Gut Mycobiome and Asthma
Abstract
1. Introduction to the Gut Mycobiome in Asthma
2. Characterizing the Gut Mycobiome in Asthma Patients
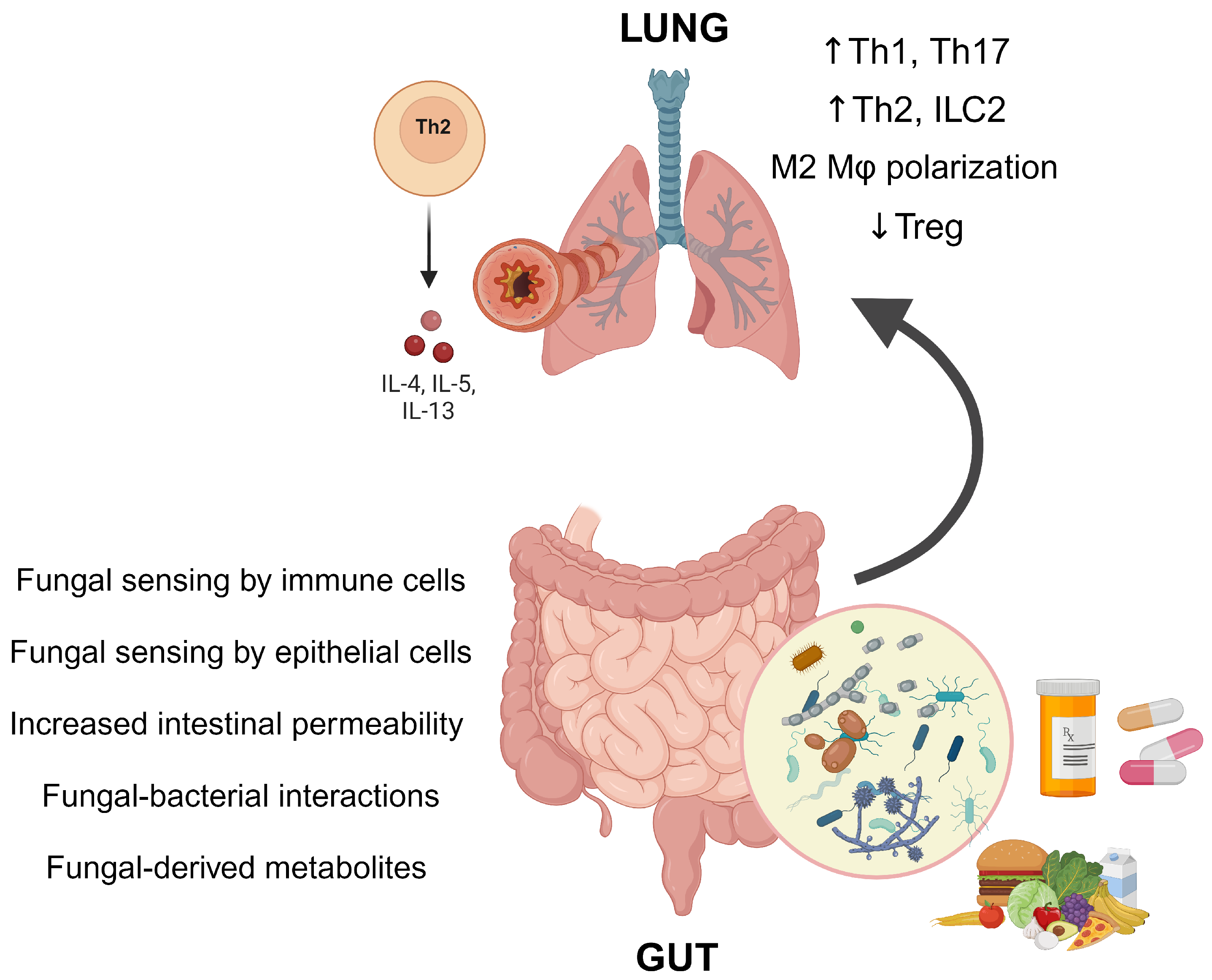
3. Mechanisms Linking the Gut Mycobiome and Asthma
4. Modulating the Gut Mycobiome in Asthma
5. Future Directions in Gut Mycobiome and Asthma Research
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Porsbjerg, C.; Melen, E.; Lehtimaki, L.; Shaw, D. Asthma. Lancet 2023, 401, 858–873. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut(-)Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef]
- Cait, A.; Hughes, M.R.; Antignano, F.; Cait, J.; Dimitriu, P.A.; Maas, K.R.; Reynolds, L.A.; Hacker, L.; Mohr, J.; Finlay, B.B.; et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018, 11, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Valverde-Molina, J.; Garcia-Marcos, L. Microbiome and Asthma: Microbial Dysbiosis and the Origins, Phenotypes, Persistence, and Severity of Asthma. Nutrients 2023, 15, 486. [Google Scholar] [CrossRef] [PubMed]
- Valero, A.; Quirce, S.; Davila, I.; Delgado, J.; Dominguez-Ortega, J. Allergic respiratory disease: Different allergens, different symptoms. Allergy 2017, 72, 1306–1316. [Google Scholar] [CrossRef]
- Wardlaw, A.J.; Rick, E.M.; Pur Ozyigit, L.; Scadding, A.; Gaillard, E.A.; Pashley, C.H. New Perspectives in the Diagnosis and Management of Allergic Fungal Airway Disease. J. Asthma Allergy 2021, 14, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Tsivileva, O.M.; Koftin, O.V.; Evseeva, N.V. Coumarins as Fungal Metabolites with Potential Medicinal Properties. Antibiotics 2022, 11, 1156. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Cai, Z.; Bartlam, M.; Wang, Y. Comparison of ITS and 18S rDNA for estimating fungal diversity using PCR-DGGE. World J. Microbiol. Biotechnol. 2015, 31, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Stiemsma, L.T.; Turvey, S.E. Asthma and the microbiome: Defining the critical window in early life. Allergy Asthma Clin. Immunol. 2017, 13, 3. [Google Scholar] [CrossRef]
- Alcazar, C.G.; Paes, V.M.; Shao, Y.; Oesser, C.; Miltz, A.; Lawley, T.D.; Brocklehurst, P.; Rodger, A.; Field, N. The association between early-life gut microbiota and childhood respiratory diseases: A systematic review. Lancet Microbe 2022, 3, e867–e880. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Levan, S.R.; Stamnes, K.A.; Lin, D.L.; Panzer, A.R.; Fukui, E.; McCauley, K.; Fujimura, K.E.; McKean, M.; Ownby, D.R.; Zoratti, E.M.; et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019, 4, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Arevalo, A.; Stiemsma, L.; Dimitriu, P.; Chico, M.E.; Loor, S.; Vaca, M.; Boutin, R.C.T.; Morien, E.; Jin, M.; et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018, 142, 424–434.e10. [Google Scholar] [CrossRef]
- Boutin, R.C.; Petersen, C.; Woodward, S.E.; Serapio-Palacios, A.; Bozorgmehr, T.; Loo, R.; Chalanuchpong, A.; Cirstea, M.; Lo, B.; Huus, K.E.; et al. Bacterial-fungal interactions in the neonatal gut influence asthma outcomes later in life. Elife 2021, 10, e67740. [Google Scholar] [CrossRef]
- Depner, M.; Taft, D.H.; Kirjavainen, P.V.; Kalanetra, K.M.; Karvonen, A.M.; Peschel, S.; Schmausser-Hechfellner, E.; Roduit, C.; Frei, R.; Lauener, R.; et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat. Med. 2020, 26, 1766–1775. [Google Scholar] [CrossRef]
