Abstract
Pleosporales represents the largest order within the class Dothideomycetes (Fungi), comprising phytopathogenic, saprobic, and endophytic taxa with a widespread presence in terrestrial and aquatic environments. Rice (Oryza sativa) is a primary economic crop in numerous tropical countries, particularly in Thailand. Studying fungal species associated with rice holds the potential to enhance our understanding of fungal diversity, lifestyles, and biology of rice, offering valuable insights for future research aimed at disease management and yield improvement. Thirty-nine pleosporalean isolates were obtained from various parts of rice plants collected across diverse regions in Chiang Rai Province, Thailand. Species identification involved a combination of morphology and molecular phylogeny, utilizing multi-locus sequence analyses of the ITS, LSU, SSU, gapdh, rpb2, tef1, and tub2 genes. The isolates were identified in 18 taxa distributed across five families and ten genera, including five new species (Bipolaris chiangraiensis, Ophiosphaerella oryzae, Paraphaeosphaeria oryzae, Pyrenochaetopsis oryzicola, and Setophoma oryzicola). Additionally, six new host records and two new geographical records are documented. Photoplates, detailed morphological descriptions, and phylogenetic trees are provided to elucidate the placement of both known and novel taxa.
1. Introduction
Rice (Oryza sativa, Poaceae) is grown in more than 100 countries, including the Middle East, the West Indies, and Latin America, with 90% of the total global production originating from Asian countries [1]. More than half of the world’s population consumes rice as a staple food, providing more than 20% of the calorie consumption globally because of its many nutrients, which contain fiber, vitamin B, proteins, and lipids [2].
In Thailand, rice is the most significant crop and is grown extensively across the country, comprising approximately half of the total cultivated area [3]. In the history of Thailand, rice is believed to have been originally cultivated as early as 3500 BC. More than 60% of the Thai population consists of farmers, with rice serving as the primary crop for the majority [4]. In 2022, Thailand exported approximately 7.7 million metric tons of rice [5], reinforcing its position as one of the world’s leading and most significant rice producers. However, production of rice in Thailand is affected by various factors.
Plants are exposed to a wide range of biotic and abiotic factors [6]. Among the biotic factors, are the microbial communities or microorganisms associated with plants. They are known as the plant microbiome or microbiota, with fungi playing essential roles, which can impact a host plant positively or negatively [7,8]. Fungi are among the most important pathogenic microorganisms in plants, especially for crop plants in agriculture; however, they are also used as biocontrol agents to prevent and manage plant diseases [9]. Rice is threatened by many pathogenic fungi leading to significant yield losses worldwide, and some of the taxa also release hazardous toxins which may be consumed [10,11]. Several fungi with different nutritional modes also exist in rice, providing many benefits to the plant [12,13,14].
The fungal order, Pleosporales was established by Luttrell [15] and legitimately proposed by Barr [16], primarily centered on Pleosporaceae, with the type species Pleospora herbarum [17]. It is the largest and most diverse order in Dothideomycetes (Ascomycota), encompassing approximately a quarter of all dothideomycetous species [18]. Pleosporalean species are distributed globally as lichenized fungi, pathogens, saprobes, endophytes, and parasites or hyperparasites on fungi and insects [19,20]. Pleosporales was recently listed with 91 families and more than 500 genera [21,22]. The sexual morphs are characterized by perithecioid ascomata, generally with a papillate apex, bitunicate, fissitunicate asci, septate ascospores of various colors and shapes, with or without a sheath [21]. Asexual morphs are mostly coelomycetous; however, sometimes they can be hyphomycetous [18,23,24,25]. This order comprises several important genera possessing both saprobic and pathogenic lifestyles on monocot plants worldwide, such as Alternaria, Bipolaris, Curvularia, and Phaeosphaeria [18,26,27,28,29,30].
Although many studies have reported fungi associated with rice worldwide, the fungal diversity on rice in Thailand is poorly understood. Hence, more investigations are required to discover and record the fungal species diversity in this strategic crop in Thailand. In this study, we focused on genera belonging to Pleosporales isolated from rice in northern Thailand.
