Kazachstania pintolopesii in Blood and Intestinal Wall of Macrophage-Depleted Mice with Cecal Ligation and Puncture, the Control of Fungi by Macrophages during Sepsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Animal Model
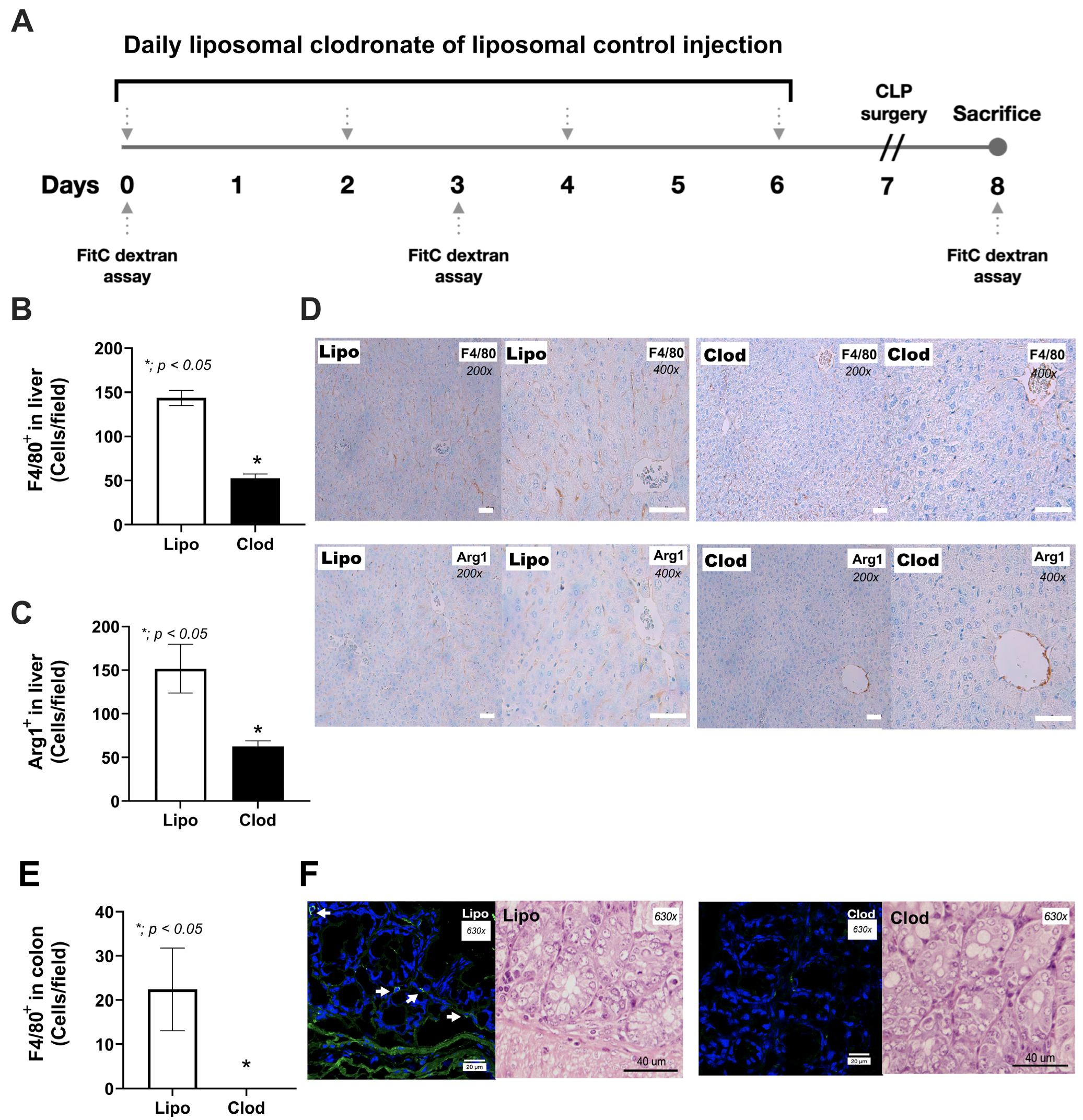
2.2. Macrophage Depletion and Sepsis Mouse Model
2.3. Detection of Bacteremia and Fungemia
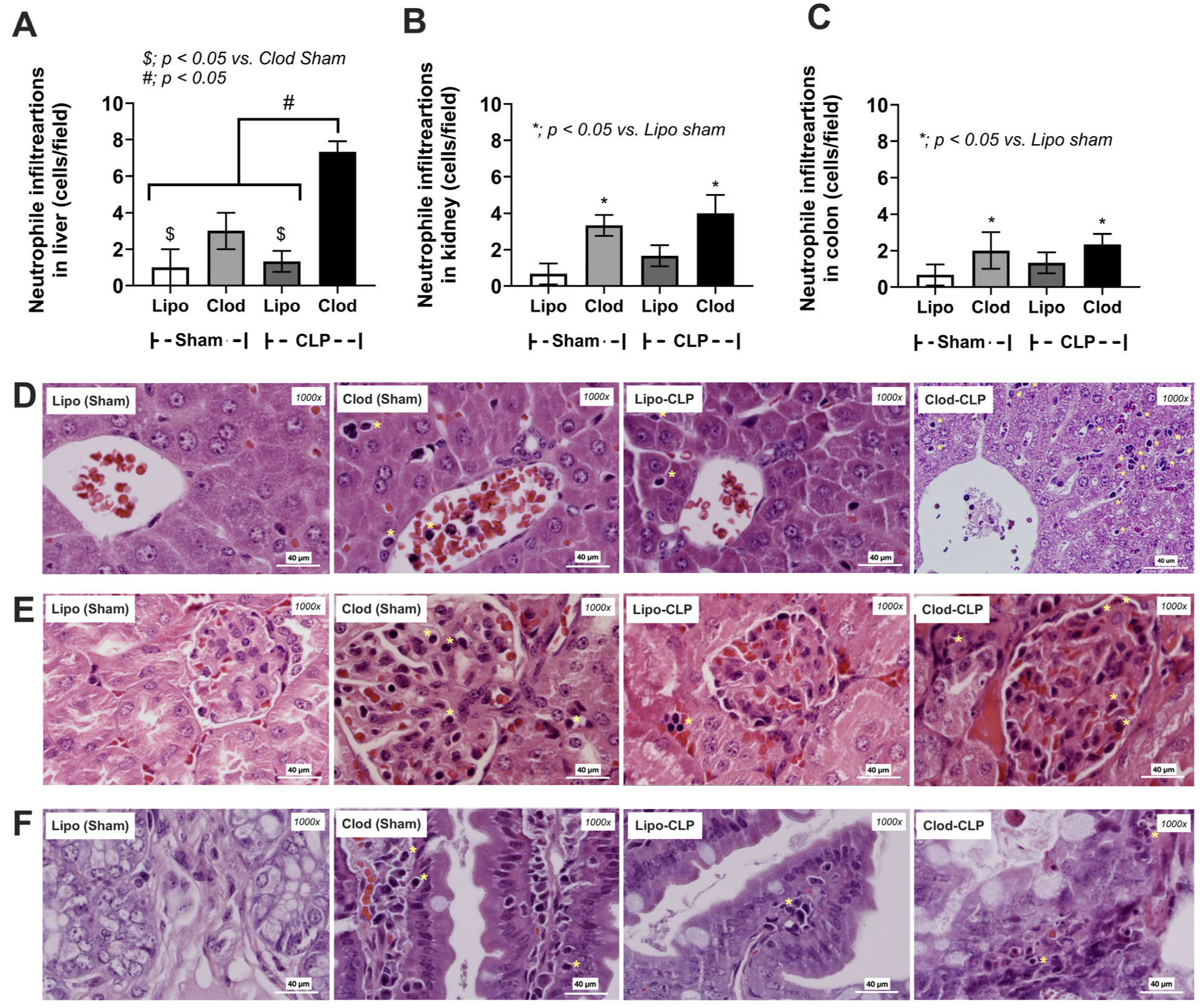
2.4. Histological Analysis
2.5. Mouse Serum Analysis and Intestinal Permeability Test (Leaky Gut)
2.6. The Fungal Burden in Colon Tissue
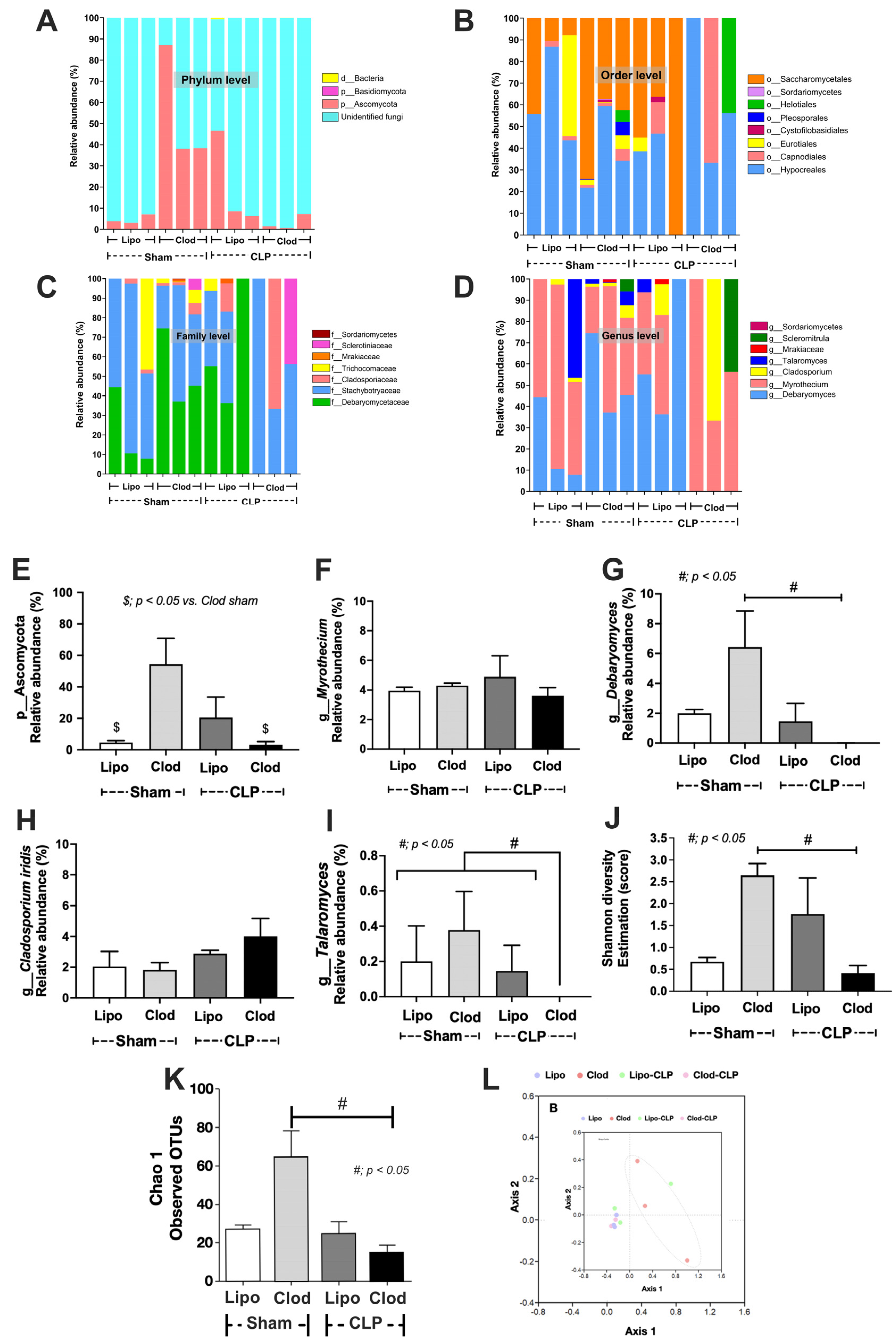
2.7. Gut Mycobiome Analysis
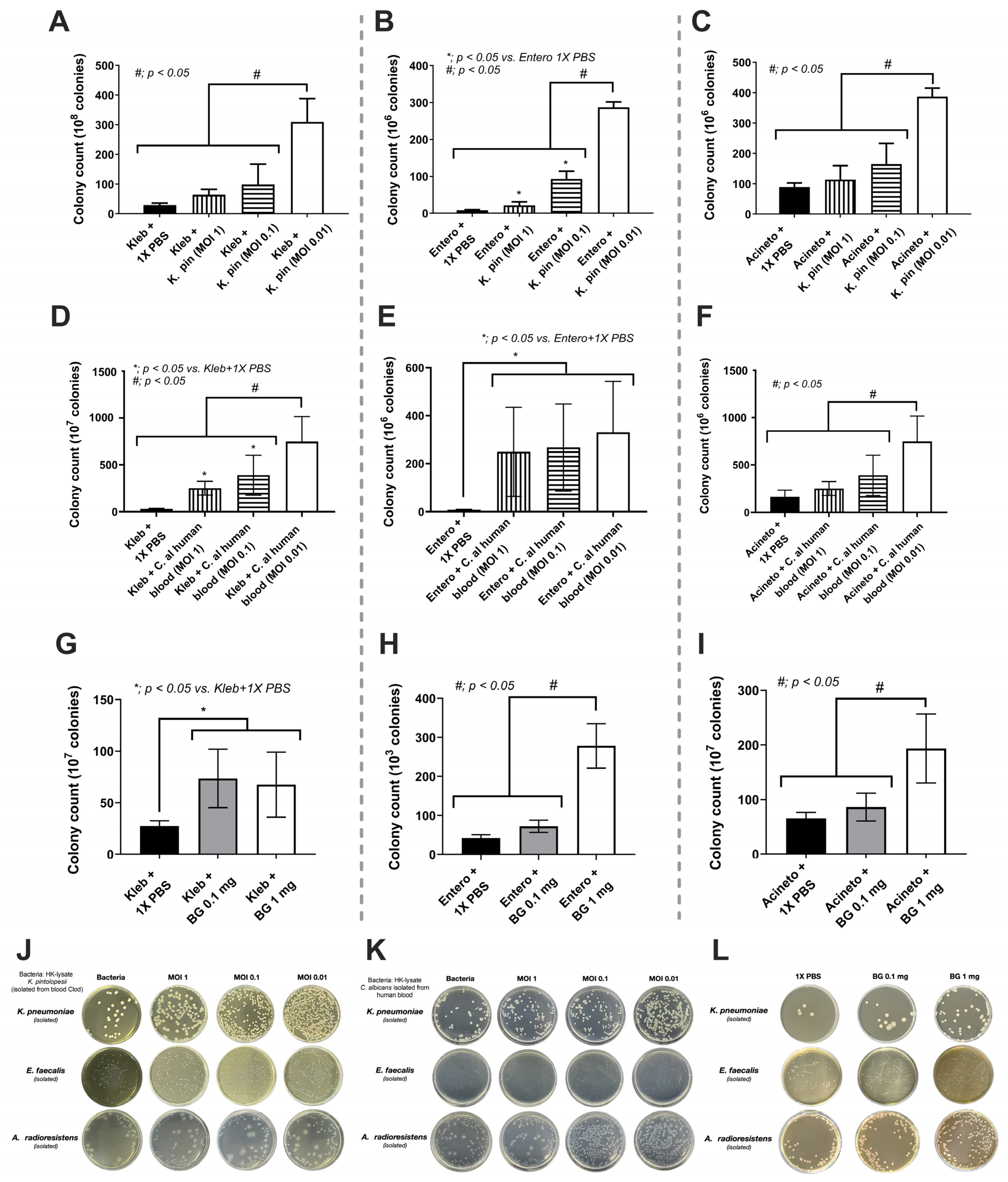
2.8. The In Vitro Experiments
2.9. Statistical Analysis
3. Results
3.1. Spontaneous Bacteremia and Fungemia after Macrophage Depletion without Sepsis, the Leaky Gut-Induced Systemic Inflammation from the Loss of Macrophage Microbial Control
3.2. More Severe Sepsis in Macrophage-Depleted Mice with Cecal Ligation and Puncture Surgery, a Possible Impact of Leaky Gut and Fungi in the Intestinal Tissue
3.3. The Overgrowth of Bacteria due to the Fungi and the Mixed Microbial Activation of Enterocytes, the Possible Influence of Gut Fungi on Sepsis Severity
4. Discussion
4.1. Macrophage Depletion Facilitated the Growth of Gut Fungi
4.2. K. pintolopesii as a Source of Fungal Molecules Influenced Intestinal Bacteria and Enterocytes
4.3. The Discrepancy between Fecal Mycobiome Analysis versus Fecal Fungi from Polymerase Chain Reaction (PCR), a Possible Role of the Internal Transcribed Spacer (ITS) Primers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sae-khow, K.; Charoensappakit, A.; Chiewchengchol, D.; Leelahavanichkul, A. High-Dose Intravenous Ascorbate in Sepsis, a Pro-Oxidant Enhanced Microbicidal Activity and the Effect on Neutrophil Functions. Biomedicines 2023, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Charoensappakit, A.; Sae-Khow, K.; Leelahavanichkul, A. Gut Barrier Damage and Gut Translocation of Pathogen Molecules in Lupus, an Impact of Innate Immunity (Macrophages and Neutrophils) in Autoimmune Disease. Int. J. Mol. Sci. 2022, 23, 8223. [Google Scholar] [CrossRef] [PubMed]
- Boulange, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, B.; Ramakrishna, K.; Dhamoon, A.S. Sepsis: The evolution in definition, pathophysiology, and management. SAGE Open Med. 2019, 7, 2050312119835043. [Google Scholar] [CrossRef] [PubMed]
- Dolin, H.H.; Papadimos, T.J.; Chen, X.; Pan, Z.K. Characterization of Pathogenic Sepsis Etiologies and Patient Profiles: A Novel Approach to Triage and Treatment. Microbiol. Insights 2019, 12, 1178636118825081. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Worasilchai, N.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Gastrointestinal Colonization of Candida Albicans Increases Serum (1→3)-β-D-Glucan, without Candidemia, and Worsens Cecal Ligation and Puncture Sepsis in Murine Model. Shock 2018, 49, 62–70. [Google Scholar] [CrossRef]
