Isolation, Morphological and Molecular–Phenological Identification of Nematophagous Fungi Inhabiting the Soils of Agricultural Lands in Southern Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling for the Isolation of NF
2.2. Isolation of Predatory Fungi
2.3. Test Object—Phytoparasitic Nematodes
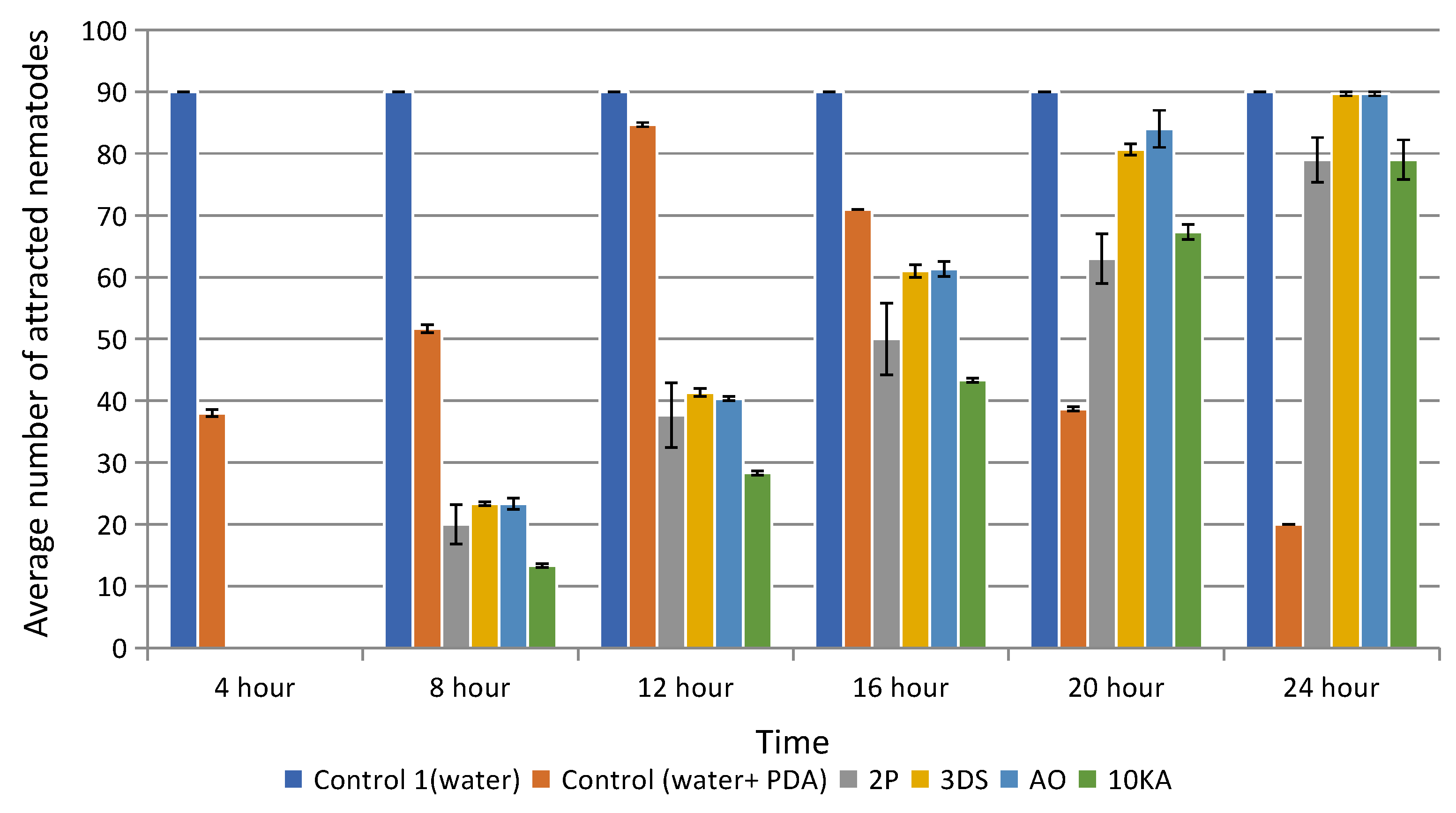
2.4. Attractive Activity of NF
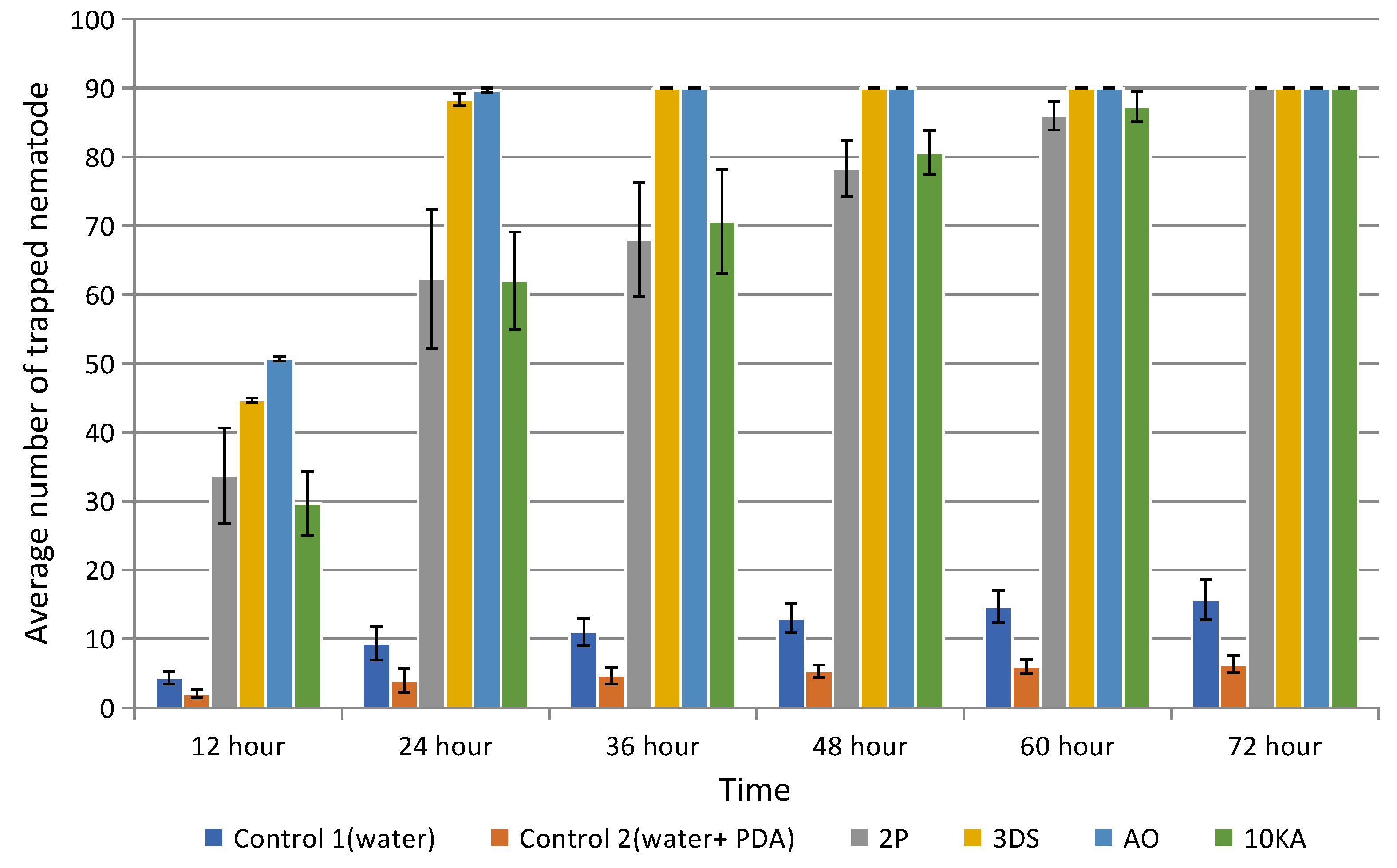
2.5. Nematophagous Activity of NF
2.6. Statistical Analysis
2.7. Identification of NF Strains
2.8. NF DNA Extraction
2.9. PCR Analysis and Amplification of the ITS Region
2.10. Determination of the Nucleotide Sequence
2.11. Analysis of Nucleotide Sequences
3. Results
3.1. Isolation of NF
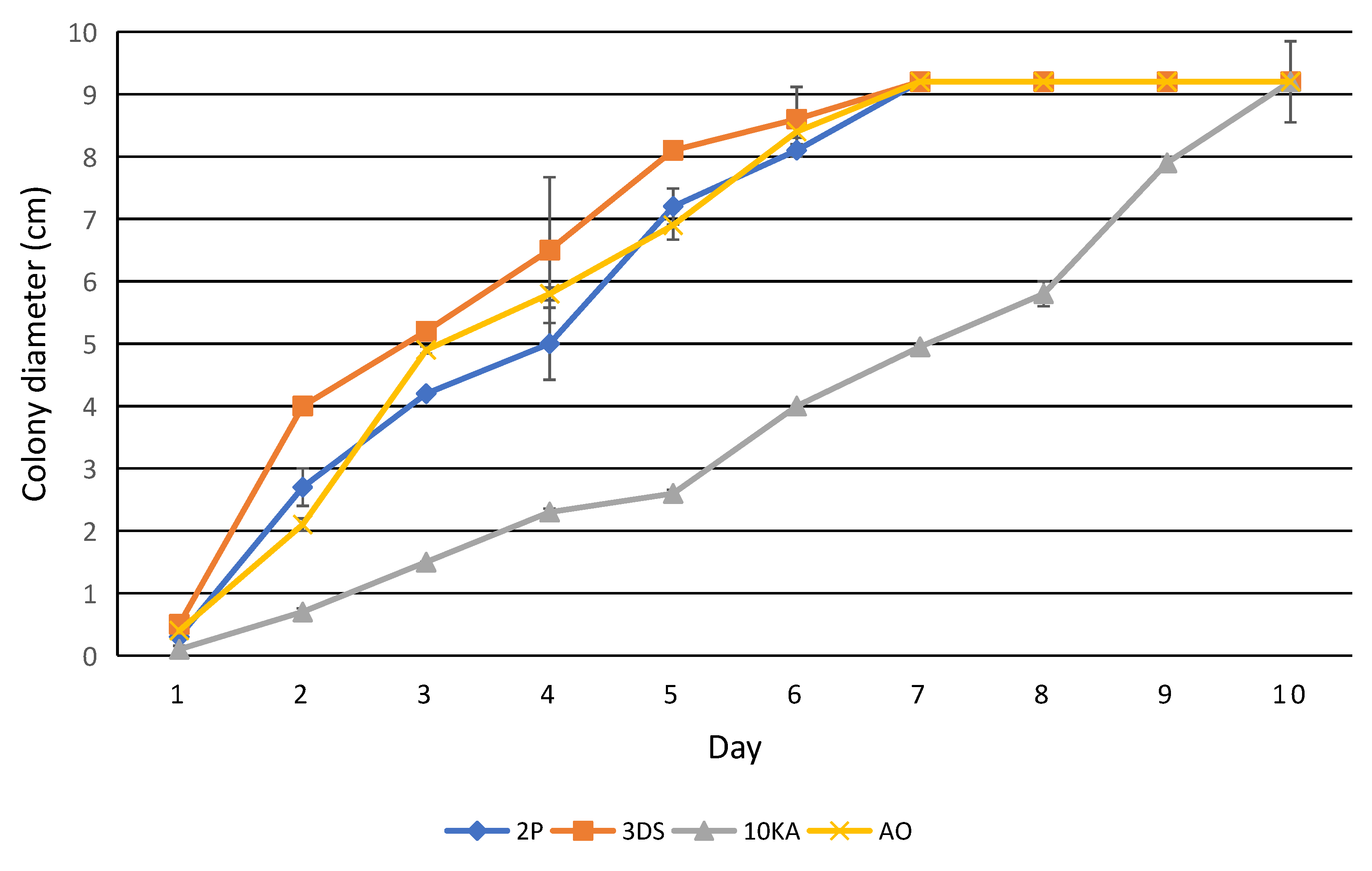
3.2. Assessment of the Growth Rate, Attractive and Trapping Activity of NF