- Zhang, F.; Aschenbrenner, D.; Yoo, J.Y.; Zuo, T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 2022, 3, e969–e983. [Google Scholar] [CrossRef]
- Huang, C.; Tang, W.; Dai, R.; Wang, P.; Shi, G.; Du, W.; Ni, Y. Disentangling the potential roles of the human gut mycobiome and metabolites in asthma. Clin. Transl. Med. 2022, 12, e1012. [Google Scholar] [CrossRef]
- Kanj, A.N.; Kottom, T.J.; Schaefbauer, K.J.; Choudhury, M.; Limper, A.H.; Skalski, J.H. Dysbiosis of the intestinal fungal microbiota increases lung resident group 2 innate lymphoid cells and is associated with enhanced asthma severity in mice and humans. Respir. Res. 2023, 24, 144. [Google Scholar] [CrossRef]
- Skalski, J.H.; Limon, J.J.; Sharma, P.; Gargus, M.D.; Nguyen, C.; Tang, J.; Coelho, A.L.; Hogaboam, C.M.; Crother, T.R.; Underhill, D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018, 14, e1007260. [Google Scholar] [CrossRef]
- Schei, K.; Simpson, M.R.; Oien, T.; Salamati, S.; Rudi, K.; Odegard, R.A. Allergy-related diseases and early gut fungal and bacterial microbiota abundances in children. Clin. Transl. Allergy 2021, 11, e12041. [Google Scholar] [CrossRef]
- Noverr, M.C.; Noggle, R.M.; Toews, G.B.; Huffnagle, G.B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 2004, 72, 4996–5003. [Google Scholar] [CrossRef]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 2020, 11, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Kanj, A.N.; Guiance, I.R.; Kottom, T.J.; Schaefbauer, K.J.; Choudhury, M.; Limper, A.H.; Skalski, J.H. The intestinal commensal fungus Wallemia mellicola enhances asthma in mice through Dectin-2. Med. Mycol. 2024, 62, myae004. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Udayanga, K.G.; Totsuka, N.; Weinberg, J.B.; Nunez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2). Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef]
- Yamada, T.; Nakashima, T.; Masuda, T.; Sakamoto, S.; Yamaguchi, K.; Horimasu, Y.; Miyamoto, S.; Iwamoto, H.; Fujitaka, K.; Hamada, H.; et al. Intestinal overgrowth of Candida albicans exacerbates bleomycin-induced pulmonary fibrosis in mice with dysbiosis. J. Pathol. 2023, 261, 227–237. [Google Scholar] [CrossRef]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 2018, 9, 1534. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, D.H.; Erofeev, A.; Sandvik, A.; Grcic, V.; Jahnsen, F.L.; Gaustad, P.; McCoy, K.D.; Macpherson, A.J.; Meza-Zepeda, L.A.; Johansen, F.E. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS ONE 2011, 6, e17996. [Google Scholar] [CrossRef]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Nials, A.T.; Uddin, S. Mouse models of allergic asthma: Acute and chronic allergen challenge. Dis. Model. Mech. 2008, 1, 213–220. [Google Scholar] [CrossRef]
- Foster, P.S.; Maltby, S.; Rosenberg, H.F.; Tay, H.L.; Hogan, S.P.; Collison, A.M.; Yang, M.; Kaiko, G.E.; Hansbro, P.M.; Kumar, R.K.; et al. Modeling T(H) 2 responses and airway inflammation to understand fundamental mechanisms regulating the pathogenesis of asthma. Immunol. Rev. 2017, 278, 20–40. [Google Scholar] [CrossRef]