- Lu, R.; Zhang, Y.G.; Xia, Y.; Sun, J. Imbalance of autophagy and apoptosis in intestinal epithelium lacking the vitamin D receptor. FASEB J. 2019, 33, 11845–11856. [Google Scholar] [CrossRef]
- Lin, M.Y.; Carmeli, Y.; Zumsteg, J.; Flores, E.L.; Tolentino, J.; Sreeramoju, P.; Weber, S.G. Prior antimicrobial therapy and risk for hospital-acquired Candida glabrata and Candida krusei fungemia: A case-case-control study. Antimicrob. Agents Chemother. 2005, 49, 4555–4560. [Google Scholar] [CrossRef]
- Lavin, Y.; Mortha, A.; Rahman, A.; Merad, M. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015, 15, 731–744. [Google Scholar] [CrossRef]
- Li, X.; Mu, G.; Song, C.; Zhou, L.; He, L.; Jin, Q.; Lu, Z. Role of M2 Macrophages in Sepsis-Induced Acute Kidney Injury. Shock 2018, 50, 233–239. [Google Scholar] [CrossRef]
- McWhorter, F.Y.; Wang, T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef] [PubMed]
- Xing, R.L.; Zhao, L.R.; Wang, P.M. Bisphosphonates therapy for osteoarthritis: A meta-analysis of randomized controlled trials. Springerplus 2016, 5, 1704. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saviola, G.; Abdi-Ali, L.; Povino, M.R.; Campostrini, L.; Sacco, S.; Dalle Carbonare, L.; Carbonare, L.D. Intramuscular clodronate in erosive osteoarthritis of the hand is effective on pain and reduces serum COMP: A randomized pilot trial-The ER.O.D.E. study (ERosive Osteoarthritis and Disodium-clodronate Evaluation). Clin. Rheumatol. 2017, 36, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Hurst, M.; Noble, S. Clodronate: A review of its use in breast cancer. Drugs Aging 1999, 15, 143–167. [Google Scholar] [CrossRef] [PubMed]
- Zeisberger, S.M.; Odermatt, B.; Marty, C.; Zehnder-Fjällman, A.H.M.; Ballmer-Hofer, K.; Schwendener, R.A. Clodronate-liposome-mediated depletion of tumour-associated macrophages: A new and highly effective antiangiogenic therapy approach. Br. J. Cancer 2006, 95, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Surawut, S.; Ondee, T.; Taratummarat, S.; Palaga, T.; Pisitkun, P.; Chindamporn, A.; Leelahavanichkul, A. The role of macrophages in the susceptibility of Fc gamma receptor IIb deficient mice to Cryptococcus neoformans. Sci. Rep. 2017, 7, 40006. [Google Scholar] [CrossRef] [PubMed]
