G., S.H. and S.C.d.O.; Data curation, M.F.M.G., D.P. and S.C.d.O.; Writing—original draft, I.M.M. and M.F.M.G.; Writing—review & editing, D.P., J.A.P., J.T.G. and S.C.d.O.; Supervision, S.C.d.O. All authors have read and agreed to the published version of the manuscript.
Funding
This article was supported by National Funds through FCT—Fundação para a Ciência e a Tecnologia, I.P., within CINTESIS, R&D Unit (reference UIDP/4255/2020).
Institutional Review Board Statement
The Ethical competent authority approved this study—CES (Comissão de Ética para a Saúde, São João Local Health Unit) and the Administration Council and Epidemiology Unit of ULSSJ—project code 67-21 and date of approval 12 May 2021.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
Micael F. M. Gonçalves thanks the FCT—Fundação para a Ciência e a Tecnologia I.P., under the project/grant UID/50006 + LA/P/0094/2020 (doi.org/10.54499/LA/P/0094/2020) and his contract 2022.00758.CEECIND/CP1720/CT0051 (doi.org/10.54499/2022.00758.CEECIND/CP1720/CT0051. The authors would like to thank Isabel Santos for the excellent technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Lyman, M.; Forsberg, K.; Sexton, D.J.; Chow, N.A.; Lockhart, S.R.; Jackson, B.R.; Chiller, T. Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann. Intern. Med. 2023, 176, 489–495. [Google Scholar] [CrossRef]
- Biswal, M.; Rudramurthy, S.M.; Jain, N.; Shamanth, A.S.; Sharma, D.; Jain, K.; Yaddanapudi, L.N.; Chakrabarti, A. Controlling a possible outbreak of Candida auris infection: Lessons learnt from multiple interventions. J. Hosp. Infect. 2017, 97, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. WHO publish fungal priority pathogens list. Lancet Microbe 2023, 4, e74. [Google Scholar] [CrossRef]
- Alvarez-Moreno, C.A.; Morales-López, S.; Rodriguez, G.J.; Rodriguez, J.Y.; Robert, E.; Picot, C.; Ceballos-Garzon, A.; Parra-Giraldo, C.M.; Le Pape, P. The Mortality Attributable to Candidemia in C. auris Is Higher than That in Other Candida Species: Myth or Reality? J. Fungi 2023, 9, 430. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gomez-Lopez, A.; Boekhout, T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar]
- Liu, F.; Hu, Z.D.; Zhao, X.M.; Zhao, W.N.; Feng, Z.X.; Yurkov, A.; Alwasel, S.; Boekhout, T.; Bensch, K.; Hui, F.L.; et al. Phylogenomic analysis of the Candida auris-Candida haemuli clade and related taxa in the Metschnikowiaceae, and proposal of thirteen new genera, fifty-five new combinations and nine new species. Persoonia 2024, 52, 22–43. [Google Scholar] [CrossRef] [PubMed]
- Suphavilai, C.; Ko, K.K.K.; Lim, K.M.; Tan, M.G.; Boonsimma, P.; Chu, J.J.K.; Goh, S.S.; Rajandran, P.; Lee, L.C.; Tan, K.Y.; et al. Detection and characterisation of a sixth Candida auris clade in Singapore: A genomic and phenotypic study. Lancet Microbe 2024, 5, 100878. [Google Scholar] [CrossRef]
- Chow, N.A.; De Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential fifth clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef]
- Khan, T.; Faysal, N.I.; Hossain, M.M.; Mah, E.M.S.; Haider, A.; Moon, S.B.; Sen, D.; Ahmed, D.; Parnell, L.A.; Jubair, M.; et al. Emergence of the novel sixth Candida auris Clade VI in Bangladesh. Microbiol. Spectr. 2024, 12, e0354023. [Google Scholar] [CrossRef] [PubMed]
- Semenya, M.D.; Aladejana, A.E.; Ndlovu, S.I. Characterization of susceptibility patterns and adaptability of the newly emerged Candida auris. Int. Microbiol. 2024, 28, 575–587. [Google Scholar] [CrossRef]
- Munoz, J.F.; Welsh, R.M.; Shea, T.; Batra, D.; Gade, L.; Howard, D.; Rowe, L.A.; Meis, J.F.; Litvintseva, A.P.; Cuomo, C.A. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics 2021, 218, iyab029. [Google Scholar] [CrossRef]
- Das, S.; Singh, S.; Tawde, Y.; Dutta, T.K.; Rudramurthy, S.M.; Kaur, H.; Shaw, T.; Ghosh, A. Comparative fitness trade-offs associated with azole resistance in Candida auris clinical isolates. Heliyon 2024, 10, e32386. [Google Scholar] [CrossRef]
- Carolus, H.; Sofras, D.; Boccarella, G.; Sephton-Clark, P.; Biriukov, V.; Cauldron, N.C.; Lobo Romero, C.; Vergauwen, R.; Yazdani, S.; Pierson, S.; et al. Acquired amphotericin B resistance leads to fitness trade-offs that can be mitigated by compensatory evolution in Candida auris. Nat. Microbiol. 2024, 9, 3304–3320. [Google Scholar] [CrossRef] [PubMed]
- Henriques, J.; Mixão, V.; Cabrita, J.; Duarte, T.I.; Sequeira, T.; Cardoso, S.; Germano, N.; Dias, L.; Bento, L.; Duarte, S.; et al. Candida auris in Intensive Care Setting: The First Case Reported in Portugal. J. Fungi 2023, 9, 837. [Google Scholar] [CrossRef] [PubMed]
- FastQC: A Quality Control Tool for High Throughput Sequence. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 27 August 2025).