- Kanj, A.; Kottom, T.; Choudhury, M.; Schaefbauer, K.; Limper, A.H.; Skalski, J. Dectin-2 Recognizes Gut Dysbiosis of the Commensal Fungi Wallemia mellicola and Is Essential for the Gut-Lung Axis Interactions Aggravating Asthma in Murine Models. Am. J. Respir. Crit. Care Med. 2022, 205, A5685. [Google Scholar]
- Robinson, M.J.; Osorio, F.; Rosas, M.; Freitas, R.P.; Schweighoffer, E.; Gross, O.; Verbeek, J.S.; Ruland, J.; Tybulewicz, V.; Brown, G.D.; et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J. Exp. Med. 2009, 206, 2037–2051. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Leonardi, I.; Semon, A.; Doron, I.; Gao, I.H.; Putzel, G.G.; Kim, Y.; Kabata, H.; Artis, D.; Fiers, W.D.; et al. Response to Fungal Dysbiosis by Gut-Resident CX3CR1(+) Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe 2018, 24, 847–856.e4. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, I.; Li, X.; Semon, A.; Li, D.; Doron, I.; Putzel, G.; Bar, A.; Prieto, D.; Rescigno, M.; McGovern, D.P.B.; et al. CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 2018, 359, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef]
- Zhang, B.; An, J.; Shimada, T.; Liu, S.; Maeyama, K. Oral administration of Enterococcus faecalis FK-23 suppresses Th17 cell development and attenuates allergic airway responses in mice. Int. J. Mol. Med. 2012, 30, 248–254. [Google Scholar] [CrossRef]
- Karimi, K.; Inman, M.D.; Bienenstock, J.; Forsythe, P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2009, 179, 186–193. [Google Scholar] [CrossRef]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.R.; Kragelund, C.; Jensen, P.O.; Keller, M.K.; Twetman, S. Probiotic Lactobacillus reuteri has antifungal effects on oral Candida species in vitro. J. Oral Microbiol. 2017, 9, 1274582. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Camara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Li, S.C.; Yeh, Y.M.; Lee, S.Y.; Kuo, H.C.; Yang, C.Y. Gut mycobiome dysbiosis and its impact on intestinal permeability in attention-deficit/hyperactivity disorder. J. Child. Psychol. Psychiatry 2023, 64, 1280–1291. [Google Scholar] [CrossRef] [PubMed]
- Benard, A.; Desreumeaux, P.; Huglo, D.; Hoorelbeke, A.; Tonnel, A.B.; Wallaert, B. Increased intestinal permeability in bronchial asthma. J. Allergy Clin. Immunol. 1996, 97, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, Z.; Molla, A.M.; Al-Habashi, H.; Muawad, W.M.; Molla, A.M.; Sharma, P.N. Intestinal permeability is increased in bronchial asthma. Arch. Dis. Child. 2004, 89, 227–229. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martin, R.; Bermudez-Humaran, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; Tian, G.; et al. The fungal community and its interaction with the concentration of short-chain fatty acids in the faeces of Chenghua, Yorkshire and Tibetan pigs. Microb. Biotechnol. 2020, 13, 509–521. [Google Scholar] [CrossRef]
- Kita, H. ILC2s and fungal allergy. Allergol. Int. 2015, 64, 219–226. [Google Scholar] [CrossRef]
- Rapeport, W.G.; Ito, K.; Denning, D.W. The role of antifungals in the management of patients with severe asthma. Clin. Transl. Allergy 2020, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Moss, R.B. Treatment options in severe fungal asthma and allergic bronchopulmonary aspergillosis. Eur. Respir. J. 2014, 43, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Tiwary, M.; Samarasinghe, A.E. Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization. Cells 2021, 10, 913. [Google Scholar] [CrossRef] [PubMed]