- Epstein-Barash, H.; Gutman, D.; Markovsky, E.; Mishan-Eisenberg, G.; Koroukhov, N.; Szebeni, J.; Golomb, G. Physicochemical parameters affecting liposomal bisphosphonates bioactivity for restenosis therapy: Internalization, cell inhibition, activation of cytokines and complement, and mechanism of cell death. J. Control Release 2010, 146, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Chono, S.; Tanino, T.; Seki, T.; Morimoto, K. Influence of particle size on drug delivery to rat alveolar macrophages following pulmonary administration of ciprofloxacin incorporated into liposomes. J. Drug Target. 2006, 14, 557–566. [Google Scholar] [CrossRef]
- Recenti, R.; Leone, G.; Simi, L.; Orfei, M.; Pinzani, P.; Pieraccini, G.; Moneti, G.; Carossino, A.M.; Franchi, A.; Bartolucci, G.; et al. Clodronate acts on human osteoclastic cell proliferation, differentiation and function in a bioreversible manner. Clin. Cases Miner. Bone Metab. 2007, 4, 146–155. [Google Scholar]
- Ravichandran, S.; Manickam, N.; Kandasamy, M. Liposome encapsulated clodronate mediated elimination of pathogenic macrophages and microglia: A promising pharmacological regime to defuse cytokine storm in COVID-19. Med. Drug Discov. 2022, 15, 100136. [Google Scholar] [CrossRef]
- Smith, P.D.; Smythies, L.E.; Shen, R.; Greenwell-Wild, T.; Gliozzi, M.; Wahl, S.M. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011, 4, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Hiengrach, P.; Panpetch, W.; Chindamporn, A.; Leelahavanichkul, A. Macrophage depletion alters bacterial gut microbiota partly through fungal overgrowth in feces that worsens cecal ligation and puncture sepsis mice. Sci. Rep. 2022, 12, 9345. [Google Scholar] [CrossRef] [PubMed]
- Angele, M.K.; Pratschke, S.; Hubbard, W.J.; Chaudry, I.H. Gender differences in sepsis: Cardiovascular and immunological aspects. Virulence 2014, 5, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Weisser, S.B.; van Rooijen, N.; Sly, L.M. Depletion and reconstitution of macrophages in mice. J. Vis. Exp. 2012, 66, e4105. [Google Scholar] [CrossRef]
- Phuengmaung, P.; Khiewkamrop, P.; Makjaroen, J.; Issara-Amphorn, J.; Boonmee, A.; Benjaskulluecha, S.; Ritprajak, P.; Nita-Lazar, A.; Palaga, T.; Hirankarn, N.; et al. Less Severe Sepsis in Cecal Ligation and Puncture Models with and without Lipopolysaccharide in Mice with Conditional Ezh2-Deleted Macrophages (LysM-Cre System). Int. J. Mol. Sci. 2023, 24, 8517. [Google Scholar] [CrossRef]
- Hiengrach, P.; Visitchanakun, P.; Tongchairawewat, P.; Tangsirisatian, P.; Jungteerapanich, T.; Ritprajak, P.; Wannigama, D.L.; Tangtanatakul, P.; Leelahavanichkul, A. Sepsis Encephalopathy Is Partly Mediated by miR370-3p-Induced Mitochondrial Injury but Attenuated by BAM15 in Cecal Ligation and Puncture Sepsis Male Mice. Int. J. Mol. Sci. 2022, 23, 5445. [Google Scholar] [CrossRef]