3.3. Molecular Phylogenetic Identification of NF
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sagitov, A.O.; Perevertin, K.A. Principles and methods of quantitative study of nematode-phytoparasite populations and prediction of their harmfulness. In Principles and Methods of Mathematical Modeling in Plant Protection; Collection of Scientific Papers of KazNIIZR: Almaty, Kazakhstan, 1989; pp. 94–112. [Google Scholar]
- Sagitov, A.O.; Shesteperov, A.A. Parasitic Nematodes of Agricultural Crops; Kainar: Almaty, Kazakhstan, 1982; p. 27. [Google Scholar]
- Dababat, A.; İmren, M.; Pridannikov, M.; Özer, G.; Zhapayev, R.; Mokrini, F.; Otemissova, A.; Yerimbetova, A.; Morgounov, A. Plant-parasitic nematodes on cereals in northern Kazakhstan. J. Plant Dis. Prot. 2020, 127, 641–649. [Google Scholar] [CrossRef]
- FAO. Available online: https://www.fao.org/4/y4011e/y4011e0p.htm#:~:text=Their%20effects%20are%20commonly%20underestimated,damage%20(Whitehead%2C%201998) (accessed on 15 November 2024).
- Chitwood, D.J. Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture–Agricultural Research Service. Pest Manag. Sci. 2003, 59, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, Z.; Jiang, Z.; Nizamani, M.M.; Zhang, H.; Liu, M.; Wei, S.; Wang, Y.; Li, K. Isolation, Identification, and Evaluation of the Predatory Activity of Chinese Arthrobotrys Species towards Economically Important Plant-Parasitic Nematodes. J. Fungi 2023, 9, 1125. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wolyn, D. Potential role for salicylic acid in induced resistance of asparagus roots to Fusarium oxysporum f. sp. asparagi. Plant Pathol. 2005, 54, 227–232. [Google Scholar] [CrossRef]
- FAO. Available online: https://www.fao.org/4/v9978e/v9978e08.htm#:~:text=Once%20nematicides%20or%20their%20degradation,fumigant%20nematicides%20in%20some%20countries (accessed on 15 November 2024).
- Gheysen, G.; Gheysen, G.; Abad, P.; Bleve, T.; Blok, V.; Fenoll, C.; Gatehouse, J.A.; Grundler, F.; Lindsey, K.; Ohl, S.; et al. Concerted efforts to develop handles for plant parasitic nematode control. Dev. Plant Genet. Breed. 2000, 6, 159–167. [Google Scholar] [CrossRef]
- Askary, T.H. Nematophagous Fungi as Biocontrol Agents of Phytonematodes. In Biocontrol Agents of Phytonematodes; CABI Digital Library: Wallingford, UK, 2015; pp. 81–125. Available online: https://www.cabidigitallibrary.org/doi/pdf/10.1079/9781780643755.0081?download=true (accessed on 6 July 2024).