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs for Antifungal Agents, Version 11.0, 2024. Available online: http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/ (accessed on 31 July 2025).
- Institute CLS: Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; CLSI doxument M27A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008.
- The European Committee on Antimicrobial Susceptibility Testing. Testing ECoAS: EUCAST Definitive Document E.Def 7.4—Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts; 2023. Available online: https://www.eucast.org/ (accessed on 31 July 2025).
- Institute CLS: Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 4th ed.; CLSI supplemenmt M27-M57S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Ceballos-Garzon, A.; Penuela, A.; Valderrama-Beltran, S.; Vargas-Casanova, Y.; Ariza, B.; Parra-Giraldo, C.M. Emergence and circulation of azole-resistant C. albicans, C. auris and C. parapsilosis bloodstream isolates carrying Y132F, K143R or T220L Erg11p substitutions in Colombia. Front. Cell Infect. Microbiol. 2023, 13, 1136217. [Google Scholar] [CrossRef]
- Rybak, J.M.; Barker, K.S.; Muñoz, J.F.; Parker, J.E.; Ahmad, S.; Mokaddas, E.; Abdullah, A.; Elhagracy, R.S.; Kelly, S.L.; Cuomo, C.A.; et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin. Microbiol. Infect. 2022, 28, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hind, C.; Furner-Pardoe, J.; Sutton, J.M.; Rahman, K.M. Understanding the mechanisms of resistance to azole antifungals in Candida species. JAC Antimicrob. Resist. 2025, 7, dlaf106. [Google Scholar] [CrossRef] [PubMed]
- Wasi, M.; Khandelwal, N.K.; Moorhouse, A.J.; Nair, R.; Vishwakarma, P.; Bravo Ruiz, G.; Ross, Z.K.; Lorenz, A.; Rudramurthy, S.M.; Chakrabarti, A.; et al. ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes. Front. Microbiol. 2019, 10, 1445. [Google Scholar] [CrossRef]
- Rybak, J.M.; Muñoz, J.F.; Barker, K.S.; Parker, J.E.; Esquivel, B.D.; Berkow, E.L.; Lockhart, S.R.; Gade, L.; Palmer, G.E.; White, T.C.; et al. Mutations in TAC1B: A novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, L.; Huang, X.; Wang, Y.; Chen, G.; Moses, M.; Zou, Y.; Xiong, S.; Xue, W.; Dong, Y.; et al. Targeting epigenetic regulators to overcome drug resistance in the emerging human fungal pathogen Candida auris. Nat. Commun. 2025, 16, 4668. [Google Scholar] [CrossRef]
- Ahmed, S.H.; El-Kholy, I.M.A.; El-Mehalawy, A.A.; Mahmoud, E.M.; Elkady, N.A. Molecular characterization of some multidrug resistant Candida Auris in egypt. Sci. Rep. 2025, 15, 4917. [Google Scholar] [CrossRef]
- Singh, S.D.; Robbins, N.; Zaas, A.K.; Schell, W.A.; Perfect, J.R.; Cowen, L.E. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009, 5, e1000532. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Coste, A.T.; Bachmann, D.; Sanglard, D.; Lamoth, F. Deciphering the Mrr1/Mdr1 Pathway in Azole Resistance of Candida auris. Antimicrob. Agents Chemother. 2022, 66, e0006722. [Google Scholar] [CrossRef]
- Yadav, A.; Singh, A.; Wang, Y.; Haren, M.H.V.; Singh, A.; de Groot, T.; Meis, J.F.; Xu, J.; Chowdhary, A. Colonisation and Transmission Dynamics of Candida auris among Chronic Respiratory Diseases Patients Hospitalised in a Chest Hospital, Delhi, India: A Comparative Analysis of Whole Genome Sequencing and Microsatellite Typing. J. Fungi 2021, 7, 81. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Guinea, J.; Arikan-Akdagli, S.; Meijer, E.F.J.; Meis, J.F.; Buil, J.B.; Dannaoui, E.; Giske, C.G.; Lyskova, P.; Meletiadis, J.; et al. How to Interpret MICs of Amphotericin B, Echinocandins and Flucytosine Against Candida Auris (Candidozyma auris) According to the Newly Established European Committee for Antimicrobial Susceptibility Testing (EUCAST) Breakpoints. Clin. Microbiol. Infect. 2025, in press. [CrossRef] [PubMed]