- Makjaroen, J.; Thim-Uam, A.; Dang, C.P.; Pisitkun, T.; Somparn, P.; Leelahavanichkul, A. A Comparison Between 1 Day versus 7 Days of Sepsis in Mice with the Experiments on LPS-Activated Macrophages Support the Use of Intravenous Immunoglobulin for Sepsis Attenuation. J. Inflamm. Res. 2021, 14, 7243–7263. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006, 2006, pdb-prot4455. [Google Scholar] [CrossRef]
- Taratummarat, S.; Sangphech, N.; Vu, C.T.B.; Palaga, T.; Ondee, T.; Surawut, S.; Sereemaspun, A.; Ritprajak, P.; Leelahavanichkul, A. Gold nanoparticles attenuates bacterial sepsis in cecal ligation and puncture mouse model through the induction of M2 macrophage polarization. BMC Microbiol. 2018, 18, 85. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Somparn, P.; Panich, T.; Chancharoenthana, W.; Wongphom, J.; Pisitkun, T.; Hirankarn, N.; Eiam-Ong, S. Serum miRNA-122 in acute liver injury induced by kidney injury and sepsis in CD-1 mouse models. Hepatol. Res. 2015, 45, 1341–1352. [Google Scholar] [CrossRef]
- Saithong, S.; Saisorn, W.; Visitchanakun, P.; Sae-Khow, K.; Chiewchengchol, D.; Leelahavanichkul, A. A Synergy Between Endotoxin and (1→3)-Beta-D-Glucan Enhanced Neutrophil Extracellular Traps in Candida Administered Dextran Sulfate Solution Induced Colitis in FcGRIIB-/- Lupus Mice, an Impact of Intestinal Fungi in Lupus. J. Inflamm. Res. 2021, 14, 2333–2352. [Google Scholar] [CrossRef] [PubMed]
- Saejong, S.; Townamchai, N.; Somparn, P.; Tangtanatakul, P.; Ondee, T.; Hirankarn, N.; Leelahavanichkul, A. MicroRNA-21 in plasma exosome, but not from whole plasma, as a biomarker for the severe interstitial fibrosis and tubular atrophy (IF/TA) in post-renal transplantation. Asian Pac. J. Allergy Immunol. 2022, 40, 94–102. [Google Scholar] [PubMed]
- Liu, Z.; Li, N.; Fang, H.; Chen, X.; Guo, Y.; Gong, S.; Niu, M.; Zhou, H.; Jiang, Y.; Chang, P.; et al. Enteric dysbiosis is associated with sepsis in patients. FASEB J. 2019, 33, 12299–12310. [Google Scholar] [CrossRef] [PubMed]
- Tungsanga, S.; Udompornpitak, K.; Worasilchai, J.; Ratana-aneckchai, T.; Wannigama, D.L.; Katavetin, P.; Leelahavanichkul, A. Candida Administration in 5/6 Nephrectomized Mice Enhanced Fibrosis in Internal Organs: An Impact of Lipopolysaccharide and (1→3)-β-D-Glucan from Leaky Gut. Int. J. Mol. Sci. 2022, 23, 15987. [Google Scholar] [CrossRef]
- Tongthong, T.; Kaewduangduen, W.; Phuengmaung, P.; Chancharoenthana, W.; Leelahavanichkul, A. Lacticaseibacillus rhamnosus dfa1 Attenuate Cecal Ligation-Induced Systemic Inflammation through the Interference in Gut Dysbiosis, Leaky Gut, and Enterocytic Cell Energy. Int. J. Mol. Sci. 2023, 24, 3756. [Google Scholar] [CrossRef]