2. Materials and Methods
2.1. Sample Collection and Specimen Examination
Rice samples (different tissue parts) of commonly cultivated varieties were collected from different locations in Chiang Rai Province, Thailand, from November 2021 to November 2022. Rice collections consisted of healthy tissues for isolating endophytes, dead plant material for obtaining saprobes, and the leaves with lesions in order to isolate fungi associated with symptoms. Details such as the collection date, location, and where possible, cultivar names were obtained and noted on each collecting bag and the samples were then transferred to the laboratory. The specimens underwent external observation using a Motic SMZ 168 stereomicroscope (Motic, Xiamen, China), and micro-morphological characteristics were studied using a compound microscope equipped with a digital camera (Nikon Eclipse 80i, Nikon, Minato City, Japan). Measurements were acquired using the Tarosoft (R) Image Frame Work program, and subsequent adjustments were made using Adobe Photoshop v. 21.1.3 (Adobe, San Jose, CA, USA).
2.2. Isolation
Fungi were isolated by using both direct and tissue isolation methods [31]. To purify the obtained strains, single spore and hyphal tip isolation techniques were employed, following the protocols outlined by Senanayake et al. [31]. Morphological characteristics were examined directly from the host or after a two-week incubation of obtained cultures. Dried herbarium specimens were archived in the Mae Fah Luang University Herbarium (Herb. MFLU), located in Chiang Rai, Thailand. Living cultures on potato dextrose agar (PDA) were deposited in the Culture Collection of Mae Fah Luang University (MFLUCC). Index Fungorum [32] and Faces of Fungi [33] numbers were obtained for novel taxa.
2.3. DNA Extraction, PCR Amplification and Sequencing
Genomic DNA was extracted from 200 mg mycelia scraped from two-week-old cultures using the OMEGA E.Z.N.A.® Forensic DNA Kit (Omega Bio-tek, Norcross, GA, USA) following the manufacturer’s protocols. PCR amplification reactions were conducted to amplify partial gene regions of internal transcribed spacers (ITS), 28S ribosomal RNA (LSU), 18S ribosomal RNA (SSU), RNA polymerase II second largest subunit (rpb2), β-tubulin (tub2), glyceraldehyde-3-phosphate dehydrogenase (gapdh), and translation elongation factor 1–alpha (tef1) using appropriate primers (Table 1). The PCR mixture comprised 12.5 μL of 2 × Power Taq PCR MasterMix (a premix and ready-to-use solution containing Taq DNA Polymerase, dNTPs, and optimized buffer), 9.5 μL of deionized water, 1 μL of each primer (10 pM), and 1 μL of genomic DNA. Positive amplicons were assessed using agarose gel electrophoresis and visualized on a cybergreen-stained agarose gel under UV light using a molecular imaging system. Positive PCR products were forwarded to SolGent Co., Daejeon, Republic of Korea, for purification and sequencing, employing the same primers.

Table 1.
Details of genes/loci and PCR primers used in this research.
2.4. Sequence Alignment
Individual loci sequences were aligned using MAFFT v. 7.036 (http://mafft.cbrc.jp/alignment/server/large.html, accessed on 21 February 2024) [40] with default settings. Before phylogenetic analyses, the alignments underwent manual editing with BioEdit v. 7.0.5.2 [41]. Newly generated nucleotide sequences were submitted to GenBank, and corresponding accession numbers were included in the appropriate entries.
2.5. Phylogenetic Analyses
The genus of all isolates was confirmed using the BLASTn tool (Basic Local Alignment Search Tool; https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 5 October 2023) at the National Center for Biotechnology Information (NCBI), and related sequence data were subsequently obtained from GenBank and recently published papers [42,43,44,45]. Maximum likelihood (ML) analysis was carried out using RAxML-HPC2 on XSEDE v.8.2.8 [46,47] in the CIPRES Science Gateway platform [48] with the GTRGAMMA evolution model and bootstrap supports with 1000 replicates. Bayesian inference (BI) analysis was conducted using MrBayes v.3.1.2 [49] by Markov chain Monte Carlo sampling (MCMC) based on four simultaneous Markov chains, with 1,000,000 generations and sampling every 100th generation. The first 25% of trees of the burn-in phase were discarded, and the remaining 75% of trees were used for calculating the posterior probability (PP). The phylogenetic trees were customized with FigTree v.1.4.0 [50] and rearranged in Adobe Illustrator CC 22.0.0 (Adobe Systems, San Jose, CA, USA).
