Abstract
Patellaria species are widely distributed in terrestrial and marine habitats and are saprobes growing on decaying wood, stems, or bark. However, studies on this genus in Mexico are limited, and only the type species Patellaria atrata Fr. has been cited. This study describes twelve new Patellaria species in Mexico supported by molecular (ITS-LSU-SSU) and morphological data. Phylogenetic analysis shows that species of this genus in Mexico are not closely related to Patellaria atrata. Moreover, the results demonstrate that the greatest species diversity is found in dry climates, particularly in xerophilous scrub.
1. Introduction
Patellaria was described by Fries in 1822 [1] and is characterized by superficial ascomata, black or dark coloration, is sessile, and has a dark green to black epithecium, which is formed by branched and rounded pseudoparaphyses, cylindrical and pedicellate asci, and clavate to allantoid septate ascospores; currently, it is represented by around 50 morphological species [2].
Patellaria species are widely distributed in terrestrial and marine habitats [3,4] and are saprobes on decaying wood, stems, or bark. Patellaria atrata Fr., as the type species, is the most widely distributed species and has been cited as growing on several woody hosts worldwide [3]. This species has apotecioid ascomata 675–1160 µm long, ascospores 30–45 × 7–10 µm, and 5–11 septa [5].
Most species of this genus have been described based on morphological features, including the shape and color of the ascomata, the shape and size of the asci, and the size and number of septa in the ascospores, and only five species have been described using morphological and molecular data: Patellaria quercus Crous & R.K. Schumach. from Germany [6], P. apiculatae Dayar. and K.D. Hyde and P. chromolaenae Mapook and K.D. Hyde from Thailand [3,7], P. microspora Ekanayaka and K.D. Hyde from the United Kingdom [8], and P. guizhouensis J.F. Zhang, Y.Y. Chen, and Z.Y. Liu from China [9]. Recent studies on the genus have revealed that P. chromolaena demonstrates antibacterial activity against E. coli [7]. Therefore, in Mexico, studies on Patellaria are scarce, and only P. atrata has been cited in Chiapas, Veracruz [10], Sonora [11,12], and Quintana Roo [13,14], and only with morphological support. This study aims to analyze specimens of the genus Patellaria collected in Mexico using morphological and molecular characteristics for species delimitation.
2. Materials and Methods
2.1. Morphological Studies
Specimens deposited in the herbarium of the fungal collection of the Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional (ENCB) were collected from decorticated wood or dead twigs or branches in the tropical dry forest, xerophilous scrub, tropical rain forest, and mangroves between 2014 and 2023; a total of 40 specimens were examined in this study. Macroscopic characteristics, such as the size and color of the ascomata, were measured using a stereomicroscope (Leica S9E, Leica, Wetzlar, Germany), and microscopical observations were made of cross-sections from the middle part of the ascomata; mounted in temporary slides; and rehydrated in 70% alcohol, 5% KOH, and water. The microscopical characters were described according to Yacharoen et al. [5], and the peridium, epithecium, asci, pseudoparaphyses, and ascospores were measured and characterized using an optical microscope (Olympus CX31, Olympus, Tokyo, Japan). The color key for the epithecium was determined using the Methuen Handbook of Colour [15].
2.2. Amplification and Sequencing
We obtained DNA from herbarium specimens. Genomic DNA was extracted using the CTAB method [16]. The DNA was quantified with a Nanodrop 2000c (Thermo, Waltham, MA, USA). We prepared dilutions from each sample at 20 ng/µL to amplify the next regions: the Internal Transcribed Spacer rDNA-ITS1 5.8S rDNA-ITS2 (ITS), the nuclear large subunit ribosomal DNA (nLSU), and the region of the mitochondrial small subunit (mtSSU) (Table 1). The reaction mixture for PCRs was used with a final volume of 15 µL containing 1× buffer, 0.8 mM of dNTPs mix, 20 pmol of each primer, 2 units of GoTaq DNA (Promega, Madison, WI, USA), and 100 ng of template DNA. The PCR products were verified by agarose gel electrophoresis. The gels were run for 1 h at 95 V cm−3 in 1.5% agarose and 1× TAE buffer (Tris Acetate-EDTA). The gel was stained with GelRed (Biotium, Fremont, CA, USA), and the bands were visualized in an Infinity 3000 transilluminator (Vilber Lourmat, Eberhardzell, Germany). The amplified products were purified with the ExoSAP Purification kit (Affymetrix, Santa Clara, CA, USA), following the manufacturer’s instructions. They were quantified and prepared for the sequence reaction using a BigDye Terminator v.3.1 (Applied Biosystems, Waltham, MA, USA). These products were sequenced in both directions with an Applied Biosystem model 3730XL (Applied BioSystems, Foster City, CA, USA) at the Instituto de Biología of the Universidad Nacional Autónoma de México (UNAM). The sequences of both strands of each of the genes were analyzed, edited, and assembled using BioEdit v. 7.0.5 [17] to generate a consensus sequence that was compared with those deposited in GenBank (2020) using BLASTN v. 2.2.9 [18].
2.3. Phylogenetic Analysis
To explore the phylogenetic relationships of the new species of Patellaria, an alignment was made based on the taxonomic sampling employed by Maharachchikumbura et al. [19] (Table 2). Each gene region was independently aligned using the online version of MAFFT v. 7 [20,21,22]. Alignment was reviewed in PhyDE v.10.0 [23], followed by minor manual adjustments to ensure character homology between taxa. The matrix was formed for ITS by 36 taxa (695 characters), nLSU by 42 taxa (850 characters), and SSU by 29 taxa (643 characters). Three partitioning schemes were established: one for the ITS, one for the nLSU, and one SSU, which were established using the option to minimize the stop codon with Mesquite v3.70 [24]. The data were analyzed using maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI). Maximum parsimony analyses were conducted in PAUP* 4.0b10 [25] using the heuristic search mode, 1000 random starting replicates, and TBR branch swapping, with MULTREES and Collapse on. Bootstrap values were estimated using 1000 bootstrap replicates under the heuristic search mode, each with 100 random starting replicates. Maximum likelihood analyses were conducted in RAxML v. 8.2.10 [26] with a GTR + G model of nucleotide substitution. To assess branch support, 10,000 rapid bootstrap replicates were run with the GTRGAMMA model. Bayesian inference was conducted in MrBayes v. 3.2.6 x64 [27] with four chains, and the best evolutionary model for alignment was sought using PartitionFinder [28,29,30]. Phylogenetic analyses were performed using MrBayes v3.2.6 x64 [27]. The information block for the matrix included two simultaneous runs of Montecarlo chains, a temperature set to 0.2, and sampling of 10 million generations (standard deviation ≤ 0.1). Chain convergence was visualized in Tracer v.1.7 [31]. The remaining trees were used to calculate a 50% majority-rule consensus topology and posterior probabilities (PP). Trees were visualized and optimized in FigTree v. 1.1.4 [32], and they were edited in Adobe Illustrator (Adobe Systems, Inc., San Jose, CA, USA).

Table 2.
The taxa used in phylogenetic analyses, along with their GenBank accession numbers for nLSU, ITS, and SSU sequence data. “-----” indicates that the sequence was unavailable in GenBank. Accession numbers for sequences generated in this study are denoted in boldface.

Table 1.
The primers used in the amplification and sequencing of the DNA fragments.
Table 1.
The primers used in the amplification and sequencing of the DNA fragments.
| Loci/Segment | Primer | Sequence 5′-3′ | T (°C) | Reference |
|---|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 58 | [33] |
| ITS4 | TCCTCCGCTTATTGATATGC | 58 | [33] | |
| nLSU | LROR | ACCCGCTGAACTTAAGC | 48 | [33] |
| LR3 | GGTCCGTGTTTCAAGAC | 48 | [33] | |
| 0mtSSU | MS1 | CAGCAGTCAAGAATATTAGTCAATG | 63 | [33] |
| MS2 | GCGGATTATCGAATTAAATAAC | 53 | [33] |
3. Results
3.1. Phylogenetic Analysis
The dataset of ITS, nLSU, and SSU combined comprises 42 taxa with 2300 characters, including gaps. The three phylogenetic analyses of the dataset, MP, ML, and BI, recovered similar topologies (Figure 1). No significant conflict (bootstrap value > 80%) was detected among the topologies obtained via the separate phylogenetic analyses. The parsimony analysis of the alignment found 942 trees of 112 steps (CI = 0.1002, HI = 0.1004, RI = 0.2062, RC = 0.1197). The best RAxML tree with a final likelihood value of −20624.050971 is presented. The matrix had 542 distinct alignment patterns, with 3.07% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.108340, C = 0.108312, G = 0.100074, T = 0.100214; substitution rates: AC = 1.000927, AG = 1.008320, AT = 1.000734, CG = 1.000831, CT = 4.000347, GT = 1.100000; gamma distribution shape parameter α = 0.002000. In the Bayesian analysis, the standard deviation between the chains stabilized at 0.002 after 5.2 million generations. No significant changes in tree topology trace or cumulative split frequencies of selected nodes were observed after about 0.25 million generations, which were discarded as 25% burn-in. The analysis produced a phylogenetic tree where Patellaria is shown as a monophyletic group (BS = 100%, BS = 100%, BI p = 1).