- van Tilburg Bernardes, E.; Pettersen, V.K.; Gutierrez, M.W.; Laforest-Lapointe, I.; Jendzjowsky, N.G.; Cavin, J.B.; Vicentini, F.A.; Keenan, C.M.; Ramay, H.R.; Samara, J.; et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat. Commun. 2020, 11, 2577. [Google Scholar] [CrossRef] [PubMed]
- Zwolinska-Wcislo, M.; Brzozowski, T.; Budak, A.; Kwiecien, S.; Sliwowski, Z.; Drozdowicz, D.; Trojanowska, D.; Rudnicka-Sosin, L.; Mach, T.; Konturek, S.J.; et al. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa. J. Physiol. Pharmacol. 2009, 60, 107–118. [Google Scholar] [PubMed]
- Turunen, J.; Paalanne, N.; Reunanen, J.; Tapiainen, T.; Tejesvi, M.V. Development of gut mycobiome in infants and young children: A prospective cohort study. Pediatr. Res. 2023, 94, 486–494. [Google Scholar] [CrossRef]
- Tejesvi, M.V.; Turunen, J.; Salmi, S.; Reunanen, J.; Paalanne, N.; Tapiainen, T. Delivery Mode and Perinatal Antibiotics Influence the Infant Gut Bacteriome and Mycobiome: A Network Analysis. J. Fungi 2023, 9, 718. [Google Scholar] [CrossRef]
- Nakajima, A.; Saraya, T.; Mori, T.; Ikeda, R.; Sugita, T.; Watanabe, T.; Fujiwara, M.; Takizawa, H.; Goto, H. Familial summer-type hypersensitivity pneumonitis in Japan: Two case reports and review of the literature. BMC Res. Notes 2013, 6, 371. [Google Scholar] [CrossRef][Green Version]
- Ventin-Holmberg, R.; Saqib, S.; Korpela, K.; Nikkonen, A.; Peltola, V.; Salonen, A.; de Vos, W.M.; Kolho, K.L. The Effect of Antibiotics on the Infant Gut Fungal Microbiota. J. Fungi 2022, 8, 328. [Google Scholar] [CrossRef]
- Patrick, D.M.; Sbihi, H.; Dai, D.L.Y.; Al Mamun, A.; Rasali, D.; Rose, C.; Marra, F.; Boutin, R.C.T.; Petersen, C.; Stiemsma, L.T.; et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: Evidence from population-based and prospective cohort studies. Lancet Respir. Med. 2020, 8, 1094–1105. [Google Scholar] [CrossRef]
- Delavy, M.; Burdet, C.; Sertour, N.; Devente, S.; Docquier, J.D.; Grall, N.; Volant, S.; Ghozlane, A.; Duval, X.; Mentre, F.; et al. A Clinical Study Provides the First Direct Evidence That Interindividual Variations in Fecal beta-Lactamase Activity Affect the Gut Mycobiota Dynamics in Response to beta-Lactam Antibiotics. mBio 2022, 13, e0288022. [Google Scholar] [CrossRef] [PubMed]
- Ciprandi, G.; Tosca, M.A.; Drago, L. Probiotics in asthma management: Fiction or truth? Expert. Rev. Clin. Immunol. 2023, 19, 457–460. [Google Scholar] [CrossRef]
- Drago, L.; Cioffi, L.; Giuliano, M.; Pane, M.; Amoruso, A.; Schiavetti, I.; Reid, G.; Ciprandi, G.; Propam Study, G. The Probiotics in Pediatric Asthma Management (PROPAM) Study in the Primary Care Setting: A Randomized, Controlled, Double-Blind Trial with Ligilactobacillus salivarius LS01 (DSM 22775) and Bifidobacterium breve B632 (DSM 24706). J. Immunol. Res. 2022, 2022, 3837418. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Jiang, P.; Liu, J.; Sun, R.; Zhu, L. Association between probiotic supplementation and asthma incidence in infants: A meta-analysis of randomized controlled trials. J. Asthma 2020, 57, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhang, Y.; He, C.; Dai, J. Probiotics supplementation in children with asthma: A systematic review and meta-analysis. J. Paediatr. Child. Health 2018, 54, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Ma, T.; Xu, N.; Jin, H.; Zhao, F.; Kwok, L.Y.; Zhang, H.; Zhang, S.; Sun, Z. Adjunctive Probiotics Alleviates Asthmatic Symptoms via Modulating the Gut Microbiome and Serum Metabolome. Microbiol. Spectr. 2021, 9, e0085921. [Google Scholar] [CrossRef] [PubMed]
- Sadrifar, S.; Abbasi-Dokht, T.; Forouzandeh, S.; Malek, F.; Yousefi, B.; Salek Farrokhi, A.; Karami, J.; Baharlou, R. Immunomodulatory effects of probiotic supplementation in patients with asthma: A randomized, double-blind, placebo-controlled trial. Allergy Asthma Clin. Immunol. 2023, 19, 1. [Google Scholar] [CrossRef]
- Fonseca, V.M.B.; Milani, T.M.S.; Prado, R.; Bonato, V.L.D.; Ramos, S.G.; Martins, F.S.; Vianna, E.O.; Borges, M.C. Oral administration of Saccharomyces cerevisiae UFMG A-905 prevents allergic asthma in mice. Respirology 2017, 22, 905–912. [Google Scholar] [CrossRef]
- Milani, T.M.S.; Sandy, C.M.; Calazans, A.; Silva, R.Q.; Fonseca, V.M.B.; Martins, F.S.; Borges, M.C. Dose-Response Effect of Saccharomyces cerevisiae UFMG A-905 on the Prevention of Asthma in an Animal Model. Probiotics Antimicrob. Proteins 2024, 16, 53–61. [Google Scholar] [CrossRef]
- Kang, Y.; Cai, Y. Future prospect of faecal microbiota transplantation as a potential therapy in asthma. Allergol. Immunopathol. 2018, 46, 307–309. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Scholl, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef]
- Singh, R.P.; Bhardwaj, A. Beta-glucans: A potential source for maintaining gut microbiota and the immune system. Front. Nutr. 2023, 10, 1143682. [Google Scholar] [CrossRef]
- Sarinho, E.; Medeiros, D.; Schor, D.; Rego Silva, A.; Sales, V.; Motta, M.E.; Costa, A.; Azoubel, A.; Rizzo, J.A. Production of interleukin-10 in asthmatic children after Beta-1-3-glucan. Allergol. Immunopathol. 2009, 37, 188–192. [Google Scholar] [CrossRef]
- Burg, A.R.; Quigley, L.; Jones, A.V.; O’Connor, G.M.; Boelte, K.; McVicar, D.W.; Orr, S.J. Orally administered beta-glucan attenuates the Th2 response in a model of airway hypersensitivity. Springerplus 2016, 5, 815. [Google Scholar] [CrossRef]
- Talbott, S.M.; Talbott, J.A.; Talbott, T.L.; Dingler, E. Beta-Glucan supplementation, allergy symptoms, and quality of life in self-described ragweed allergy sufferers. Food Sci. Nutr. 2013, 1, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Biagini Myers, J.M.; Brandt, E.B.; Ryan, P.H.; Lindsey, M.; Mintz-Cole, R.A.; Reponen, T.; Vesper, S.J.; Forde, F.; Ruff, B.; et al. Beta-Glucan exacerbates allergic asthma independent of fungal sensitization and promotes steroid-resistant T(H)2/T(H)17 responses. J. Allergy Clin. Immunol. 2017, 139, 54–65.e8. [Google Scholar] [CrossRef]
- Hoving, L.R.; Katiraei, S.; Heijink, M.; Pronk, A.; van der Wee-Pals, L.; Streefland, T.; Giera, M.; Willems van Dijk, K.; van Harmelen, V. Dietary Mannan Oligosaccharides Modulate Gut Microbiota, Increase Fecal Bile Acid Excretion, and Decrease Plasma Cholesterol and Atherosclerosis Development. Mol. Nutr. Food Res. 2018, 62, e1700942. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.F.; Waters, C.M.; LeMessurier, K.S.; Samarasinghe, A.E.; Song, C.Y.; Malik, K.U.; Lew, D.B. Airway Epithelial Repair by a Prebiotic Mannan Derived from Saccharomyces cerevisiae. J. Immunol. Res. 2017, 2017, 8903982. [Google Scholar] [CrossRef] [PubMed]