3. Results
Out of 53 rice samples collected from 12 locations, 39 fungal isolates were obtained, all identified as belonging to the Pleosporales order. Five new species (Bipolaris chiangraiensis, Ophiosphaerella oryzae, Paraphaeosphaeria oryzae, Pyrenochaetopsis oryzicola, and Setophoma oryzicola) were discovered through micro-morphology and phylogeny analyses, along with six new host records and two new geographical records.
Taxonomy and Phylogeny
Pleosporaceae Nitschke
Pleosporaceae introduced by Nitschke [51] is the largest family in the Pleosporales and is considered the representative of the order [18,21,22,23,52]. The family includes pathogenic or saprobic species on wood and dead herbaceous stems and leaves, as well as some pathogens in humans [53,54]. The sexual morph of this family is recognized by its perithecial ascomata which is usually black and sometimes setose with bitunicate and cylindrical asci and phragmosporous ascospores [21,55]. The asexual morph can be coelomycetous or hyphomycetous with phialidic, annellidic or sympodial blastic conidiogenous cells. The most common genera are Alternaria, Bipolaris, and phoma-like genera, which exist as saprobes or parasites on various host plants [23,56].
Bipolaris chiangraiensis S. Absalan, S. Lumyong & K. D. Hyde, sp. nov. (Figure 1).

Figure 1.
Bipolaris chiangraiensis (MFLU 24-0090, holotype). (a) Appearance of conidial mass on panicle of Oryza sativa; (b) Close-up of conidiophores and conidia on the substrate; (c) Top and reverse of colony on PDA; (d) Conidiophore; (e) Conidiogenous cells and conidia; (f) Base of conidiophore; (g) Unipolar germinating conidium; (h–l) Conidia from immature to mature. Scale bars: (d–e) = 50 μm; (f–l) = 20 μm.
Index Fungorum number: IF902533; Facesoffungi number: FoF 16746
Etymology: Named after Chiang Rai Province from where it was collected.
Saprobic on panicle. Sexual morph: Not observed. Asexual morph: Hyphae 3–5 μm wide, branched, septate, hyaline to pale brown. Conidiophores 140–420 × 5.5–9 μm ( = 280 × 7 μm, n = 10), septate, mostly simple and straight, infrequently branched, micronematous to semi-macronematous, considerably swollen at apex, brown to dark brown. Conidiogenous cells 9–19 × 5–6.5 μm ( = 11 × 5.5 μm, n = 6), terminal or intercalary, sympodial, subcylindrical to slightly swollen, pale brown to brown. Conidia 54–93 × 12–30 μm ( = 70 × 19 μm, n = 20), 5–9-distoseptate, smooth-walled, straight, rarely curved, fusiform, obovoid, sub-cylindrical to broadly swollen, unipolar germination, brown to dark brown, basal cells pale brown.
Culture characteristics: Colonies on PDA reaching 55–58 mm diameter after a week at 27 °C, irregular margin, sparse aerial mycelium, cottony, dark grey to dark olivaceous grey; reverse dark grey.
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, Tha Sut subdistrict, on panicle of Oryza sativa, 17 December 2021, Sahar Absalan, TS102, (MFLU 24-0090, holotype); (ex-type living culture MFLUCC 24-0016).
GenBank numbers: ITS = PQ368326, tef1 = PQ412470, gapdh = PQ412512
Notes: According to phylogenetic analyses, isolate MFLUCC 24-0016 is closely related to Bipolaris marantae with 73% ML and 0.89 BYPP support (Figure 2). However, pairwise DNA sequence comparison revealed that our strain is distinct with 3% base pair differences across 550 nucleotides in the ITS region and 2% base pair differences across 339 nucleotides in gapdh region. The conidia of B. marantae are 80–150 × 12.5–22.5 μm, mostly 5–7 distoseptate and bipolar germination. B. chiangraiensis has smaller conidia, mostly 5–9 distoseptate, and unipolar germination. B. marantae, was isolated from leaf spots on Maranta leuconeura (Marantaceae) in Brazil [57].