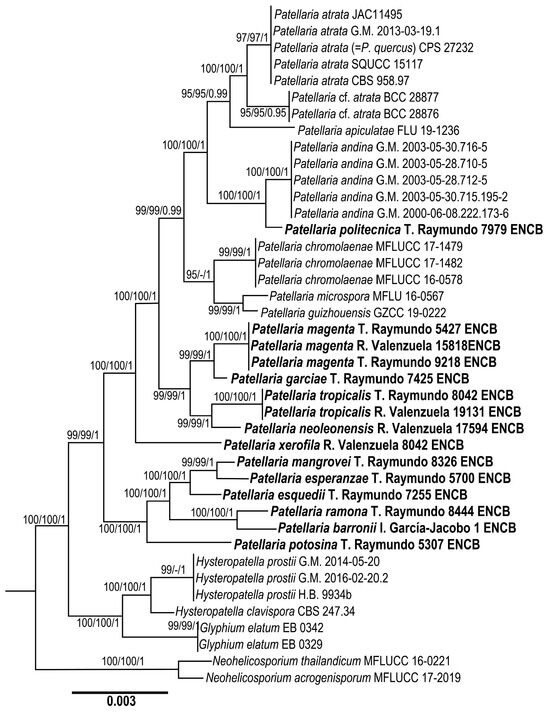
Figure 1.
Maximum likelihood phylogeny based on the concatenated ITS, nLSU, and SSU sequence alignment. Maximum parsimony and Bayesian analyses recovered identical topologies with respect to the relationships among the main clades of members of Patellaria. For each node, the following values are provided: the maximum parsimony bootstrap (%)/maximum likelihood bootstrap (%) and posterior confidence (p-value). The scale bar represents the expected number of nucleotide substitutions per site.
3.2. Taxonomy
Patellaria barronii García-Jacobo, Raymundo, and R. Valenz. sp. nov.
MycoBank: 855485.
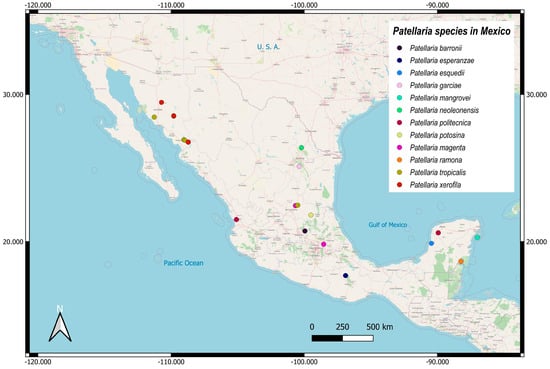
Figure 2.
The distribution of new Patellaria species.
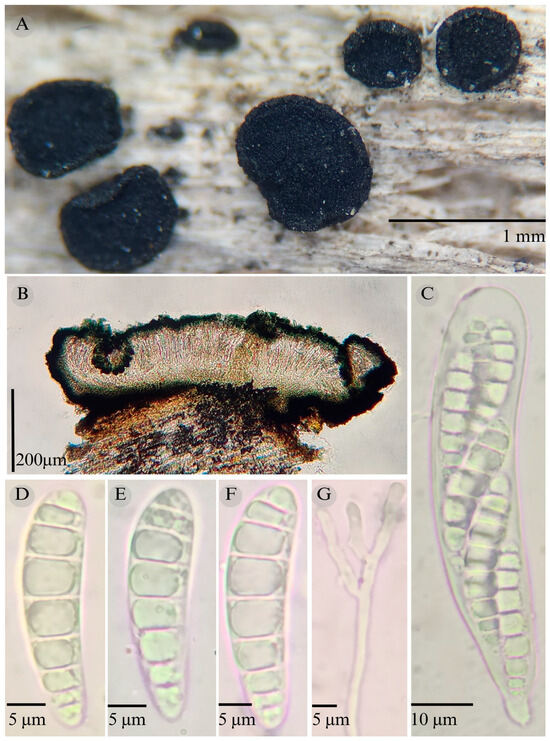
Figure 3.
Patellaria barronii García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–F) ascospores, and (G) pseudoparaphysis.
TYPE: Mexico. Querétaro, Bernal municipality, Cerca del Gallito, Peña de Bernal, 20°44′45.0″ N, 99°56′44.5″ W, 2145 m, 24 December 2023, S. García 1 (ENCB-Holotype).
Etymology: In honor of Dr. Isaac Silva Barrón, who supported the study of mycology in Querétaro.
Sexual morph: Ascomata of 500–730 × 500–560 µm, apothecioid, superficial, sessile, circular, flattened, with a thin carbonaceous rim, exposing a black hymenium at the center; peridium 33–40 µm wide ( = 37, n = 10), composed of two layers, outer layer of 20–22 µm ( = 21.2, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 4–6.5 × 4–6 µm wide, inner layer of 15 to 25 µm ( = 20, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 35 to 40 µm ( = 37, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a green jungle (25E7) to black thick epithecium over the asci when mounted in KOH ( = 20 µm, n = 10); bitunicate asci, 96–101.3 × 14.8–15.3 µm, ( = 101.33 × 15.3, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short unilobed pedicel, eight-spored, bitunicate; ascospores (28) 31–35 (36) × 6–8 µm, ( = 31.46 × 6.33, n = 30), clavate, narrowed at the lower end, 6–8 (9) septa, hyaline refractive in a pale green tone. Asexual morph: unknown.
Notes: This species is distinguished by the ascospores size (28–36 × 5–8) and number of septa (6–8). Morphologically, a similar species is Patellaria xerofila; both grow in the same type of vegetation (xerophilous scrub). However, P. xerofila differs by the dark turquoise pigmentation in KOH and ascospores 28–30 × 6–7 µm with septa (5–7). Phylogenetically, it is related to P. ramona, but differs by having larger asci ( = 101.33 µm vs. = 88.5 µm), larger ascospores ( = 31.46 µm vs. = 21.11 µm), and more septa (6–8 vs. 4–6). Therefore, P. barronii is described as a new species based on phylogeny and morphology comparison.
Patellaria esperanzae García-Jacobo and Raymundo sp. nov.
MycoBank: 855490.
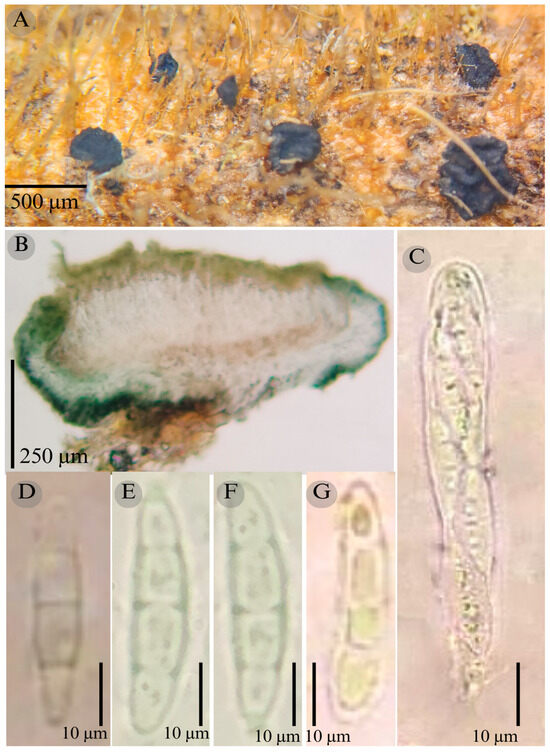
Figure 4.
Patellaria esperanzae García-Jacobo, and Raymundo. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, and (D–G) ascospores.
TYPE: Mexico. Oaxaca, Dto. Ixtlán de Juárez, Santiago Comaltepec Municipality, La Esperanza, 17°35′28.1″ N, 96°53′52.2″ W, 1398 m, 15 May 2015; T. Raymundo 5700 (ENCB, Holotype)
Etymology: The epithet refers to the locality “La Esperanza”, where this species was collected.
Sexual morph: Ascomata of 400–600 × 400–600 µm, apothecioid, superficial, sessile, circular, flattened, with a thick carbonaceous rim, exposing a black hymenium at the center; exciple, 45–52 µm wide ( = 47.5, n = 3), composed of two layers, outer layer of 10–15 µm ( = 12.5, n = 3), pseudoparenchymatous, black, composed of thick-walled cells 5–5 × 5–6 µm wide, inner layer of 30–35 µm ( = 33, n = 3), textura prismatica, composed of thick-walled cells; subhymenium of 25–40 µm wide ( = 30.71, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 2–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming an olive green (27E) to black thick epithecium over the asci when mounted in KOH ( = 25, n = 3); bitunicate asci, 70–95 × 10 µm, ( = 83 × 10, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short pedicel, eight-spored, bitunicate; ascospores 16–20 × 4–5 ( = 18.1 × 4.3, n = 30), subfusiform, three septa, hyaline to greenish. Asexual morph: unknown.
Habitat: Gregarious on dead branches of tree ferns in the genus Cyathea.
Taxonomical notes: This species is distinguished by having ascomata of 400–600 × 400–600 µm and for the size of the ascospores (16–20 × 4–5), which are fusiform and three septa. Morphologically, it is similar to Patellaria andina; they have the same number of septa in the ascospores, but they differ by the size of the ascospores ( = 18.1 vs. = 22–25 µm). P. esperanzae and P. andina are not closely related in the phylogenetic analysis.
In the present phylogenetic analysis, P. esperanzae is related to P. mangrovei. However, P. esperanzae differs from P. mangrovei by the epithecium color (olive green (27E) vs. olive brown (4E8)) and by having smaller ascospores (18.1 × 4.3 µm vs. 29.36 × 6.83 µm) and fewer septa (3 vs. 6–8). Additionally, P. esperanzae grows on tree ferns of the genus Cyathea in tropical montane cloud forest in Oaxaca, while P. mangrovei grows on Rhizophora mangle in a mangrove community in Quintana Roo.
Patellaria esquedii García-Jacobo, Raymundo, and R. Valenz. sp. nov.
MycoBank: 855493.