- Soria, I.; Lopez-Relano, J.; Vinuela, M.; Tudela, J.I.; Angelina, A.; Benito-Villalvilla, C.; Diez-Rivero, C.M.; Cases, B.; Manzano, A.I.; Fernandez-Caldas, E.; et al. Oral myeloid cells uptake allergoids coupled to mannan driving Th1/Treg responses upon sublingual delivery in mice. Allergy 2018, 73, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Toriumi, H.; Takahashi, D.; Kamasaki, T.; Fujioka, Y.; Nagatoishi, S.; Li, J.; Liu, Y.; Hosokawa, T.; Tsumoto, K.; et al. Safe and efficient oral allergy immunotherapy using one-pot-prepared mannan-coated allergen nanoparticles. Biomaterials 2023, 303, 122381. [Google Scholar] [CrossRef] [PubMed]
- Lew, D.B.; LeMessurier, K.S.; Palipane, M.; Lin, Y.; Samarasinghe, A.E. Saccharomyces cerevisiae-Derived Mannan Does Not Alter Immune Responses to Aspergillus Allergens. Biomed. Res. Int. 2018, 2018, 3298378. [Google Scholar] [CrossRef]
- Laue, H.E.; Coker, M.O.; Madan, J.C. The Developing Microbiome from Birth to 3 Years: The Gut-Brain Axis and Neurodevelopmental Outcomes. Front. Pediatr. 2022, 10, 815885. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- El Aidy, S.; Hooiveld, G.; Tremaroli, V.; Backhed, F.; Kleerebezem, M. The gut microbiota and mucosal homeostasis: Colonized at birth or at adulthood, does it matter? Gut Microbes 2013, 4, 118–124. [Google Scholar] [CrossRef]
- Sun, Y.; Zuo, T.; Cheung, C.P.; Gu, W.; Wan, Y.; Zhang, F.; Chen, N.; Zhan, H.; Yeoh, Y.K.; Niu, J.; et al. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology 2021, 160, 272–286.e11. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Boonyuen, N.; Ranaweera, C.B.; de Zoysa, H.K.S.; Padmathilake, R.E.; Nifla, F.; Dai, D.Q.; Liu, Y.; Suwannarach, N.; Kumla, J.; et al. OMICS and Other Advanced Technologies in Mycological Applications. J. Fungi 2023, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Mac Aogain, M.; Narayana, J.K.; Tiew, P.Y.; Ali, N.; Yong, V.F.L.; Jaggi, T.K.; Lim, A.Y.H.; Keir, H.R.; Dicker, A.J.; Thng, K.X.; et al. Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 2021, 27, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.L.; Chen, Z.; Shankar, V.; Tyberg, M.; Vicencio, A.; Burk, R. Lower airway microbiota and mycobiota in children with severe asthma. J. Allergy Clin. Immunol. 2018, 141, 808–811.e7. [Google Scholar] [CrossRef] [PubMed]
- Vargas, S.L.; Ponce, C.A.; Gallo, M.; Perez, F.; Astorga, J.F.; Bustamante, R.; Chabe, M.; Durand-Joly, I.; Iturra, P.; Miller, R.F.; et al. Near-universal prevalence of Pneumocystis and associated increase in mucus in the lungs of infants with sudden unexpected death. Clin. Infect. Dis. 2013, 56, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Eddens, T.; Campfield, B.T.; Serody, K.; Manni, M.L.; Horne, W.; Elsegeiny, W.; McHugh, K.J.; Pociask, D.; Chen, K.; Zheng, M.; et al. A Novel CD4(+) T Cell-Dependent Murine Model of Pneumocystis-driven Asthma-like Pathology. Am. J. Respir. Crit. Care Med. 2016, 194, 807–820. [Google Scholar] [CrossRef] [PubMed]