- Xu, Z.; Zhang, L.; Han, R.; Ding, C.; Shou, H.; Duan, X.; Zhang, S. A Candidemia Case Caused by a Novel Drug-Resistant Candida auris with the Y132F Mutation in Erg11 in Mainland China. Infect. Drug Resist. 2023, 16, 3065–3072. [Google Scholar] [CrossRef]
- Ben Abid, F.; Salah, H.; Sundararaju, S.; Dalil, L.; Abdelwahab, A.H.; Salameh, S.; Ibrahim, E.B.; Almaslmani, M.A.; Tang, P.; Perez-Lopez, A.; et al. Molecular characterization of Candida auris outbreak isolates in Qatar from patients with COVID-19 reveals the emergence of isolates resistant to three classes of antifungal drugs. Clin. Microbiol. Infect. 2023, 29, 1083.e1081–1083.e1087. [Google Scholar] [CrossRef]
- Yan, L.; Zhao, F.; Zhang, K.; Wang, P.; Hu, S. Isolation of Fluconazole-Resistant South Asian Clade Candida auris in Beijing, China. Am. J. Trop. Med. Hyg. 2025, 113, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Proctor, D.M.; Dangana, T.; Sexton, D.J.; Fukuda, C.; Yelin, R.D.; Stanley, M.; Bell, P.B.; Baskaran, S.; Deming, C.; Chen, Q.; et al. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat. Med. 2021, 27, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Sexton, D.J.; Bentz, M.L.; Welsh, R.M.; Derado, G.; Furin, W.; Rose, L.J.; Noble-Wang, J.; Pacilli, M.; McPherson, T.D.; Black, S.; et al. Positive Correlation Between Candida auris Skin-Colonization Burden and Environmental Contamination at a Ventilator-Capable Skilled Nursing Facility in Chicago. Clin. Infect. Dis. 2021, 73, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Snell, H.M.; Cordier, C.; Desnos-Olivier, M.; Dellière, S.; Aissaoui, N.; Sturny-Leclère, A.; Da Silva, E.; Eblé, C.; Rouveau, M.; et al. First Patient-to-Patient Intrahospital Transmission of Clade I Candida auris in France Revealed after a Two-Month Incubation Period. Microbiol. Spectr. 2022, 10, e0183322. [Google Scholar] [CrossRef]
- Al-Siyabi, T.; Al Busaidi, I.; Balkhair, A.; Al-Muharrmi, Z.; Al-Salti, M.; Al’Adawi, B. First report of Candida auris in Oman: Clinical and microbiological description of five candidemia cases. J. Infect. 2017, 75, 373–376. [Google Scholar] [CrossRef]
- Southwick, K.; Ostrowsky, B.; Greenko, J.; Adams, E.; Lutterloh, E.; Denis, R.J.; Patel, R.; Erazo, R.; Fernandez, R.; Bucher, C.; et al. A description of the first Candida auris-colonized individuals in New York State, 2016–2017. Am. J. Infect. Control 2022, 50, 358–360. [Google Scholar] [CrossRef]
- Simon, S.P.; Li, R.; Silver, M.; Andrade, J.; Tharian, B.; Fu, L.; Villanueva, D.; Abascal, D.G.; Mayer, A.; Truong, J.; et al. Comparative Outcomes of Candida auris Bloodstream Infections: A Multicenter Retrospective Case-Control Study. Clin. Infect. Dis. 2023, 76, E1436–E1443. [Google Scholar] [CrossRef]
- Chow, N.A.; Gade, L.; Tsay, S.V.; Forsberg, K.; Greenko, J.A.; Southwick, K.L.; Barrett, P.M.; Kerins, J.L.; Lockhart, S.R.; Chiller, T.M.; et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: A molecular epidemiological survey. Lancet Infect. Dis. 2018, 18, 1377–1384. [Google Scholar] [CrossRef]
- Kekana, D.; Naicker, S.D.; Shuping, L.; Velaphi, S.; Nakwa, F.L.; Wadula, J.; Govender, N.P. Candida auris Clinical Isolates Associated with Outbreak in Neonatal Unit of Tertiary Academic Hospital, South Africa. Emerg. Infect. Dis. 2023, 29, 2044–2053. [Google Scholar] [CrossRef]