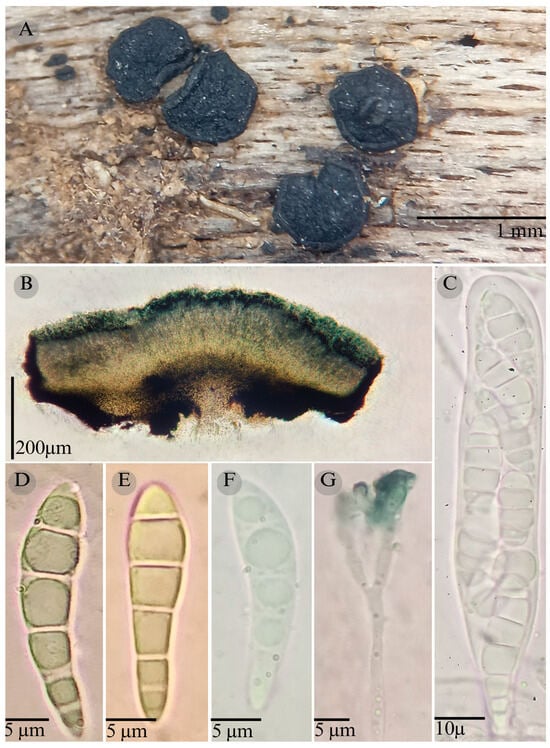
Figure 5.
Patellaria esquedii García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–F) ascospores, and (G) pseudoparaphysis.
TYPE: Mexico. Campeche, Imí municipality, Km. 179 de la Carretera Champotón–Mérida, Kopomá, 20°37′21″ N, 89°56′23″ W, 18 m, 19 January 2018, T. Raymundo 7255 (ENCB, Holotype).
Etymology: The epithet honors Dr. Martín Esqueda for his important contributions to Mexican mycology.
Sexual morph: Ascomata of 600–1000 × 400–700 µm, apothecioid, superficial, sessile, circular, flattened, with a thick carbonaceous rim, exposing a black hymenium at the center; peridium 35–40 µm wide ( = 37.3, n = 10), composed of two layers, outer layer of 20–24 µm ( = 12.5, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 4–5 × 6 µm wide, inner layer of 11–13 µm ( = 12, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 35–40 µm ( = 37.5, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1.5–3.5 µm wide, thick-walled, filiform to cylindrical, septa, branched, hyaline pseudoparaphyses, rounded at the apex, forming a green jungle (25F8) to black thick epithecium over the asci when mounted in KOH ( = 22.3, n = 10); bitunicate asci, 85–98 × 13–17 µm ( = 91 × 13.83, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short unilobed pedicel, eight-spored, bitunicate; ascospores (27) 30–32 × 6 µm ( = 29.8 × 6, n = 30), obclavate, slightly curved, narrowed at the ends, 5–6 septa, hyaline to greenish. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in the tropical rain forest.
Additional specimens: Campeche, Imí municipality, Km. 179 de la Carretera Champotón–Mérida, Kopomá, 20°37′21″ N, 89°56′23″ W, 18 m, 19 Jan 2018, R. Valenzuela 15715 (ENCB).
Notes: P. esquedii is characterized by the size of the asci 85–98 × 13–17 (short pedicellate), the size of the ascospores (27–32 × 6), and the number of septa (5–6) in the ascospores and its obclavate shape.
In this study, P. esquedii forms a sister taxon to Patellaria ramona and P. mangrovei, from which it differs in the shape of the ascospores: clavate in P. ramona and P. mangrovei, and obclavate in P. esquedii. Additionally, the ascospores of P. esquedii have 5–6 septa, whereas P. mangrovei has 5–8 septa and P. ramona has 4–6.
Patellaria garciae García-Jacobo, Raymundo, Martínez-González, and R. Valenz. sp. nov.
MycoBank: 855489.
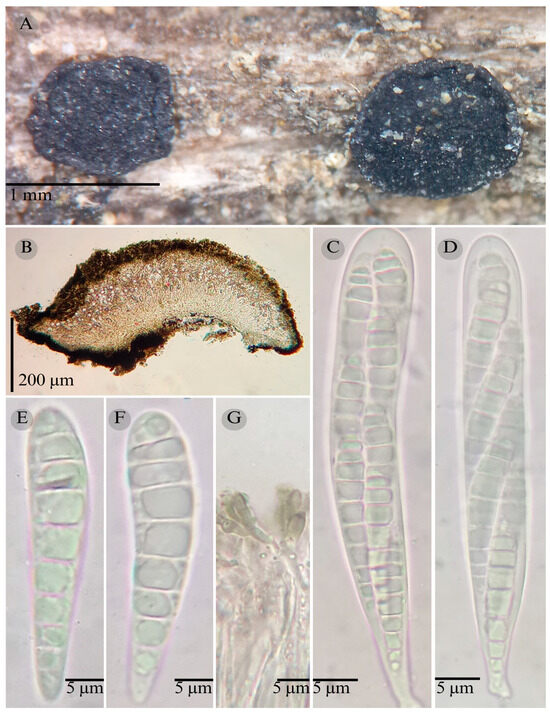
Figure 6.
Patellaria garciae García-Jacobo, Raymundo, Martínez-González, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C,D) asci, (E,F) ascospores, and (G) pseudoparaphysis.
TYPE: Mexico. Coahuila, Arteaga municipality, El Renacer de la Sierra, 25°12′33″ N, 100°22′52″ W, 3177 m, 17 March 2017, T. Raymundo 7425 (ENCB, Holotype).
Etymology: In honor of Dr. Jesús García for important contributions to Mexican mycology.
Sexual morph: Ascomata of 700–800 × 700–750 µm, apothecioid, superficial, sessile, circular, flattened with a thin carbonaceous rim, exposing a black hymenium at the center; peridium 50 µm wide ( = 5 0 µm, n = 3), composed of two layers, outer layer of 25 µm ( = 25 µm, n = 3), pseudoparenchymatous, black, composed of thick-walled cells 4–6 × 4–6 µm wide, inner layer of 25 µm ( = 25 µm, n = 3), textura prismatica, composed of thick-walled cells; subhymenium 30 µm (n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, filiform to cylindrical, septa, branched, hyaline pseudoparaphyses, rounded at the apex, forming a spruce green (25F3) to black thick epithecium over the asci when mounted in KOH ( = 30 µm, n = 3); bitunicate asci 100–115 × 15 µm ( = 108 × 15 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short bilobed pedicel, eight-spored, bitunicate; ascospores, 35–40 (42) × 6–7 (8) µm ( = 39.31 × 6.76 µm, n = 30), clavate, slightly curved, narrowed at the lower end, 8–9 septa, hyaline refractive in a pale green tone. Asexual morph: unknown.
Hábitat: Gregarious on decaying wood in coniferous forests.
Additional specimens: Coahuila, Arteaga Municipality, El Renacer de la Sierra, 25°12′33″ N, 100°22′ 52″ W, 3177 m, 17 March 2017, T. Raymundo 7425 (ENCB).
Notes: Differs from P. atrata by having shorter asci (100–115 × 15 µm), a short pedicel, and smaller ascospores (35–42 × 6–8), with fewer septa (8–9 vs. 5–11).
Patellaria neoleonensis is similar to P. garciae in the number of septa (8–9 vs. 8–10) but differs by having long pedicellate asci and a dark turquoise epithecium reaction in KOH. These species also differ because P. neoleonensis grows in xerophytic scrub and P. garciae grows in coniferous forests. Phylogenetically, P. garciae is closely related to P. magenta, from which it differs morphologically by having smaller asci ( = 108 × 15 µm vs. = 120 × 13 µm) and a different ascospore shape: in P. magenta, the ascospores are clavate but conic at the apex, while in P. garciae, they are clavate and slightly curved. They also differ in the number of septa (8–9 vs. 7–13). Additionally, P. magenta grows on unidentified wood in tropical dry forests in Hidalgo, San Luis Potosí, and Sonora, whereas P. garciae grows in coniferous forests in Coahuila.
Patellaria magenta García-Jacobo, Raymundo, Mart.-Pineda, and R. Valenz. sp. nov.
MycoBank: 855481.
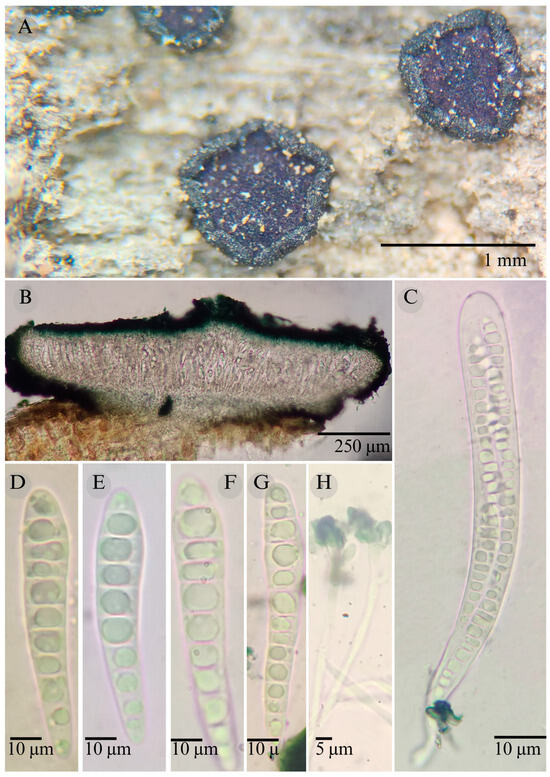
Figure 7.
Patellaria magenta García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–G) ascospores, and (H) pseudoparaphysis.
TYPE: Mexico. Sonora, Álamos municipality, Sierra de Álamos-Río Cuchujaqui Biosphere Reserve, Promontorios, 27°00′54.1″ N, 109°02′10.5″ W, 602 m, 7 October 2013, T. Raymundo 5427 (ENCB, Holotype).