- Al-Ani, L.K.T.; De Freitas Soares, F.E.; Sharma, A.; De Los Santos-Villalobos, S.; Valdivia-Padilla, A.V.; Aguilar-Marcelino, L. Strategy of nematophagous fungi in determining the activity of plant parasitic nematodes and their prospective role in sustainable agriculture. Front. Fungal Biol. 2022, 3, 863198. [Google Scholar] [CrossRef]
- Singh, K.P.; Jaiswal, R.K.; Kumar, N.; Kumar, D. Nematophagous Fungi Associated with Root Galls of Rice Caused by Meloidogyne graminicola and its Control by Arthrobotrys dactyloides and Dactylaria brochopaga. J. Phytopathol. 2007, 155, 193–197. [Google Scholar] [CrossRef]
- Phani, V.; Khan, M.R.; Dutta, T.K. Plant-parasitic nematodes as a potential threat to protected agriculture: Current status and management options. Crop Prot. 2021, 144, 105573. [Google Scholar] [CrossRef]
- Chaerani, N.; Hariyadi, T.Z.P.; Dewi, N. Plant parasitic nematodes infesting three minor legumes (velvet bean, lablab bean, and jack bean). AIP Conf. Proc. 2022, 262, 020016. [Google Scholar] [CrossRef]
- Nordbring-Hertz, B.; Jansson, H.B.; Tunlid, A. Nematophagous fungi. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; pp. 1–11. [Google Scholar] [CrossRef]
- Lopez-Llorca, L.V.; Jansson, H.; Vicente, J.G.M.; Salinas, J. Nematophagous fungi as root endophytes. In Microbial Root Endophytes; Springer: Berlin/Heidelberg, Germany, 2007; pp. 191–206. [Google Scholar] [CrossRef]
- Lopez-Llorca, L.V.; Maciá-Vicente, J.G.; Jansson, H. Mode of action and interactions of nematophagous fungi. In Integrated Management and Biocontrol of Vegetable and Grain Crops Nematodes; Springer: Dordrecht, The Netherlands, 2007; pp. 51–76. [Google Scholar] [CrossRef]
- De Freitas Soares, F.E.; Sufiate, B.L.; De Queiroz, J.H. Nematophagous fungi: Far beyond the endoparasite, predator and ovicidal groups. Agric. Nat. Resour. 2018, 52, 1–8. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Lopez-Llorca, L.V.; Jansson, H.; Salinas, J.; Persmark, L.; Asensio, L. Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytologist. 2002, 154, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Llorca, L.; Bordallo, J.; Salinas, J.; Monfort, E.; López-Serna, M. Use of light and scanning electron microscopy to examine colonization of barley rhizosphere by the nematophagous fungus Verticillium chlamydosporium. Micron 2002, 33, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.; Gronquist, M.R.; Schwarz, E.M.; Nath, R.D.; Lee, C.; Gharib, S.; Schroeder, F.C.; Sternberg, P.W. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLife 2017, 6, e20023. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Vilcinskas, A. Metabolites from nematophagous fungi and nematocidal natural products from fungi as an alternative for biological control. Part I: Metabolites from nematophagous ascomycetes. Appl. Microbiol. Biotechnol. 2016, 100, 3799–3812. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M.; Askary, T.H. Fungal and bacterial nematicides in integrated nematode management strategies. Egypt. J. Biol. Pest Control 2018, 28, 74. [Google Scholar] [CrossRef]
- Adetunji, C.O.; Egbuna, C.; Tijjani, H.; Adom, D.; Al-Ani, L.K.T.; Patrick-Iwuanyanwu, K.C. Homemade preparations of natural biopesticides and applications. In Natural Remedies for Pest, Disease and Weed Control; Egbuna, C., Sawicka, B., Eds.; Elsevier Science: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2019; pp. 179–186. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef]