- Lobatón, T.; Bessissow, T.; De Hertogh, G.; Lemmens, B.; Maedler, C.; Van Assche, G.; Vermeire, S.; Bisschops, R.; Rutgeerts, P.; Bitton, A.; et al. The Modified Mayo Endoscopic Score (MMES): A New Index for the Assessment of Extension and Severity of Endoscopic Activity in Ulcerative Colitis Patients. J. Crohns Colitis 2015, 9, 846–852. [Google Scholar] [CrossRef]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 2020, 11, 465–480. [Google Scholar] [CrossRef]
- Panpetch, W.; Chancharoenthana, W.; Bootdee, K.; Nilgate, S.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Lactobacillus rhamnosus L34 Attenuates Gut Translocation-Induced Bacterial Sepsis in Murine Models of Leaky Gut. Infect. Immun. 2018, 86, 10–1128. [Google Scholar] [CrossRef]
- Thim-Uam, A.; Surawut, S.; Issara-Amphorn, J.; Jaroonwitchawan, T.; Hiengrach, P.; Chatthanathon, P.; Wilantho, A.; Somboonna, N.; Palaga, T.; Pisitkun, P.; et al. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci. Rep. 2020, 10, 777. [Google Scholar] [CrossRef]
- Sae-Khow, K.; Charoensappakit, A.; Visitchanakun, P.; Saisorn, W.; Svasti, S.; Fucharoen, S.; Leelahavanichkul, A. Pathogen-Associated Molecules from Gut Translocation Enhance Severity of Cecal Ligation and Puncture Sepsis in Iron-Overload β-Thalassemia Mice. J. Inflamm. Res. 2020, 13, 719–735. [Google Scholar] [CrossRef]
- Saithong, S.; Worasilchai, N.; Saisorn, W.; Udompornpitak, K.; Bhunyakarnjanarat, T.; Chindamporn, A.; Tovichayathamrong, P.; Torvorapanit, P.; Chiewchengchol, D.; Chancharoenthana, W.; et al. Neutrophil Extracellular Traps in Severe SARS-CoV-2 Infection: A Possible Impact of LPS and (1→3)-β-D-glucan in Blood from Gut Translocation. Cells 2022, 11, 1103. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 31, 315–322. [Google Scholar]
- Arfken, A.M.; Frey, J.F.; Summers, K.L. Temporal Dynamics of the Gut Bacteriome and Mycobiome in the Weanling Pig. Microorganisms 2020, 8, 868. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xia, Y.; He, F.; Zhu, C.; Ren, W. Intestinal mycobiota in health and diseases: From a disrupted equilibrium to clinical opportunities. Microbiome 2021, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Kounatidis, D.; Christodoulatos, G.S.; Panagopoulos, F.; Karampela, I.; Dalamaga, M. Mycobiome and Cancer: What Is the Evidence? Cancers 2021, 13, 3149. [Google Scholar] [CrossRef]
- Hughes, S.A.; Shewry, P.R.; Gibson, G.R.; McCleary, B.V.; Rastall, R.A. In vitro fermentation of oat and barley derived beta-glucans by human faecal microbiota. FEMS Microbiol. Ecol. 2008, 64, 482–493. [Google Scholar] [CrossRef]