Etymology: The epithet refers to the color of the hymenium.
Sexual morph: Ascomata of 600–930 × 660–1000 µm, discoidal, superficial, sessile, circular, flattened, with a thick and involute carbonaceus rim, exposing a black to dark ruby (12F8) hymenium at the center; peridium 37–56 µm wide ( = 46.5 µm, n = 10), composed of two layers, outer layer 20–27 µm wide ( = 23.6 µm, n = 10) pseudoparenchymatous, black, composed of thick-walled cells 5–5 × 6 µm wide, inner layer 20–29 µm wide ( = 24.25 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium 25–37 µm wide ( = 30 µm, n = 30), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a green jungle (25E7) to thick black epithecium over the asci when mounted in KOH; bitunicate asci 95–145 × 12–20 µm, ( = 120 × 13 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, with a long bilobed pedicel, eight-spored, bitunicate; ascospores (30) 32–37 (54) × 6–8 µm, ( = 36 × 6 µm, n = 30), clavate, conic at the apex, narrowed at the lower end, (7) 8–10 (13) septa, and hyaline refractive in green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in tropical dry forests.
Additional specimens: Hidalgo, Tepeapulco municipality, Zona Arqueológica El Xihuingo, 19°48′44″ N, 98°33′12″ W, 2499 m, 9 June 2024, P.M. Alvarez-Cortés 520 (ENCB). San Luis Potosí, Villa Hidalgo municipality, Km. 88 Carretera San Luis– Río Verde, 22°31′11″ N, 100°39′43″ W, 1572 m, 18 June 2023, T. Raymundo 9218 (ENCB). Sonora, Álamos municipality, Sierra de Álamos-Río Cuchujaqui Biosphere Reserve, Promontorios, 27°00′54.1″ N, 109°02′10.5″ W, 602.2 m, 7 October 2014, R. Valenzuela 15818 (ENCB); 26 October 2018, T. Raymundo 8071 (ENCB); R. Valenzuela 18751 (ENCB).
Notes: Patellaria magenta is characterized by its purple epithecium color and the size (30–54 × 6–8) and number of septa on its ascospores (7–13). P. neoleonensis is similar in the ascospore size (32–50 × 6–8) and the long pedicellate asci but differs in the KOH reaction (dark turquoise: 24F8) and has fewer septa (8–10).
Patellaria magenta is phylogenetically related to P. tropicalis and P. neoleonensis, which forms a sister clade with P. magenta. P. tropicalis and P. magenta can be distinguished by the size of the asci ( = 120 × 13 µm vs. = 85.12), the size of the ascospores ( = 36 × 6 µm vs. = 28.3 × 6.12 µm), and the number of septa (7–13 vs. 5–7).
Patellaria mangrovei García-Jacobo, M. Mtz.–Pineda, R. Valenz, and Raymundo sp. nov.
MycoBank: 855488.
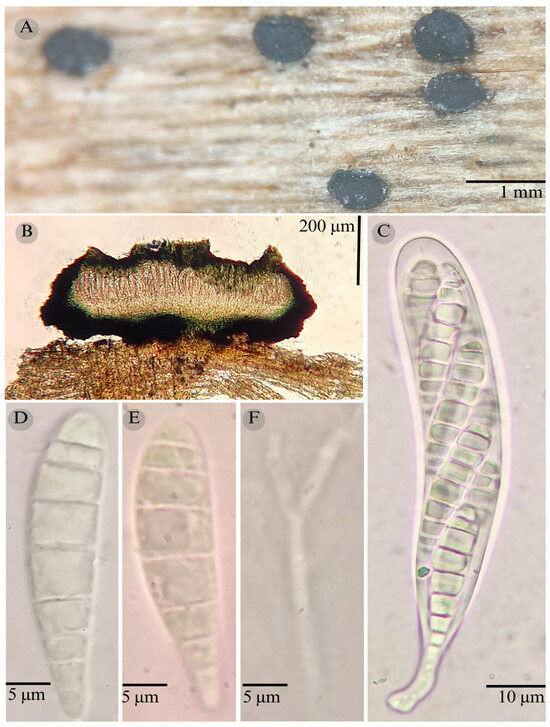
Figure 8.
Patellaria mangrovei García-Jacobo, M. Mtz.–Pineda, R. Valenz, and Raymundo. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D,E) ascospores, and (F) pseudoparaphysis.
TYPE: Mexico. Quintana Roo, San Miguel de Cozumel municipality, Parque Ecológico Punta Sur, Cozumel Island Biosphere Reserve, 20°18′01″ N, 87°00′0″ W, 1 m, 15 October 2019, T. Raymundo 8326 (ENCB, Holotype).
Etymology: Name refers to the fact that the species grows on mangroves of genus Rhizophorae.
Sexual morph: Ascomata of 500–700 × 500–600 µm, apothecioid, superficial, sessile, circular, flattened, with a thin carbonaceous rim, exposing a black hymenium at the center; peridium 40–43 µm wide ( = 42 µm, n = 3), composed of two layers, outer layer of 20–22 µm ( = 20.5 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 4–5 × 4–5 µm wide, inner layer of 20–23 µm ( = 22.6 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 60 µm (n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming an olive brown (4E8) to black thick epithecium over the asci when mounted in KOH ( = 20, n = 3); bitunicate asci 75–90 × 14–16 µm, ( = 83 × 15 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, short pedicellate, eight-spored, bitunicate; ascospores (25) 29–30 (35) × 6–7 (9) µm, ( = 29.36 × 6.83 µm, n = 30), clavate, slightly curved, narrowed at the lower end, 5–8 septa, hyaline refractive in a pale green tone. Asexual morph: unknown.
Habitat: Gregarious to solitary on decaying wood of Rhizophora mangle L.
Notes: Patellaria mangrovei is distinguished from other Patellaria species by the epithecium color in the presence of KOH (olive brown). This species grows on decaying wood of Rhizophora mangle. Patellaria potosina is similar in that it has 5–8 septate ascospores, but in this species, 8 septa is rare. It differs by the reaction in KOH (nautical blue: 24E7) and the ascospore size (25–35 × 6–9 vs. 31–37 × 5–7), and shape; P. mangrovei ascospores are clavate, and those of P. potosina are obclavate.
With regard to other mangrove species, P. apiculatae has been reported growing on Rhizophora apiculata. These species can be distinguished by morphological features, such as the color of the epithecium in KOH (olive brown vs. bluish-black), the size of the asci ( = 83 × 15 µm vs. = 62 × 16 µm), with P. mangrovei having larger asci, and the number of septa in the ascospores (5–8 vs. 5–6). Furthermore, these species are not closely related in terms of their phylogeny.
Patellaria neoleonensis García-Jacobo, Raymundo and R. Valenz. sp. nov.
MycoBank: 855491.
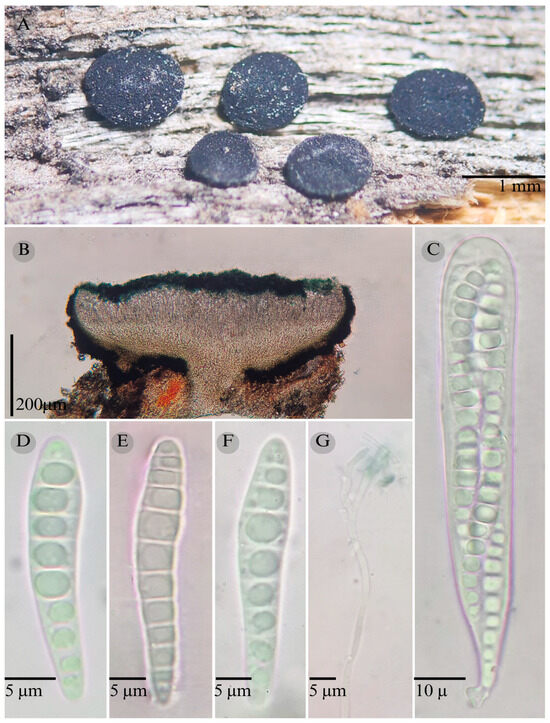
Figure 9.
Patellaria neoleonensis García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–F) ascospores, and (G) pseudoparaphysis.
TYPE: Mexico. Nuevo León, Sabinas Hidalgo municipality, La Cuchilla, 26°29′07″ N, 100°12′52″ W, 338 m, 22 October 2017, R. Valenzuela 17594 (ENCB, Holotype).
Etymology: The epithet refers to the state where this species was collected.
Sexual morph: Ascomata of 830–1000 × 700–1000 µm, apothecioid, superficial, sessile, circular, flattened, exposing a black hymenium at the center, with a thick, involute, and striate carbonaceous rim; peridium 45–62 µm wide ( = 55 µm, n = 10), composed of two layers, outer layer of 25–35 µm ( = 28.6 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 5–5 × 6 µm wide, inner layer of 20 to 37 µm ( = 27.3 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium 20–35 µm wide ( = 28.33 µm, n = 3) greenish blue, composed of thick-walled cells; hamathecium 1.5–3.5 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a dark turquoise (24F8) to black thick epithecium over the asci when mounted in KOH ( = 21, n = 10); bitunicate asci 93–154 × 12–20 µm, ( = 109.7 × 14.7 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short bilobed pedicel, eight-spored; ascospores (32) 38–42 (50) × 6–8 µm, ( = 39.87 × 6.6 µm, n = 30), clavate, narrowed at the lower end, 8–10 septa, hyaline refractive in a pale green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in xerophilous scrub.