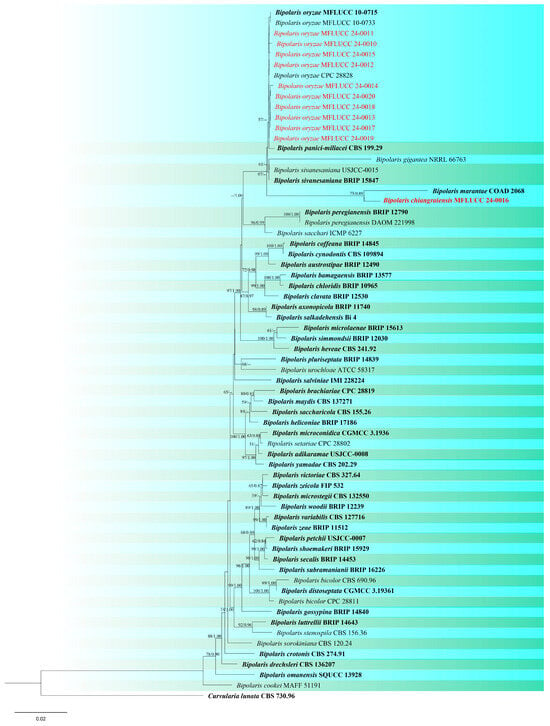
Figure 2.
Phylogenetic tree of Bipolaris generated from a maximum likelihood analysis based on concatenated ITS, gapdh, and tef1 sequence data. Sixty-six strains were included in the combined analyses, which comprised 2013 characters after alignment. Nodes on the tree with bootstrap values ≥50% for IQ-Tree and ≥0.7 for Bayesian posterior probabilities (BYPP) are indicated. Isolates from the current study are in red, while ex-type strains are in bold black. The tree is rooted with Curvularia lunata (CBS 730.96), and the scale bar represents the expected number of nucleotide substitutions per site.
Bipolaris oryzae (Breda de Haan) Shoemaker, Canadian Journal of Botany 37(5): 883 (1959). (Figure S1).
Index Fungorum number: IF482518; Facesoffungi number: FoF 14290
For description see Manamgoda et al. [29].
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, Tha Sut subdistrict, on panicle of Oryza sativa (Poaceae), 17 December 2021, Sahar Absalan, TS97, (MFLU 24-0089); (living culture MFLUCC 24-0015). See Table 2 for details of other strains.

Table 2.
Details of the species obtained in this study.
GenBank numbers: MFLUCC 24-0010: ITS = PQ368316, tef1 = PQ412460, gapdh = PQ412502; MFLUCC 24-0011: ITS = PQ368317, tef1 = PQ412461, gapdh = PQ412503; MFLUCC 24-0012: ITS = PQ368318, tef1 = PQ412462, gapdh = PQ412504; MFLUCC 24-0013: ITS = PQ368319, tef1 = PQ412463, gapdh = PQ412505; MFLUCC 24-0014: ITS = PQ368320, tef1 = PQ412464, gapdh = PQ412506; MFLUCC 24-0015: ITS = PQ368321, tef1 = PQ412465, gapdh = PQ412507; MFLUCC 24-0017: ITS = PQ368322, tef1 = PQ412466, gapdh = PQ412508; MFLUCC 24-0018: ITS = PQ368323, tef1 = PQ412467, gapdh = PQ412509; MFLUCC 24-0019: ITS = PQ368324, tef1 = PQ412468, gapdh = PQ412510; MFLUCC 24-0020: ITS = PQ368325, tef1 = PQ412469, gapdh = PQ412511
Notes: Ten strains clustered with B. oryzae in the combined three-gene analysis. The type strain (MFLUCC 10-0715) was originally isolated from Oryza sativa [58], as were all of the strains in this study. There were no nucleotide differences between the type strain and most of our strains, except for isolate MFLUCC 24-0010 with one nucleotide difference in gapdh, and one in tef1 loci, and isolate MFLUCC 24-0015 and MFLUCC 24-0014 with one and two different nucleotides in gapdh locus respectively. The strains were isolated from different varieties of rice collected from four districts, Mueang Chiang Rai, Phan, Mae Sai, and Mae Chan. Bipolaris oryzae is best known as a significant phytopathogen [59]. However, we isolated eight strains as saprobes from panicles and stems, and two strains were obtained as endophytes from leaves (Table 2).