- Ren, D.; Li, C.; Qin, Y.-Q.; Yin, R.-L.; Du, S.-W.; Ye, F.; Liu, H.-F.; Wang, M.; Sun, Y.; Li, X.; et al. Lactobacilli Reduce Chemokine IL-8 Production in Response to TNF- α and Salmonella Challenge of Caco-2 Cells. BioMed Res. Int. 2013, 2013, 925219. [Google Scholar] [CrossRef]
- Gilbert, A.S.; Wheeler, R.T.; May, R.C. Fungal Pathogens: Survival and Replication within Macrophages. Cold Spring Harb. Perspect. Med. 2014, 5, a019661. [Google Scholar] [CrossRef]
- Heung, L.J. Monocytes and the Host Response to Fungal Pathogens. Front. Cell Infect. Microbiol. 2020, 10, 34. [Google Scholar] [CrossRef]
- Austermeier, S.; Kasper, L.; Westman, J.; Gresnigt, M.S. I want to break free—Macrophage strategies to recognize and kill Candida albicans, and fungal counter-strategies to escape. Curr. Opin. Microbiol. 2020, 58, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhan, J.; Pei, G.; Zeng, R. Depletion of macrophages with clodronate liposomes partially attenuates renal fibrosis on AKI-CKD transition. Ren. Fail. 2023, 45, 2149412. [Google Scholar] [CrossRef] [PubMed]
- Ponzoni, M.; Pastorino, F.; Di Paolo, D.; Perri, P.; Brignole, C. Targeting Macrophages as a Potential Therapeutic Intervention: Impact on Inflammatory Diseases and Cancer. Int. J. Mol. Sci. 2018, 19, 1953. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.G. Depleting Macrophages In Vivo with Clodronate-Liposomes. Methods Mol. Biol. 2018, 1784, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Kameka, A.M.; Haddadi, S.; Jamaldeen, F.J.; Moinul, P.; He, X.T.; Nawazdeen, F.H.; Bonfield, S.; Sharif, S.; van Rooijen, N.; Abdul-Careem, M.F. Clodronate treatment significantly depletes macrophages in chickens. Can. J. Vet. Res. 2014, 78, 274–282. [Google Scholar] [PubMed]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J.; Ward, J.M.; Brayton, C.; Gorelick, P.; Walsh, T.J. Multigene phylogenetic analysis of pathogenic candida species in the Kazachstania (Arxiozyma) telluris complex and description of their ascosporic states as Kazachstania bovina sp. nov., K. heterogenica sp. nov., K. pintolopesii sp. nov., and K. slooffiae sp. nov. J. Clin. Microbiol. 2005, 43, 101–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, H.; Wei, Y.; Jia, H.; Chen, D.; Tang, X.; Wang, J.; Chen, M.; Guo, Y. Immune activation of characteristic gut mycobiota Kazachstania pintolopesii on IL-23/IL-17R signaling in ankylosing spondylitis. Front. Cell Infect. Microbiol. 2022, 12, 1035366. [Google Scholar] [CrossRef]
- Amornphimoltham, P.; Yuen, P.S.T.; Star, R.A.; Leelahavanichkul, A. Gut Leakage of Fungal-Derived Inflammatory Mediators: Part of a Gut-Liver-Kidney Axis in Bacterial Sepsis. Dig. Dis. Sci. 2019, 64, 2416–2428. [Google Scholar] [CrossRef]