Additional studied specimens: Nuevo León, Sabinas Hidalgo municipality, La Cuchilla, 26°29′07″ N, 100°12′52″ W, 338, 22 October 2017, T. Raymundo 7050 (ENCB); R. Valenzuela 17597 (ENCB).
Notes: Patellaria neoleonensis can be differentiated by the color of its reaction in KOH (spruce green: 25F3) and grows in xerophilous scrub. Patellaria garciae is similar, having clavate ascospores and 8–9 septa but differs by growing in coniferous forests and its KOH reaction (dark turquoise: 24F8). In this work, this species is phylogenetically related to P. tropicalis, and they differ morphologically in having larger asci ( = 85.12 µm vs. = 109.7 µm), larger ascospores ( = 39.87 vs. = 28.3 µm), and more septa (8–10 vs. 5–7).
Patellaria politecnica García-Jacobo, Raymundo, Mart.-Pineda, and R. Valenz. sp. nov. Figure 1, Figure 2 and Figure 10.
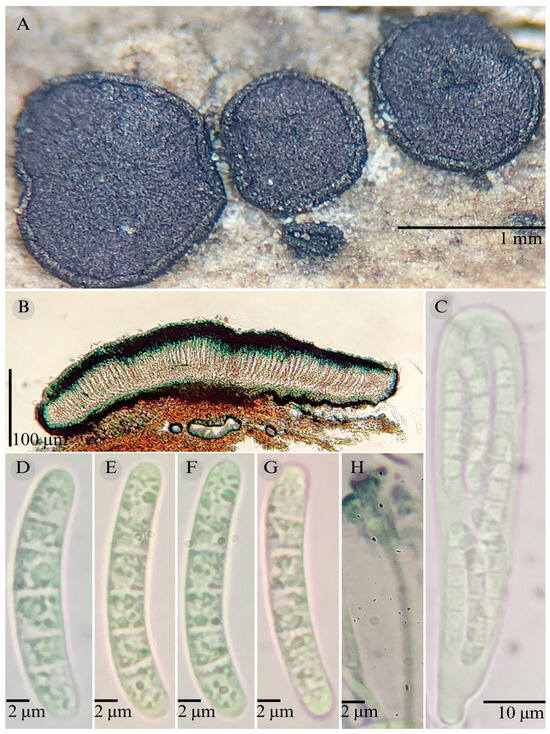
Figure 10.
Patellaria politecnica García-Jacobo, Raymundo, Mart.-Pineda, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–G) ascospores, and (H) pseudoparaphysis.
MycoBank: 855487.
TYPE: Mexico. Nayarit. San Blas municipality, Km 27 carretera San Blas–Tepic, 21°33′09″ N, 105°05′32″ W, 577 m, 29 September 2018, T. Raymundo 7979 (ENCB, Holotype).
Etymology: This species is dedicated to the Instituto Politécnico Nacional for its support in the study of Dothideomycetes in Mexico.
Sexual morph: Ascomata of 700–860 × 500–800 µm, apothecioid, superficial, sessile, circular, flattened, with a thin carbonaceous rim, exposing a black hymenium at the center; peridium 25–30 µm wide ( = 28.3 µm, n = 10), composed of two layers, outer layer of 10–17 µm ( = 14.33 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 4–5–5 × 6 µm wide, inner layer of 12 to 25 µm ( = 16.7 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 10 µm (n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a nautical blue (24E7) to black thick epithecium over the asci when mounted in KOH ( = 18.3 µm, n = 10); bitunicate asci of 55–76 × 10–15 µm ( = 65.18 × 12.60 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, with a short unilobed pedicel, eight-spored, bitunicate; ascospores 21–26(27) 32 × 4–5 µm ( = 25 × 4.04 µm, n = 30), allantoid, (3) 4–6 septa, and hyaline refractive in a pale green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in the tropical rain forest.
Additional specimens: Campeche, Kopomá municipality, Km. 179 Carretera Champotón–Mérida, 20°37′21″ N, 89°56′23″ W, 18 m, 19 January 2018, T. Raymundo 7256 (ENCB). Nayarit. San Blas municipality, Km. 27 carretera San Blas–Tepic, 21°33′09″ N, 105°05′32″ W, 577 m, 29 September 2018, R. Valenzuela 18679 (ENCB), M. Sánchez 1392 (ENCB).
Notes: Patellaria politecnica differs from other Patellaria species by having a thinner subhymenium (10 µm) and distinctively sized ascospores (21–32 × 4–5 µm). The allantoid ascospores found in P. chromolaenae are differentiated from those of P. politecnica by the number of septa (4–6 vs. 5–6) and the hymenium color, which is black in P. chromolaenae and litmus bluish red in P. politecnica. P. politecnica is phylogenetically close to P. andina, a species described from Argentina that grows on decorticated branches of Prosopis alpataco. P. politecnica differs morphologically from P. andina by having larger ascomata (700–860 × 500–800 µm vs. 250–500 µm) and differently shaped ascospores (allantoid vs. cylindrical). Thay also differ in the number of septa in the ascospores (4–6 vs. 3).
Patellaria potosina García-Jacobo, Raymundo and R. Valenz. sp. nov.
MycoBank: 855482.
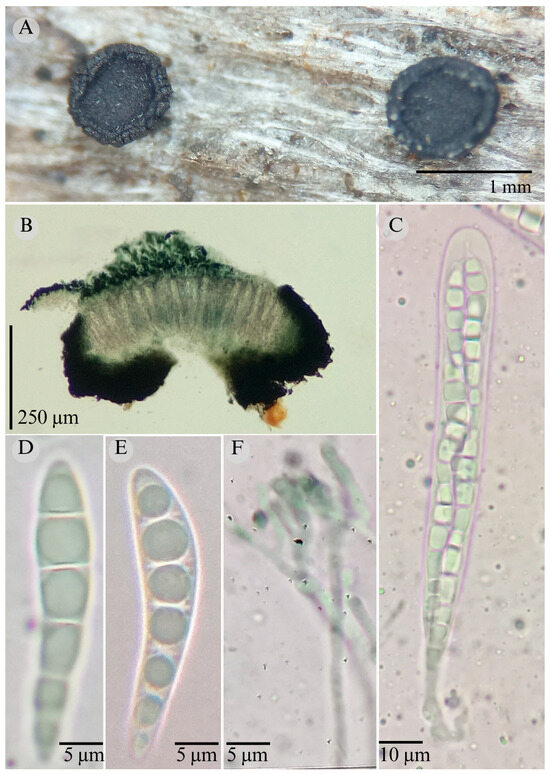
Figure 11.
Patellaria potosina García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D,E) ascospores, (F) pseudoparaphysis.
TYPE: Mexico. San Luis Potosí, Rayón municipality, Km. 65 Carretera Cd. Valles–Río Verde, 21°51′51″ N, 99°30′01″ W, 1123 m, 14 September 2015; T. Raymundo 5307 (ENCB, Holotype).
Etymology: The epithet refers to the state where the species was collected.
Sexual morph: Ascomata of 550–750 × 550–730 µm, discoidal, superficial, sessile, circular, flattened, with a thin, striate carbonaceus rim, involute, exposing a black hymenium at the center; peridium 35–45 µm wide ( = 40 µm, n = 10), composed of two layers, outer layer of 15–20 µm ( = 18 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 3.5–5 × 4–5 µm wide, inner layer of 20 µm ( = 20 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 40–55 µm ( = 45 µm, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1.5–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses forming a nautical blue (24E7) to black thick epithecium over the asci when mounted in KOH ( = 22.5, n = 10); bitunicate asci 75–116 × 11–15 µm ( = 91.42 × 12.56 µm, n = 10), a cylindrical, clavate, apically rounded, generally straight appearance, with a long bilobed pedicel, eight-spored, bitunicate; ascospores (30)31–33(38) × 5–7 µm ( = 31.48 × 5.98 µm, n = 30), obclavate, slightly curved, narrowed at the lower end, 5–6 (8) septa, and hyaline refractive in green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in xerophilous scrub.
Additional specimens: San Luis Potosí. Rayón municipality, Km. 65 de la Carretera Cd. Valles–Río Verde, 21°51′51″ N, 99°30′01″ W, 1123 m, 14 September 2015; R. Valenzuela 15715 (ENCB).
Notes: P. potosina can be recognized by asci 75–116 × 11–15, with bulbous pedicel; ascospores (30)31–33(38) × 5–7 µm, clavate, and 5–8 septa.
Patellaria mangrovei is similar to P. potosina in that it has ascospores and 5–8 septa but differs in its habitat, with P. mangrovei growing on Rhizhophora mangle and P. potosina growing in the Quercus forest, also called “Chaparral”. P. mangrovei also differs in its KOH reaction (olive brown) and the ascospore shapes, which are clavate in P. mangrovei and obclavate in P. potosina. This species shows phylogenetic affinities to P. barronii and P. ramona and can be distinguished from P. barronii by the epithecium color in KOH (nautical blue (24E7) vs. green jungle (25E7)) and the shape of the pedicel in the asci, which is bilobed in P. potosina and unilobed in P. barronii. They can also be differentiated by the shape and number of septa (5–6 vs. 6–8) of the ascospores, which are obclavate in P. potosina and clavate in P. barronii. P. potosina differs from P. ramona by the pedicel in the asci, which is unilobed in P. ramona, and in the size (The figure refers to the map where the distribution of each species is indicated The figure refers to the map where the distribution of each species is indicated ( = 31.48 × 5.98 µm vs. = 21.11 × 6 µm), shape, and septa of ascospores (clavate and 4–6 septa in P. ramona). Additionally, P. ramona grows in the tropical rain forest.
Patellaria ramona García-Jacobo and Raymundo sp. nov.
MycoBank: 855492.