- Sharma, D.; Paul, R.A.; Rudramurthy, S.M.; Kashyap, N.; Bhattacharya, S.; Soman, R.; Shankarnarayan, S.A.; Chavan, D.; Singh, S.; Das, P.; et al. Impact of FKS1 Genotype on Echinocandin In Vitro Susceptibility in Candida auris and In Vivo Response in a Murine Model of Infection. Antimicrob. Agents Chemother. 2022, 66, e0165221. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef]
- Lewis, J.S., 2nd; Wiederhold, N.P.; Hakki, M.; Thompson, G.R., 3rd. New Perspectives on Antimicrobial Agents: Isavuconazole. Antimicrob. Agents Chemother. 2022, 66, e0017722. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef]
- Frías-De-león, M.G.; Hernández-Castro, R.; Vite-Garín, T.; Arenas, R.; Bonifaz, A.; Castañón-Olivares, L.; Acosta-Altamirano, G.; Martínez-Herrera, E. Antifungal resistance in Candida auris: Molecular determinants. Antibiotics 2020, 9, 568. [Google Scholar] [CrossRef]
- Chaabane, F.; Graf, A.; Jequier, L.; Coste, A.T. Review on Antifungal Resistance Mechanisms in the Emerging Pathogen Candida auris. Front. Microbiol. 2019, 10, 2788. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J. Associations between Genomic Variants and Antifungal Susceptibilities in the Archived Global Candida auris Population. J. Fungi 2024, 10, 86. [Google Scholar] [CrossRef]
- Hirayama, T.; Miyazaki, T.; Tanaka, R.; Kitahori, N.; Yoshida, M.; Takeda, K.; Ide, S.; Iwanaga, N.; Tashiro, M.; Takazono, T.; et al. TAC1b mutation in Candida auris decreases manogepix susceptibility owing to increased CDR1 expression. Antimicrob. Agents Chemother. 2025, 69, e0150824. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Zhang, H.; Liu, L.L.; Guo, L.N.; Liu, W.J.; Liu, Y.L.; Li, D.D.; Zhao, Y.; Zhu, R.Y.; Li, Y.; et al. Genome-wide analysis of in vivo-evolved Candida auris reveals multidrug-resistance mechanisms. Mycopathologia 2024, 189, 35. [Google Scholar] [CrossRef]
- Massic, L.; Doorley, L.A.; Jones, S.J.; Richardson, I.; Siao, D.D.; Siao, L.; Dykema, P.; Hua, C.; Schneider, E.; Cuomo, C.A.; et al. Acquired Amphotericin B Resistance Attributed to a Mutated ERG3 in Candidozyma auris. ASM J. 2025, e00601-25. [Google Scholar] [CrossRef]
- Kodedova, M.; Sychrova, H. Changes in the Sterol Composition of the Plasma Membrane Affect Membrane Potential, Salt Tolerance and the Activity of Multidrug Resistance Pumps in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0139306. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Jacobs, J.L.; Dennis, E.K.; Taimur, S.; Rana, M.; Patel, D.; Gitman, M.; Patel, G.; Schaefer, S.; Iyer, K.; et al. Candida auris Pan-Drug-Resistant to Four Classes of Antifungal Agents. Antimicrob. Agents Chemother. 2022, 66, e0005322. [Google Scholar] [CrossRef] [PubMed]
- Biagi, M.J.; Wiederhold, N.P.; Gibas, C.; Wickes, B.L.; Lozano, V.; Bleasdale, S.C.; Danziger, L. Development of high-level echinocandin resistance in a patient with recurrent candida auris candidemia secondary to chronic candiduria. Open Forum Infect. Dis. 2019, 6, ofz262. [Google Scholar] [CrossRef]
- Miyazaki, T.; Yamauchi, S.; Inamine, T.; Nagayoshi, Y.; Saijo, T.; Izumikawa, K.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 2010, 54, 1639–1643. [Google Scholar] [CrossRef]
- Chen, Y.L.; Brand, A.; Morrison, E.L.; Silao, F.G.; Bigol, U.G.; Malbas, F.F., Jr.; Nett, J.E.; Andes, D.R.; Solis, N.V.; Filler, S.G.; et al. Calcineurin controls drug tolerance, hyphal growth, and virulence in Candida dubliniensis. Eukaryot. Cell 2011, 10, 803–819. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).