- Doolotkeldieva, T.; Bobushova, S.; Muratbekova, A.; Schuster, C.; Leclerque, A. Isolation, Identification, and Characterization of the Nematophagous Fungus Arthrobotrys oligospora from Kyrgyzstan. Acta Parasitol. 2021, 66, 1349–1365. [Google Scholar] [CrossRef]
- Noweer, E.; Aboul-Eid, H. Biological control of root-knot nematode Meloidogyne incognita infesting cucumber Cucumis sativus L. cvs. Alfa by the nematode-trapping fungus Dactylaria brochopaga under field conditions. Agric. Biol. J. N. Am. 2013, 4, 435–440. [Google Scholar] [CrossRef]
- Tepper, E.Z.; Shilnikova, V.K.; Pereverzeva, G.I. Practical Training in Microbiology: Textbook. Manual; Drofa: Moscow, Russia, 2004. [Google Scholar]
- Duddington, C. Notes on the technique of handling predacious fungi. Trans. Br. Mycol. Soc. 1955, 38, 97–103. [Google Scholar] [CrossRef]
- Cooke, R.; Godfrey, B. A key to the nematode-destroying fungi. Trans. Br. Mycol. Soc. 1964, 47, 61–74. [Google Scholar] [CrossRef]
- Chronis, D.; Chen, S.; Lang, P.; Tran, T.; Thurston, D.; Wang, X. In vitro Nematode Infection on Potato Plant. Bio-Protocol 2014, 4, e1016. [Google Scholar] [CrossRef]
- Soprunov, F.F. Predaceous Fungi-Hyphomycetes and Their Application in the Control of Pathogenic Nematodes; Publishing House of the Academy of Sciences of the Turkmen SSR: Ashkabad, Turkmenistan, 1958. [Google Scholar]
- Teplyakova, T.V. Bioecological Aspects of the Study and Use of Predatory Fungi-Hyphomycetes. Novosibirsk. 1999, p. 252. Available online: https://earthpapers.net/bioekologicheskie-aspekty-izucheniya-i-ispolzovaniya-hischnyh-gribov-gifomitsetov (accessed on 14 September 2024).
- Badotti, F.; De Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Clarridge, J.E., 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 14 September 2024).
- Cayrol, J.C. Possibilities of biological control of Meloidogyne nematode pest with the use of the ’R 350’ nematophagous fungus (Arthrobotrys irregularis). Def. Veg. 1981, 35, 279–282. [Google Scholar]
- Cayrol, J.C.; Frankowski, J.P. Une méthode de lutte biologique contre lês nématodes à galles des racines appartenat au genre Meloidogyne. PHM Rev. Hortic. 1979, 193, 15–23. Available online: https://hal.inrae.fr/hal-02730841v1 (accessed on 14 September 2024).
- Zhang, J.; Fu, B.; Lin, Q.; Riley, I.T. Colonization of Beauveria bassiana 08F04 in root-zone soil and its biocontrol of cereal cyst nematode (Heterodera filipjevi). PLoS ONE 2020, 15, e0232770. [Google Scholar] [CrossRef]
- Yao, Y.; Huo, J.; Gao, W.; Wang, W. Biocontrol efficacy of endophytic fungus, Acremonium sclerotigenum, against Meloidogyne incognita under in vitro and in vivo conditions. Biologia 2023, 78, 3305–3313. [Google Scholar] [CrossRef]
- Rahman, M.U.; Chen, P.; Zhang, X.; Fan, B. Predacious strategies of nematophagous fungi as bio-control agents. Agronomy 2023, 13, 2685. [Google Scholar] [CrossRef]
- Li, H.; Wang, R.; Zhang, K.-Q.; Xu, J. Fungi–nematode interactions: Diversity, ecology, and biocontrol prospects in agriculture. J. Fungi 2020, 6, 206. [Google Scholar] [CrossRef]





| Location/Habitats | Site | GPS Coordinates |
|---|---|---|