- van Woerden, H.C.; Gregory, C.; Brown, R.; Marchesi, J.R.; Hoogendoorn, B.; Matthews, I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: A community based case control study. BMC Infect. Dis. 2013, 13, 69. [Google Scholar] [CrossRef]
- Sharma, A.; Laxman, B.; Naureckas, E.T.; Hogarth, D.K.; Sperling, A.I.; Solway, J.; Ober, C.; Gilbert, J.A.; White, S.R. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J. Allergy Clin. Immunol. 2019, 144, 1214–1227.e7. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, M.G.; Chishimba, L.; Niven, R.M.; Bromley, M.; Simpson, A.; Smyth, L.; Denning, D.W.; Bowyer, P. Corticosteroid treatment is associated with increased filamentous fungal burden in allergic fungal disease. J. Allergy Clin. Immunol. 2018, 142, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Narayana, J.K.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Microbiomics-focused Data Integration: A Fresh Solve for the Rubik’s Cube of Endophenotyping? Am. J. Respir. Crit. Care Med. 2022, 206, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Leal Rodriguez, C.; Shah, S.A.; Rasmussen, M.A.; Thorsen, J.; Boulund, U.; Pedersen, C.T.; Castro-Mejia, J.L.; Poulsen, C.E.; Poulsen, C.S.; Deng, L.; et al. The infant gut virome is associated with preschool asthma risk independently of bacteria. Nat. Med. 2024, 30, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.J.M.; Mommers, M.; Penders, J.; Arts, I.C.W.; Thijs, C. Intestinal archaea inversely associated with childhood asthma. J. Allergy Clin. Immunol. 2019, 143, 2305–2307. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.; Matricardi, P.M. Worms, asthma, and the hygiene hypothesis. Lancet 2006, 367, 1556–1558. [Google Scholar] [CrossRef] [PubMed]
- Alcantara-Neves, N.M.; de SG Britto, G.; Veiga, R.V.; Figueiredo, C.A.; Fiaccone, R.L.; da Conceicao, J.S.; Cruz, A.A.; Rodrigues, L.C.; Cooper, P.J.; Pontes-de-Carvalho, L.C.; et al. Effects of helminth co-infections on atopy, asthma and cytokine production in children living in a poor urban area in Latin America. BMC Res. Notes 2014, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- Senaratna, C.V.; Perera, P.K.; Arulkumaran, S.; Abeysekara, N.; Piyumanthi, P.; Hamilton, G.S.; Nixon, G.M.; Rajakaruna, R.S.; Dharmage, S.C. Association of helminth infestation with childhood asthma: A nested case-control study. Int. J. Infect. Dis. 2023, 128, 272–277. [Google Scholar] [CrossRef]
- Barelli, C.; Donati, C.; Albanese, D.; Pafco, B.; Modry, D.; Rovero, F.; Hauffe, H.C. Interactions between parasitic helminths and gut microbiota in wild tropical primates from intact and fragmented habitats. Sci. Rep. 2021, 11, 21569. [Google Scholar] [CrossRef]
- Ray, A.; Das, J.; Wenzel, S.E. Determining asthma endotypes and outcomes: Complementing existing clinical practice with modern machine learning. Cell Rep. Med. 2022, 3, 100857. [Google Scholar] [CrossRef] [PubMed]
- Georas, S.N.; Wright, R.J.; Ivanova, A.; Israel, E.; LaVange, L.M.; Akuthota, P.; Carr, T.F.; Denlinger, L.C.; Fajt, M.L.; Kumar, R.; et al. The Precision Interventions for Severe and/or Exacerbation-Prone (PrecISE) Asthma Network: An overview of Network Organization, Procedures, and Interventions. J. Allergy Clin. Immunol. 2022, 149, 488–516.e9. [Google Scholar] [CrossRef] [PubMed]