- Chancharoenthana, W.; Sutnu, N.; Visitchanakun, P.; Sawaswong, V.; Chitcharoen, S.; Payungporn, S.; Schuetz, A.; Schultz, M.J.; Leelahavanichkul, A. Critical roles of sepsis-reshaped fecal virota in attenuating sepsis severity. Front. Immunol. 2022, 13, 940935. [Google Scholar] [CrossRef]
- Underhill, D.M.; Braun, J. Fungal microbiome in inflammatory bowel disease: A critical assessment. J. Clin. Investig. 2022, 132, e155786. [Google Scholar] [CrossRef]
- Mason, K.L.; Erb Downward, J.R.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.C. The interplay between gut bacteria and the yeast Candida albicans. Gut Microbes 2021, 13, 1979877. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, S.; Coen, M.; Leidi, A.; Schrenzel, J. The human gut mycobiome and the specific role of Candida albicans: Where do we stand, as clinicians? Clin. Microbiol. Infect. 2022, 28, 58–63. [Google Scholar] [CrossRef] [PubMed]
- James, S.A.; Parker, A.; Purse, C.; Telatin, A.; Baker, D.; Holmes, S.; Durham, J.; Funnell, S.G.P.; Carding, S.R. The Cynomolgus Macaque Intestinal Mycobiome Is Dominated by the Kazachstania Genus and K. pintolopesii Species. J. Fungi 2022, 8, 1054. [Google Scholar] [CrossRef] [PubMed]
- van Esse, H.P.; Van’t Klooster, J.W.; Bolton, M.D.; Yadeta, K.A.; van Baarlen, P.; Boeren, S.; Vervoort, J.; de Wit, P.J.; Thomma, B.P. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 2008, 20, 1948–1963. [Google Scholar] [CrossRef]
- van Esse, H.P.; Bolton, M.D.; Stergiopoulos, I.; de Wit, P.J.; Thomma, B.P. The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 2007, 20, 1092–1101. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef]
- Lobionda, S.; Sittipo, P.; Kwon, H.Y.; Lee, Y.K. The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms 2019, 7, 271. [Google Scholar] [CrossRef]
- de Oliveira, G.L.V.; Cardoso, C.R.B.; Taneja, V.; Fasano, A. Editorial: Intestinal Dysbiosis in Inflammatory Diseases. Front. Immunol. 2021, 12, 727485. [Google Scholar] [CrossRef]
- Issara-Amphorn, J.; Surawut, S.; Worasilchai, N.; Thim-Uam, A.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Hirankarn, N.; Pisitkun, P.; Leelahavanichkul, A. The Synergy of Endotoxin and (1→3)-β-D-Glucan, from Gut Translocation, Worsens Sepsis Severity in a Lupus Model of Fc Gamma Receptor IIb-Deficient Mice. J. Innate Immun. 2018, 10, 189–201. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Finkelman, M.; Worasilchai, N.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-β-D-glucan. PLoS ONE 2017, 12, e0181439. [Google Scholar] [CrossRef] [PubMed]