Curvularia chiangmaiensis Y. Marín, Senwanna & Crous, in Marin-Felix, et al. Mycosphere 8(9): 1565 (2017). (Figure S2).
Index Fungorum number: IF822082; Facesoffungi number: FoF 12887
For description see Marin-Felix et al. [60].
Material examined: Thailand, Chiang Rai Province, Phan District, Leaf spot on Oryza sativa, 16 July 2022, Sahar Absalan, NS169, (MFLU 24-0196); (living culture MFLUCC 24-0024).
GenBank numbers: ITS = PQ358372, tef1 = PQ412459, gapdh = PQ412487
Notes: Based on phylogeny, our isolate is closely related to the type strain of Curvularia chiangmaiensis (CPC 28829) with 65% ML and 0.87 BYPP support (Figure 3). It is challenging to identify most Curvularia species from one another by employing solely morphological characteristics. For precise species identification, the use of both morphological and molecular methods should be considered [61,62]. The pairwise DNA sequence comparison and morphological differences were not noticeable; therefore, we identified this isolate as C. chiangmaiensis based on phylogenetic analysis and morphological comparison. Our strain (MFLUCC 24-0024) differs slightly from CPC 28829 in the shape and size of conidia [60].


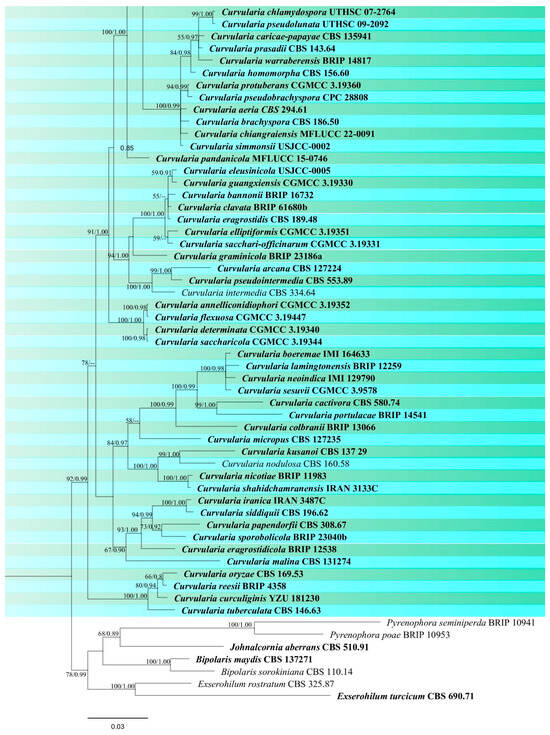
Figure 3.
Phylogenetic tree of Curvularia generated from a maximum likelihood analysis based on concatenated ITS, gapdh, and tef1 sequence data. A total of 177 strains were included in the combined analyses, which comprised 1791 characters after alignment. Nodes on the tree supported by bootstrap values ≥50% for IQ-Tree and ≥0.7 for Bayesian posterior probabilities (BYPP) are indicated. Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with other members of Pleosporaceae (Bipolaris maydis, B. sorokiniana, Exserohilum turcicum, E. rostratum, Johnalcornia aberrans, Pyrenophora poae, and P. seminiperda), and the scale bar represents the expected number of nucleotide substitutions per site.
Curvularia geniculata (Tracy & Earle) Boedijn, Bull. Jard. Bot. Buitenzorg 13 (1): 129 (1923). (Figure S3).
Index Fungorum number: IF265873; Facesoffungi number: FoF 16747
For description see Kusai et al. [63].
Material examined: Thailand, Chiang Rai Province, Wiang Chai District, Winag Chai subdistrict, on panicle of Oryza sativa, 9 November 2021, Sahar Absalan, NS34-1, (MFLU 24-0092); (living culture MFLUCC 24-0022).