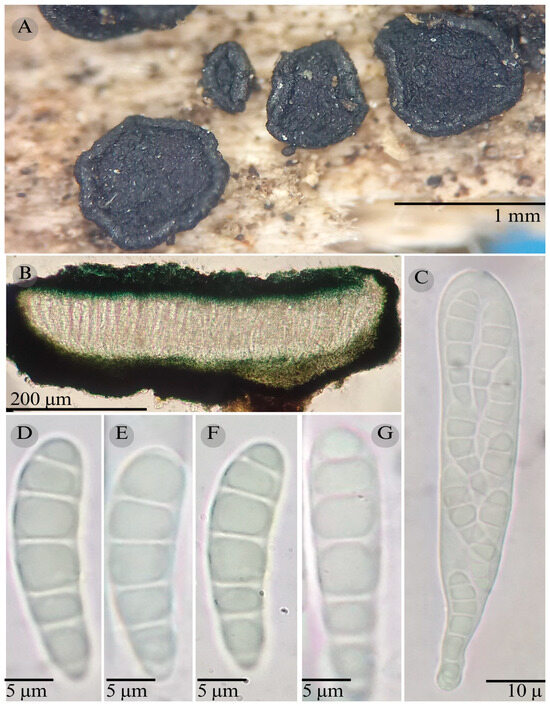
Figure 12.
Patellaria ramona García-Jacobo and Raymundo. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, and (D–G) ascospores.
TYPE: Mexico. Quintana Roo, Oxtanka archaeological zone, Othón P. Blanco municipality, 18°36′29″ N, 88°14′03″ W, 16 m, 7 February 2021, T. Raymundo 8444 (ENCB, Holotype).
Etymology: The epithet refers to the common name of the host where this species grows (Ramón (Brosimium sp.)).
Sexual morph: Ascomata of 450–700 × 575–650 µm, apothecioid, superficial, sessile, circular, flattened, with a thin carbonaceous rim, exposing a black hymenium at the center; peridium 25–30 µm wide ( = 27.5 µm, n = 10), composed of two layers, outer layer of 10–15 µm ( = 12.5 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 5 × 6 µm wide, inner layer of 15 µm ( = 15 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 35–40 µm ( = 37.5 µm, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1.5–3.5 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses rounded at the apex, forming a dark turquoise (24F8) to black thick epithecium over the asci when mounted in KOH ( = 15, n = 3); bitunicate asci 75–96 × 15–17 µm ( = 88.5 µm × 15.8, n = 10), cylindrical, clavate, apically rounded, generally straight, with a long unilobed pedicel, eight-spored; ascospores (22) 25–29 (30) × 6 µm ( = 21.11 × 6 µm, n = 30), clavate, slightly curved, narrowed at the lower end, 4–6 septa, hyaline refractive in a pale green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in Brosimium sp.
Additional specimens: Quintana Roo, Othón P. Blanco municipality, Oxtanka archeological zone, “El Ramonal” 18°36′29″ N, 88°14′03″ W, 16 m, 7 February 2021, R. Valenzuela 19244 (ENCB).
Notes: Patellaria ramona is distinguished by the size of its asci (75–96 × 15–17) with a short pedicell; ascospores are 22–29 × 6, 4–6 septa.
P. esquedii is both morphologically and ecologically similar, as it also grows in tropical rain forests. Morphologically, they differ in the color of the epithecium in KOH (dark turquoise (24F8) vs. green jungle (E7)), the length of the asci pedicels (short in P. ramona and long in P. esquedii), and the shape of the ascospores (clavate in P. ramona and obclavate in P. esquedii). Another distinguishing feature is the number of septa in the ascospores (4–6 in P. ramona and 5–6 in P. esquedii).
Patellaria tropicalis García-Jacobo, Raymundo and R. Valenz. sp. nov.
MycoBank: 855483.
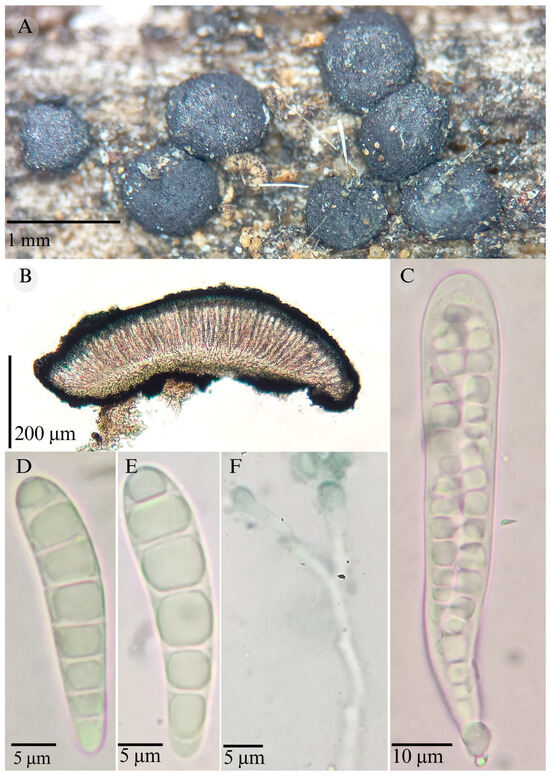
Figure 13.
Patellaria tropicalis García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D,E) ascospores, and (F) pseudoparaphysis.
TYPE: Mexico. Sonora, Promontorios, Sierra de Álamos-Río Cuchujaqui Biosphere Reserve 27°00′54.1″ N, 109°02′10.5″ W, 602 m, 3 October 2016, T. Raymundo 8042 (ENCB, Holotype).
Etymology: The epithet refers to the vegetation type where the species was collected (tropical dry forest).
Sexual morph: Ascomata of 530–760 × 500–800 µm, discoidal, superficial, sessile, circular, flattened, with a thin carbonaceous rim, exposing a black to dark ruby (12F8) hymenium at the center; peridium 25–40 µm wide ( = 39.6 µm, n = 10), composed of two layers, outer layer of 15–25 µm ( = 18.8 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 5–6 × 5–6.5 µm wide, inner layer of 15–40 µm ( = 17 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 17 to 30 µm ( = 20.28 µm, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1.5–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a deep green (25D8) to black thick epithecium over the asci when mounted in KOH ( = 20, n = 10); bitunicate asci 72–110 × 12–15 µm ( = 85.12 × 14.15 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, with a unilobed short pedicel, eight-spored, bitunicate; ascospores (24)26–30(37) × 6–7 µm ( = 28.3 × 6.12 µm, n = 30), clavate, slightly curved, narrowed at the lower end, 5–7 septa, and hyaline refractive in a pale green tone. Asexual morph: unknown.
Habitat: Gregarious on decaying wood in the tropical dry forest.
Additional specimens: Sonora, Rancho La Sierrita, Reserva de la Biósfera Sierra de Álamos-Río Cuchujaqui, 28°30′53.2″ N, 111°15′50.5″ W, 528 m, 8 October 2017, T. Raymundo 5486 (ENCB), D. Castro–Bustos 141 (ENCB),; Octubre 27 2018, R. Valenzuela 18730 (ENCB); Promontorios, Sierra de Álamos-Río Cuchujaqui Biosphere Reserve, 27°00′54.1″ N, 109°02′10.5″ W, 602 m, 7 October 2014, T. Raymundo 5404 (ENCB), T. Raymundo 5449 (ENCB); 26 October 2018, T. Raymundo 8070 (ENCB). San Luis Potosí, Sierra del Abra Tanchipa Biosphere Reserve, 22°33′05″ N, 100°29′44″ W, 1522 m, 12 October 2023, R. Valenzuela 19131 (ENCB); 18 December 2023, R. Valenzuela 19250 (ENCB).
Taxonomical notes:
Differs from other Patellaria species by the size of its asci, 72–110 × 12–15, with short pedicellate; and ascospore size, 24–37 × 6–7 µm, which are clavate, and 5–7 septa.
P. xerofila is similar; it has ascospores with 5–7 septa. These species differ in their KOH reaction (dark turquoise vs. deep green) and in the type of vegetation in which they grow; P. tropicalis is found in the tropical dry forest and P. xerofila in xerophilous scrub. For the species that grow on the same type of vegetation, P. magenta is differentiated by its epithecium color (black in P. tropicalis vs. dark magenta (13F) in P. magenta)), the size of the asci ( = 85.12 µm vs. = 120 × 13 µm), and the size ( = 28.3 × 6.12 µm vs. = 36 × 6 µm) and shape of the ascospores, which are clavate and conic at the apex in P. magenta and clavate in P. tropicalis.
However, these species are not closely related phylogenetically.
Patellaria xerofila García-Jacobo, Raymundo, and R. Valenz. sp. nov.
MycoBank: 855486.
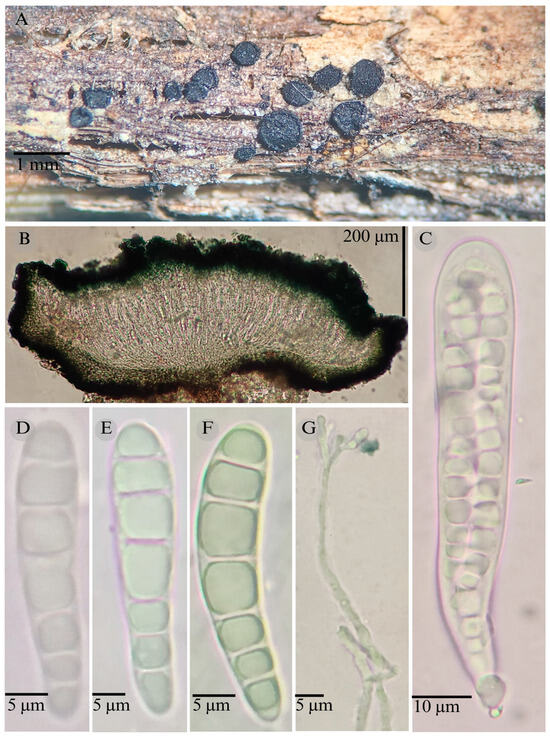
Figure 14.