| Kyzylorda region/Fruit garden | A-1, A-2, A-3 | 44°49′15.5″ N 65°30′27.9″ E |
| Kyzylorda region/Vegetable garden | A-4, A-5, A-6 | 44°49′15.5″ N 65°30′27.9″ E |
| Chu district, Orazaly village, Zhambyl region/Potato field | B-1, B-2, B-3 | 43°27′4436 N 74°15′4509 E |
| Chu district, Orazaly village, Zhambyl region/Barley grown in soil | B-4, B-5, B-6 | 43°27′4436 N 74°15′4509 E |
| Chu district, Orazaly village, Zhambyl region/Cucumbers grown in soil | B-7, B-8, B-9 | 43°27′4436 N 74°15′4509 E |
| Chu district, Orazaly village, Zhambyl region/Garlic grown in soil | B-10, B-11, B-12 | 43°27′4436 N 74°15′4509 E |
| Kazakh Research Institute of Potato and Vegetable Growing, Almaty region/Potato field | C-1, C-2, C-3 | 43°15′8993 N 76°44′878 0E |
| Kazakh Research Institute of Potato and Vegetable Growing, Almaty region/Alfalfa grown in soil | C-4, C-5, C-6 | 43°15′8993 N 76°44′8780 E |
| Kazakh Research Institute of Potato and Vegetable Growing, Almaty region/Cabbage grown in soil | C-7, C-8, C-9 | 43°15′8993 N 76°44′8780 E |
| Kazakh Research Institute of Potato and Vegetable Growing, Almaty region/Corn | C-10, C-11, C-12 | 43°15′8993 N 76°44′8780 E |
| Name Cultures | Result Identification in BLAST | ||
|---|---|---|---|
| Accession GeneBank | Strain Name | % Ident | |
| AO (PP740634) | OQ248188.1 | Orbilia oligospora strain FA806 | 98.98% |
| MF782733.1 | Orbilia oligospora strain 34314aDRJ | 98.81% | |
| KY463695.1 | Orbilia oligospora voucher BBA 69390 | 98.81% | |
| 2P (PP748482) | MK156709.1 | Duddingtonia flagrans isolate F0236 | 100.00% |
| KT215213.1 | Arthrobotrys flagrans strain CBS 583 91 | 100.00% | |
| KP257593.1 | Duddingtonia flagrans isolate SDH 035 | 100.00% | |
| 3DS (PP740516) | MN014034.1 | Arthrobotrys sp. isolate TWF592 | 100.00% |
| OQ244205.1 | Orbilia oligospora strain WA219 | 100.00% | |
| OQ244200.1 | Orbilia oligospora strain WA165 | 100.00% | |
| 10KA (PP740690) | KT215210.1 | Arthrobotrys superba strain CBS 109 52 | 99.83% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanalbek, G.; Zhanuzak, A.; Faleev, D.; Nusupov, A.; Mukhatayeva, K.; Boguspaev, K.-K. Isolation, Morphological and Molecular–Phenological Identification of Nematophagous Fungi Inhabiting the Soils of Agricultural Lands in Southern Kazakhstan. J. Fungi 2025, 11, 42. https://doi.org/10.3390/jof11010042
Kanalbek G, Zhanuzak A, Faleev D, Nusupov A, Mukhatayeva K, Boguspaev K-K. Isolation, Morphological and Molecular–Phenological Identification of Nematophagous Fungi Inhabiting the Soils of Agricultural Lands in Southern Kazakhstan. Journal of Fungi. 2025; 11(1):42. https://doi.org/10.3390/jof11010042
Chicago/Turabian StyleKanalbek, Gulzat, Akniyet Zhanuzak, Dmitry Faleev, Aidos Nusupov, Karlygash Mukhatayeva, and Kenzhe-Karim Boguspaev. 2025. "Isolation, Morphological and Molecular–Phenological Identification of Nematophagous Fungi Inhabiting the Soils of Agricultural Lands in Southern Kazakhstan" Journal of Fungi 11, no. 1: 42. https://doi.org/10.3390/jof11010042
APA StyleKanalbek, G., Zhanuzak, A., Faleev, D., Nusupov, A., Mukhatayeva, K., & Boguspaev, K.-K. (2025). Isolation, Morphological and Molecular–Phenological Identification of Nematophagous Fungi Inhabiting the Soils of Agricultural Lands in Southern Kazakhstan. Journal of Fungi, 11(1), 42. https://doi.org/10.3390/jof11010042