GenBank numbers: ITS = PQ358371, tef1 = PQ412458, gapdh = PQ412486
Notes: Based on the morphological and molecular data isolate MFLUCC 24-0022 was identified as a representative of C. geniculata with 88% ML and 0.98 BYPP support (Figure 3). This species is a cosmopolitan fungus that is most prevalent in tropical countries and can occur on a variety of host plant species [45]. Curvularia geniculata is one of the most common Curvularia species found on rice and has been reported in various countries such as Bangladesh, Brunei Darussalam, China, India, Japan, Malaysia, Papua New Guinea, Tanzania, and Uganda. [26,64,65,66]. It was reported as a saprobe on wild bananas in Thailand [67], but there are no records from rice.
Curvularia plantarum M. Raza, K. D. Hyde & L. Cai, in Raza, e al., Fungal Diversity 99: 61 (2019). (Figure S4).
Index Fungorum number: IF556664; Facesoffungi number: FoF 06154
For description see Raza et al. [68].
Material examined: Thailand, Chiang Rai Province, Phan District, on the stem of Oryza sativa, 28 June 2022, Sahar Absalan, PA155, (MFLU 24-0091); (living culture MFLUCC 24-0023).
GenBank numbers: MFLUCC 24-0021: ITS = PQ358369, tef1 = PQ412456, gapdh = PQ375101; MFLUCC 24-0023: ITS = PQ358370, tef1 = PQ412457, gapdh = PQ375102.
Notes: According to the phylogram (Figure 3), isolates MFLUCC 24-0021 and MFLUCC 24-0023 were identified as Curvularia plantarum which was originally reported from Saccharum officinarum in China [68]. Ferdinandez et al. [44] isolated this species from Oryza sativa for the first time from Sri Lanka, which is micro-morphologically more similar to our strains. Our two isolates (MFLUCC 24-0021 and MFLUCC 24-0023) were obtained from panicle spots during the mature stage and stem after harvest, respectively. However, both specimens were collected from the same district (Table 2). C. plantarum is reported for the first time from Thailand.
Phaeosphaeriaceae M.E. Barr
Phaeosphaeriaceae is one of the largest families in Dothideomycetes [21] comprising 84 genera [22], typified by Phaeosphaeria. Barr [69] established Phaeosphaeriaceae and delineated the defining characteristics of this family as either saprobic, pathogenic, or hyperparasitic, typically occurring on herbaceous stems, monocotyledonous leaves, culms, or flowers, and occasionally on woody substrates. Members of Phaeosphaeriaceae frequently occur on plants in Poaceae [70,71]. The sexual morphs have immersed, erumpent or superficial, globose or conical ascomata with short papilla, small to medium, bitunicate asci, and hyaline, yellow or brown, narrowly or broadly obovoid, aseptate or septate ascospores [18,72]. Asexual morph is characterized by its pycnidial conidiomata which are scattered, solitary or gregarious, immersed to superficial, globose to subglobose, glabrous or setose, light brown to dark brown or black, papillate with enteroblastic or holoblastic, annellidic or phialidic conidiogenous cells. Conidia can vary in shape, hyaline to brown, aseptate or septate, some with appendage or mucilaginous sheath [71,73].
Phaeosphaeria musae Sawada, Special Publication College of Agriculture National Taiwan University 8: 66 (1959). (Figure 4 and Figure 5).
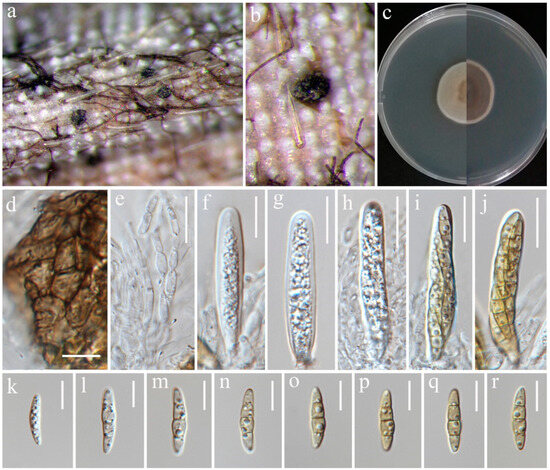
Figure 4.