Patellaria xerofila García-Jacobo, Raymundo, and R. Valenz. (A) Ascomata, (B) longitudinal section of ascomata, (C) asci, (D–F) ascospores, and (G) pseudoparaphysis.
TYPE: Mexico. Sonora, San Miguel Horcasitas municipality, 29°29′09″ N, 110°43′03″ W, 397 m, 19 October 2013, R. Valenzuela 15286 (ENCB, Holotype).
Etymology: The epithet refers to the type of vegetation where the species grows: xerophilous scrub.
Sexual morph: Ascomata of 510–660 × 510–633 µm, apothecioid, superficial, sessile, circular, flattened, with a thick carbonaceous rim, exposing a black hymenium at the center; peridium 29–45 µm wide ( = 36 µm, n = 10), composed of two layers, outer layer of 15–30 µm ( = 17.5 µm, n = 10), pseudoparenchymatous, black, composed of thick-walled cells 4–6.5 × 4–6 µm wide, inner layer of 12 to 25 µm ( = 16.7 µm, n = 10), textura prismatica, composed of thick-walled cells; subhymenium of 14 to 45 µm ( = 25 µm, n = 3), hyaline to greenish blue, composed of thick-walled cells; hamathecium 1–3 µm wide, thick-walled, filiform to cylindrical, septate, branched, hyaline pseudoparaphyses, rounded at the apex, forming a dark turquoise (24F8) to black thick epithecium over the asci when mounted in KOH ( = 17.5 µm = 10); bitunicate asci 82–110 × 11–15 µm ( = 92.9 × 13.03 µm, n = 10), cylindrical, clavate, apically rounded, generally straight, short pedicellate, eight-spored; ascospores (26)28–31(39) × 5–7 µm ( = 30.9 × 6.02 µm, n = 30), clavate, slightly curved, narrowed at the lower end, 5–7 septa, and hyaline refractive in a pale green tone.
Habitat: Gregarious on decaying wood in the tropical dry forest and xerophilous scrub.
Additional specimens: San Luis Potosí, Rayón municipality, Km. 65 de la Carretera Cd. Valles–Río Verde, 21°51′51″ N, 99°30′01″ W, 1123 m, 14 September 2015, R. Valenzuela 15715 (ENCB). Sonora, San Javier Muncipality, Km. 135 Carretera Hermosillo–Yecora, 28°35′59″ N, 109°48′02″ W, 507 m, 3 October 2016, R. Valenzuela 16965 (ENCB); 16976 (ENCB); 5 October 2016, R. Valenzuela 17124 (ENCB); T. Raymundo 6339 (ENCB); Álamos municipality, El Cajón, Sierra de Álamos-Río Cuchujaqui Biosphere Reserve, 26°51′08″ N, 108°42′24″ W, 355 m, 14 October 2013, T. Raymundo, 4819 (ENCB).
Taxonomical notes: P. xerofila is distinguished by the size of its asci, 82–110 × 11–15, and the size (26–39 × 5–7) and shape of its ascospores, which are clavate and have 6–7 septa.
P. tropicalis is similar in the number of septa but differs mainly in the epithecium reaction in KOH. However, P. tropicalis grows in tropical dry forests, and P. xerofila grows in xerophilous scrub. Phylogenetically, this species is related to P. barronii, P. ramona, P. esquedii, P. esperanzae, and P. mangrovei, which forms a sister clade. Species in the sister clade grow in different vegetation types, and only P. barronii grows in xerophilous scrub like P. xerofila. These species show morphological differences, like the epithecium color in KOH that is dark turquoise (24F8) in P. xerofila and green jungle (25E7) in P. barronii. They also differ in the number of septa (5–7 vs. 6–8).
- Key to Patellaria species forming ascomata supported by molecular and morphological data
- 1.
- Species with allantoid ascospores and 3–6 septa ……………………….…………………………………………………………2
- 1.
- Species with subfusiform, obclavate, or clavate ascospores; 3–13 septa……………………………………………………………………………3
- 2.
- Ascomata < 700 µm in diam; ascospores 27–34 × 5.2–6 µm, allantoid, 3–7 septa; growing in dead stems of Chromolaenae odorata from Thailand …………………………………………………………………P. chromolaenae
- 2.
- Ascomata > 700 µm in diam; ascospores 21–32 × 4–5 µm, allantoid, 3–6 septa; growing in dead wood of Mexico ……………………………………………………………………P. politecnica
- 3.
- Ascospores < 20 µm long, subfusiform or claviform …………………………………………………………………………………4
- 3.
- Ascospores > 20 µm long, obclavate or claviform …………………………………………………………………………………5
- 4.
- Subfusiform ascospores with three septa growing in the stem of the genus Cyathea ……………………………………………………………………P. esperanzae
- 4.
- Ellipsoid ascospores with eight septa growing in dead wood ……………………………………………………………………P. microspora
- 5.
- Obclavate ascospores, 5–6 septa …………………………………………………………………………………6
- 5.
- Clavate ascospores, 4–13 septa …………………………………………………………………………………7
- 6.
- Discoid ascomata, smooth and thick rim, green jungle coloration in KOH (25F8); unilobed pedicellate asci, ascospores 30–32 × 5–7 µm ( = 31 × 6, n = 30); growing in tropical rain forest ………………………………………………………………………P. esquedii
- 6.
- Discoid ascomata, striate and thin rim, coloration nautical blue in KOH (24E7); bilobed pedicel asci, ascospores 30–33 × 6–7 µm ( = 32 × 6, n = 30); growing in Quercus forest “Chaparral” ………………………………………………………………………P. potosina
- 7.
- Clavate ascospores, slightly curved, 4–8 septa, unilobed asci …………………………………………………………………………………8
- 7.
- Clavate ascospores, 8–10 (13) septa, bilobed asci …………………………………………………………………………………15
- 8.
- Ascospores < 30 µm long …………………………………………………………………………………9
- 8.
- Ascospores > 30 µm long …………………………………………………13
- 9.
- Species growing in temperate forests …………………………………………………………………………P. atrata
- 9.
- Species growing in tropical forests or mangroves…………………………………………………………………10
- 10.
- Species growing in mangroves ………………………………………………………………………………11
- 10.
- Species growing in tropical forests; ascospores 26–30 µ ………………………………………………………………………………12
- 11.
- Ascomata with thick rim and turquoise pigmentation in KOH (24F8), ascospores = 26 × 7, 5–6 septa; growing in Rhizophora apiculata ……………………………………………………………………P. apiculate
- 11.
- Ascomata with a thin rim and olive brown pigmentation in KOH (4E8), ascospores = 29 × 7, n = 30, 5–6 septa; growing in Rhizophora mangle ……………………………………………………………………P. mangrovei
- 12.
- Ascomata with thick rim and turquoise pigmentation in KOH (24F8), asci, short pedicellate, ascospores = 28 × 6, n = 30, 4–6 septa; growing in tropical rain forests………………………………………………………………P. ramona
- 12.
- Ascomata with a thin rim with deep green pigmentation in KOH (25D8), asci with long pedicellate, ascospores = 28 × 6, n = 30, 5–7 septa; growing in tropical dry forests ………………………………………………………………P. tropicalis
- 13.
- Ascospores with 5–6 septa ……………………………………………………………P. ghizhuensis
- 13.
- Ascospores with six or more septa……………………………………………………………………14
- 14.
- Ascomata with a thin rim, green jungle pigmentation in KOH (25F8), asci with short pedicellate, ascospores 31–35 × 6–8 µm, = 33 × 7 µm, 6–8 septa; …………………………………………………………P. barronii
- 14.
- Ascomata with thick rim turquoise pigmentation in KOH (24F8), asci long with pedicellate, ascospores 28–30 × 6–7 µm, = 30 × 6 µm, 5–7 septa…………………………………………………………………P. xerofila
- 15.
- Species growing in temperate coniferous forest, ascomata with thin and smooth rims, green jungle pigmentation in KOH(CLA), ascospores with clavate blunt apex ……………………………………………………………………P. garciae
- 15.
- Species in tropical dry forest, ascomata with thick and striate rim, English green pigmentation in KOH, ascospores clavate with conic apex……………………………………………………………………16
- 16.
- Ascospores 32–40 × 6–7 µm, = 36 × 6.2 µm, n = 30, 8–10 (13) septa, asci with long pedicellate, ……………………………………………………………………P. purpurea
- 16.
- Ascospores 38–44 × 6–7 µm, = 40 × 6.6 µm, n = 30, 8–10 septa, asci with short pedicellate………………………………………P. neolonensis
4. Discussion
In Mexico, specimens with discoidal, superficial, black ascomata, smaller than 1 mm, green color in KOH, clavate asci, and clavate ascospores with 5–11 septa have been identified as Patellaria atrata. Méndez-Mayboca described Patellaria atrata growing on sarcocaule shrubland in Puerto Peñasco, Sonora, with ascospores measuring 20–27 × 6–8 µm and 4–5 transverse septa [6]. Chacon and Tapia reported P. atrata from Palenque, Chiapas y Perote, Veracruz [5]. However, in recent polyphasic studies, a polyphasic study on fungi has revealed five species described with morphological and molecular data: P. apiculatae Dayar. and K.D. Hyde and P. chromolaenae Mapook and K.D. Hyde from Thailand [3,11], P. microspora Ekanayaka and K.D. Hyde from the United Kingdom [11], and P. ghizhouensis J.F. Zhang, Y.Y. Chen, and Z.Y. Liu from China [13]. A detailed morphological revision was made, focusing on taxonomically significant characters such as the peridium, pseudoparaphyses, asci, and ascospores (Table 3) of 37 specimens deposited in a fungal collection at the herbarium of Escuela Nacional de Ciencias Biológicas since 2014. This analysis identified 12 morphospecies, of which none coincided with the description of the type species P. atrata. Hence, a phylogenetic analysis was integrated in Figure 1, in which Patellaria was recovered as a monophyletic group within Patellariaceae.