- Maspero, J.; Adir, Y.; Al-Ahmad, M.; Celis-Preciado, C.A.; Colodenco, F.D.; Giavina-Bianchi, P.; Lababidi, H.; Ledanois, O.; Mahoub, B.; Perng, D.W.; et al. Type 2 inflammation in asthma and other airway diseases. ERJ Open Res. 2022, 8, 00576-2021. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Brightling, C. Bronchiectasis, the Latest Eosinophilic Airway Disease: What about the Microbiome? Am. J. Respir. Crit. Care Med. 2022, 205, 860–862. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]

| Study | Region | Design | Methods | Outcome | Gut Mycobiome Associated with Outcome |
|---|---|---|---|---|---|
| Pediatrics | |||||
| Fujimura et al., 2016 [15] | Detroit, MI, United States | Cohort (n = 298 with available stool samples) | Stool sample collected at age 1–11 months. ITS2 gene amplifications | Parental report of physician-diagnosed asthma at 4 years | Increased: Candida, Rhodotorula, Debaryomyces, Meyerozyma, Nigrospora, Saccharomyces, Pyrenochaetopsis, Phanerochaete Decreased: Malassezia |
| Arrieta et al., 2018 [17] | Rural Ecuador | Case-control (n = 27 with atopic wheeze; n = 70 healthy controls) | Stool sample collected at age 3 months. 18S gene amplification | Maternally reported atopic wheeze at 5 years | Increased: Pichia kudriavzevii Decreased: Saccharomyces and Saccharomycetales |
| Depner et al., 2020 [19] | Rural Austria, Finland, France, Germany, and Switzerland | Cohort (n = 930) | Stool sample collected at age 2–12 months. ITS1 gene amplifications | Physician-diagnosed asthma OR parental reports of bronchitis at 6 years | Decreased: Alternaria (at 2 months) Bacterial but not fungal maturation is protective for asthma. Alternaria at 2 months was associated with bacterial maturation |
| Schei et al., 2021 [24] | Norway | Secondary data analysis of RCT (n = 180) | Stool sample collected at four time points between age 0–24 months. ITS1 gene amplifications | Parental report of physician-diagnosed asthma at 6 years | Increased: Fungal abundance at 2 years |
| Adults | |||||
| Huang et al., 2022 [21] | Shanghai, China | Case-control (n = 26 with asthma on ICS; n = 12 with asthma not on ICS; n = 21 healthy controls) | Stool sample collected at time of interview. ITS gene amplifications | Asthma on ICS vs. healthy controls | Increased: Russula, Sebacina, Nectria, and Wallemia in those with asthma on ICS |
| Kanj et al., 2023 [22] | United States | Cross sectional (n = 24 with physician-diagnosed asthma) | Stool sample collected at time of interview. Candida gene amplifications | Severe asthma exacerbation in the past year | Increased: Candida to bacterial ratio |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanj, A.N.; Skalski, J.H. Gut Mycobiome and Asthma. J. Fungi 2024, 10, 192. https://doi.org/10.3390/jof10030192
Kanj AN, Skalski JH. Gut Mycobiome and Asthma. Journal of Fungi. 2024; 10(3):192. https://doi.org/10.3390/jof10030192
Chicago/Turabian StyleKanj, Amjad N., and Joseph H. Skalski. 2024. "Gut Mycobiome and Asthma" Journal of Fungi 10, no. 3: 192. https://doi.org/10.3390/jof10030192
APA StyleKanj, A. N., & Skalski, J. H. (2024). Gut Mycobiome and Asthma. Journal of Fungi, 10(3), 192. https://doi.org/10.3390/jof10030192