Sexual morph of Phaeosphaeria musae (MFLU 24-0316, new host record). (a,b) Appearance of ascomata on the panicle of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Peridium; (e) Pseudoparaphyses; (f–j) Asci; (k–r) Ascospores. Scale bars: (d–r) = 10 μm.
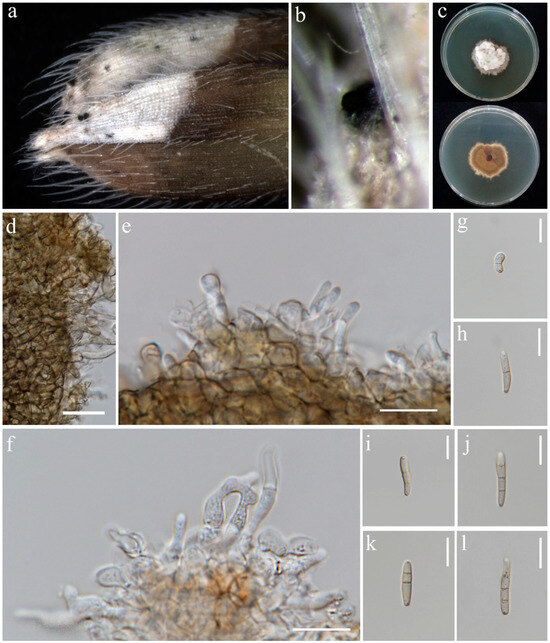
Figure 5.
Asexual morph of Phaeosphaeria musae (MFLU 24-0100). (a,b) Appearance of conidiomata on the panicle of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Peridial wall; (e,f) Conidiogenous cells; (g–l) Conidia. Scale bars: (d) = 20 µm; (e–l) = 10 µm.
Index Fungorum number: IF336200; Facesoffungi number: FoF00262
Saprobic on dead panicle of Oryza sativa. Sexual morph: Ascomata visible as black dots on the surface of the host, scattered, solitary, semi-immersed to superficially raised, ovoid, dark brown to black. Peridium 10–17 μm wide, comprising 2–3 layers of brown to dark brown cell walls of textura angularis. Hamathecium contains numerous 1.5–3 μm wide, cellular, distinctly septate, rarely branching, hyaline pseudoparaphyses. Asci 35–57 × 7.5–10.5 μm (= 44 × 8 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical to slightly clavate, with pointed base, and rounded apex, having indistinct ocular chamber. Ascospores 18–23 × 3–5.5 μm (= 21 × 5 μm, n = 30), fusiform, 3-septate, enlarged at the second cell, slightly constricted at the septa, slightly curved, sometimes straight, rough-walled, yellowish-grey. Asexual morph: Conidiomata pycnidial, solitary, scattered, most of the structure visible on the surface to semi-immersed, globose to ovoid with ostiole, dark brown to black. Peridial wall 15–28 μm wide, comprising several layers of brown, pseudoparenchymatous cells, arranged in a textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 2–4 × 3.5–5.5 μm (= 2.5 × 4.5 μm, n = 10), phialidic, smooth, ampulliform or urceolate, hyaline. Conidia 8–22 × 2–3.5 μm (= 18 × 3 μm, n = 30), solitary, cylindrical, hyaline to pale brown, 1–3-septate, sometimes slightly curved, not constricted at the septa, smooth-walled with guttules.
Culture characteristics: Colonies on PDA reaching 12–13 mm diameter after a week at 27 °C, dense, circular, velvety, pale grey with brown center and yellowish white margin; reverse grey with white margin.
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, on panicle of Oryza sativa, 10 November 2021, Sahar Absalan, RS19-1, (MFLU 24-0316); (living culture MFLUCC 24-0044). Thailand, Chiang Rai Province, Mae Chan District, on panicle of O. sativa, 11 April 2022, Sahar Absalan, M2C136, (MFLU 24-0100); (living culture MFLUCC 24-0046).