Table 3.
Characters of Patellaria species included in this analysis. XS: Xerophilous scrub, TCMF: Tropical montane cloud forest, TRF: Tropical rain forest, CF: Coniferous forest, TDF: Tropical dry forest, M: Mangrove.
In our phylogenetic analysis using three datasets (LSU, SSU, and ITS), Mexican specimens clustered in two clades. The first clade included specimens classified as Patellaria atrata from Oregon, USA, phylogenetically related to P. quercus, the only Patellaria species described as an asexual morph. Only the conidial phase was defined as conidiomata solitary, black, and erumpent, with 1–3 ostiole with brown setae, conidiophores, hyaline, smooth, and slightly pigmented at the base, 1–5 septa, and measuring 10–25 × 3–4 µm [10]. Patellaria apiculatae was registered as growing on Rhizophora apiculata. The only species in this clade is P. politecnica, related to P. andina and registered in Argentina. However, P. politecnica can be distinguished from alantoid ascospores with 3–6 septa. At the base of this clade are P. microspora and P. guizhouensis, the first from the UK, and P. guizhouensis from Guizhou province in China.
Most Mexican Patellaria species are in clade B, where P. purpurea is related to P. garciae; both have claviform ascospores larger than 40 µm and 8–10 septa. They differ morphologically because P. purpurea has ascomata with an epithecium purple and green jungle in KOH and grows in tropical dry forest and xerophilous scrub in Sonoran forest. In contrast, the other species have ascomata with black epithecium and spruce green in KOH and grow in a boreal coniferous forest in the mountain range in Arteaga, Coahuila. These species are related to P. chroolaenae. However, these species and P. politecnica have alantoid ascospores, which distinguishes them from the other species of the genus. P. tropicalis is phylogenetically related to P. neolenensis, and both grow in dry climates. However, they differ by having ascomata shorter than 800 µm in diameter. Asci with a long unilobed pedicell, 24–37 × 6–7 µm, and ascospores with 5–7 septa. Ascomata larger than 800 µm diameter, and short pedicellate, bilobed asci, claviform ascospores 32.50 × 6–8 µm, and 8–10 septa, respectively. P. xerofila grows in dry climates, too, and it differs by forming a thin and striate peridium, while P. tropicalis shows a smooth and thinner peridium. Moreover, the reaction in KOH of P. xerofila is dark turquoise (24F8), and in P. tropicalis, it is deep green (25D8).
In clade II, P. mangrovei is related to P. esperanzae, contrasting in morphology and habitat. P. mangrovei originates from mangrove communities in Biosphere Reserve Isla Cozumel in the Mexican Caribbean, growing on Rhizophora mangle and exhibiting KOH olive brown pigments (4E8) in KOH, with ascospores measuring 29–30 × 6–7 with 5–8 septa. Furthermore, P. esperanzae grows on tree ferns of the genus Cyathea, and in KOH, its pigments are green olive (27E5), subfusiform ascospores are less than 20 µm, and there are only three septa. Patellaria esquedii grows in the tropical rain forests on the coastal plains of the Gulf of Mexico and in the Yucatán Peninsula. It differs from P. mangrovei and P. esperanzae in ascospore size (25–29 × 6 vs. 31–35 × 5–8 µm) and has fewer septa (4–6 vs. 6–8). This species is closely related to P. potosina and differs in the ascospore shape, which is clavate, and in the number of septa (5–6).
5. Conclusions
This study provides new insights into the genus in Mexico and emphasizes the key characteristics for identifying Mexican species, some of which are restricted to specific types of vegetation, underscoring the importance of conservation strategies in these areas. This study also provides reference information for the study of the genus in Mexico and other regions of the world.
The most relevant morphological characteristics in the identification of Patellaria species are the size of the asci, ascospore shape, and the number of septa, but there are other characters, such as texture and thickness of the peridium and the length and shape of the pedicel, which can be informative in the morphological delimitation of species of the genus.
Additionally, most species are found in xerophilous scrub and tropical dry forests, which shows the affinity of these species for dry climates. This finding provides valuable insights for future studies of the genus in Mexico.
The morphology must not be considered an optional supplement to molecular phylogeny but rather an essential component for species compression and diversification.
Author Contributions
I.G.-J., T.R. and R.V. conceived this study, C.R.M.-G. helped with phylogenetic analysis, I.G.-J., T.R., R.V. and M.M.-P. described new species. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financed by Instituto Politécnico Nacional and their projects: SIP-20240029; SIP20240034; SIP-20240367).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Acknowledgments
All authors thank the authorities of La Esperanza, Abra Tanchipa Biosphere Reserve, Sierra de Álamos Río Cuchujaqui Biosphere Reserve, Sierra de Arteaga, and the Fundación de Parques y Museos de Cozumel (FPMC) for the facilities provided.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fries, E.M. Systema Mycologicum; Gryphiswaldiae: Sumtibus Emesti, Mauritius, 1822; Volume 2, p. 275. [Google Scholar] [CrossRef]
- Species Fungorum. Available online: http://www.speciesfungorum.org/Index.htm (accessed on 30 September 2024).
- Kutorga, E.; Hawksworth, D.L. A re-assessment of the genera referred to the family Patellariaceae (Ascomcota). Syst. Ascomycetum 1997, 15, 1–110. [Google Scholar]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chonunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Yacharoen, S.; Tian, Q.; Chommunti, P.; Boonmee, S.; Chukeatirote, E.; Bhat, J.D.; Hyde, K.D. Patellariaceae revisited. Mycosphere 2015, 6, 290–326. [Google Scholar] [CrossRef]
- Hernandez-Restrepo, M.; Schumacher, R.K.; Wingfield, M.J.; Ishtiaq, A.; Cai, L.; Duong, T.A.; Edwards, J.; Gene, J.; Groenewald, J.Z.; Sana, J.; et al. Fungal systematics and evolution: FUSE 2. Sydowia 2016, 68, 193–230. [Google Scholar]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Lücking, R.; Boonmee, S.; Bhat, J.D.; Liu, N.G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar] [CrossRef]
- Zhang, J.F.; Liu, J.K.; Hyde, K.D.; Chen, Y.; Ran, H.; Liu, Z. Ascomycetes from karst landscapes of Guizhou Province, China. Fungal Divers. 2023, 122, 1–160. [Google Scholar] [CrossRef]
- Chacón, S.; Tapia, F. Algunas especies saprobias de dothideomycetes y lecanoromycetes (Pezizomycotina: Ascomycota) en México. Rev. Mex. Biodivers. 2016, 87, 1169–1176. [Google Scholar] [CrossRef]
- Méndez-Mayboca, F.; Checa, J.; Esqueda, M.; Chacón, S. New records of Loculoascomycetes from natural protected areas in Sonora, Mexico. Mycotaxon 2010, 111, 19–30. [Google Scholar] [CrossRef]
- Esqueda, M.; Coronado, M.L.; Gutiérrez, A.; Lizárraga, M.; Raymundo, T.; Valenzuela, R. Hongos de la Reserva de la Biósfera El Pinacate y Gran desierto de Altar, 1st ed.; Centro de Investigación en Alimentación y Desarrollo (CIAD): Hermosillo, Mexico, 2013; p. 23. [Google Scholar]
- García-Martínez, Y.; Heredia Abarca, G.; Guzmán-Guillermo, J.; Valenzuela, R.; Raymundo, T. Hongos asociados al mangle rojo Rhizophora mangle (Rhizophoraceae) en la Reserva de la Biosfera Isla Cozumel, Quintana Roo, México. Acta Bot. Mex. 2021, 128. [Google Scholar] [CrossRef]
- Raymundo, T.; Martínez-Pineda, M.; Reyes, P.H.; Cobos-Villagrán, A.; Martínez, Y.A.G.; Tun, A.A.; Valenzuela, R. Ascomicetos de la Reserva de la Biosfera Isla Cozumel, México. Act. Bot. Mex. 2021, 128. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978; pp. 1–254. [Google Scholar]
- Martínez-González, C.R.; Ramírez-Mendoza, R.; Jiménez-Ramírez, J.; Gallegos-Vázquez, C.; Luna-Vega, I. Improved method for genomic DNA extraction for Opuntia Mill. (Cactaceae). Plant Methods 2017, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Wanasinghe, D.N.; Cheewangkoon, R.; Al-Sadi, A.M. Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. 2021, 106, 229–268. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Muller, K.; Quandt, D.; Muller, J.; Neinhuis, C. PhyDER-Phylogenetic Data Editor. Program Distributed by the Authors, Ver. 10.0. 2005. Available online: https://www.phyde.de (accessed on 3 September 2024).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.31. 2017. Available online: http://mesquiteproject.org (accessed on 3 September 2024).
- Swofford, D.L. Phylogenetic Analysis Using Parsimony (PAUP*) 4.0; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, P.B.; Calcott, B.; Mayer, C.; Lanfear, R. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evol. Biol. 2015, 15, 13. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v.1.4.2. A Graphical Viewer of Phylogenetic Trees. 2014. Available online: https://tree.bio.ed.ac.uk/software/figtree (accessed on 3 September 2024).
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 135–322. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).