- Hiengrach, P.; Visitchanakun, P.; Finkelman, M.A.; Chancharoenthana, W.; Leelahavanichkul, A. More Prominent Inflammatory Response to Pachyman than to Whole-Glucan Particle and Oat-β-Glucans in Dextran Sulfate-Induced Mucositis Mice and Mouse Injection through Proinflammatory Macrophages. Int. J. Mol. Sci. 2022, 23, 4027. [Google Scholar] [CrossRef] [PubMed]
- Leelahavanichkul, A.; Worasilchai, N.; Wannalerdsakun, S.; Jutivorakool, K.; Somparn, P.; Issara-Amphorn, J.; Tachaboon, S.; Srisawat, N.; Finkelman, M.; Chindamporn, A. Gastrointestinal Leakage Detected by Serum (1→3)-β-D-Glucan in Mouse Models and a Pilot Study in Patients with Sepsis. Shock 2016, 46, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Savidge, T.C.; Newman, P.G.; Pan, W.H.; Weng, M.Q.; Shi, H.N.; McCormick, B.A.; Quaroni, A.; Walker, W.A. Lipopolysaccharide-induced human enterocyte tolerance to cytokine-mediated interleukin-8 production may occur independently of TLR-4/MD-2 signaling. Pediatr. Res. 2006, 59, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.L.; Jechorek, R.P.; Olmsted, S.B.; Erlandsen, S.L. Effect of LPS on epithelial integrity and bacterial uptake in the polarized human enterocyte-like cell line Caco-2. Circ. Shock 1993, 40, 276–288. [Google Scholar] [PubMed]
- Lafontaine, D.L.; Tollervey, D. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2001, 2, 514–520. [Google Scholar] [CrossRef]
- Lai, S.; Yan, Y.; Pu, Y.; Lin, S.; Qiu, J.G.; Jiang, B.H.; Keller, M.I.; Wang, M.; Bork, P.; Chen, W.H.; et al. Enterotypes of the human gut mycobiome. Microbiome 2023, 11, 179. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiengrach, P.; Chindamporn, A.; Leelahavanichkul, A. Kazachstania pintolopesii in Blood and Intestinal Wall of Macrophage-Depleted Mice with Cecal Ligation and Puncture, the Control of Fungi by Macrophages during Sepsis. J. Fungi 2023, 9, 1164. https://doi.org/10.3390/jof9121164
Hiengrach P, Chindamporn A, Leelahavanichkul A. Kazachstania pintolopesii in Blood and Intestinal Wall of Macrophage-Depleted Mice with Cecal Ligation and Puncture, the Control of Fungi by Macrophages during Sepsis. Journal of Fungi. 2023; 9(12):1164. https://doi.org/10.3390/jof9121164
Chicago/Turabian StyleHiengrach, Pratsanee, Ariya Chindamporn, and Asada Leelahavanichkul. 2023. "Kazachstania pintolopesii in Blood and Intestinal Wall of Macrophage-Depleted Mice with Cecal Ligation and Puncture, the Control of Fungi by Macrophages during Sepsis" Journal of Fungi 9, no. 12: 1164. https://doi.org/10.3390/jof9121164
APA StyleHiengrach, P., Chindamporn, A., & Leelahavanichkul, A. (2023). Kazachstania pintolopesii in Blood and Intestinal Wall of Macrophage-Depleted Mice with Cecal Ligation and Puncture, the Control of Fungi by Macrophages during Sepsis. Journal of Fungi, 9(12), 1164. https://doi.org/10.3390/jof9121164