GenBank numbers: MFLUCC 24-0044: ITS = PQ358508, LSU = PQ373151, SSU = PQ373161, tef1 = PQ412471; MFLUCC 24-0045: ITS = PQ358509, LSU = PQ373152, SSU = PQ373162, tef1 = PQ412472; MFLUCC 24-0046: ITS = PQ358510, LSU = PQ373153, SSU = PQ373163, tef1 = N/A
Notes: All three strains have a close phylogenetic affinity to the ex-type strain of Phaeosphaeria musae (MFLUCC 11-0151) with 60% ML and 0.99 BYPP support (Figure 6). Isolates MFLUCC 24-0044 and MFLUCC 24-0045 were found as sexual morphs and isolated from the post-harvest rice panicle. The asexual morph (strain MFLUCC 24-0046) was isolated from a symptomatic rice panicle (variety Pathum Thani 60) (Table 2). To our knowledge, this is the first report of the asexual morph of P. musae, and a new host record from rice. Herein, we establish the sexual–asexual connection of Phaeosphaeria musae.
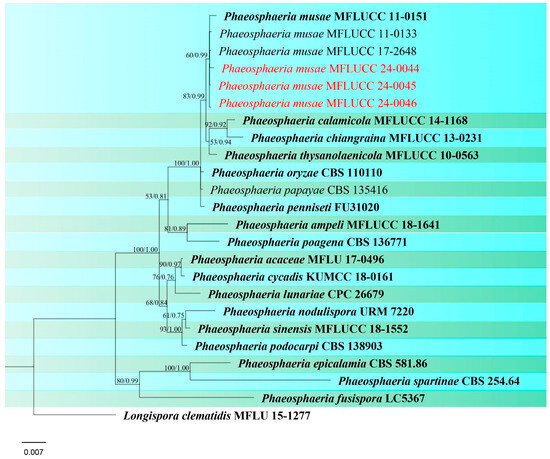
Figure 6.
Phylogenetic tree of Phaeosphaeria generated from a maximum likelihood analysis based on concatenated ITS, LSU, SSU, and tef1 sequence data. A total of 24 strains were included in the combined analyses, which comprised 2986 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for RAxML and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Longispora clematidis (MFLU 15-1277), and the scale bar represents the expected number of nucleotide substitutions per site.
Setophoma poaceicola Goonas., Thambug. & K. D. Hyde, Mycosphere 8(4): 763 (2017). (Figure 7).
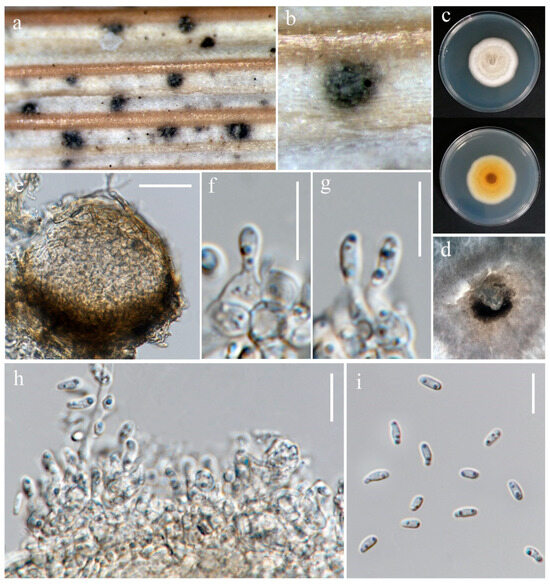
Figure 7.
Setophoma poaceicola (MFLU 24-0318). (a,b) Appearance of conidiomata on the stem of Oryza sativa; (c) Top and reverse of the colony on PDA; (d) Conidiomata superficial on PDA after four weeks; (e) Section through pycnidium. (f–h) Conidiogenous cells. (i) Conidia. Scale bars: (e) = 30 μm; (f–i) = 20 μm.
Index Fungorum number: IF552993; Facesoffungi number: FoF 03212
Saprobic on dead stem of Oryza sativa. Sexual morph: Not observed. Asexual morph: Conidiomata 90–150 μm diameter, pycnidial, semi-immersed, globose to subglobose, with an opening at apex, with dark brown pycnidial wall. Conidiophores reduced to conidiogenous cells lining the inner cavity. Conidiogenous cells 3–5 × 2.5–4.5 μm (= 4 × 4.5 μm, n = 10), hyaline, aseptate, smooth, ampulliform or sometimes irregular. Conidia 4–6.5 × 2–2.5 μm (= 5.5 × 2 μm, n = 30), smooth, cylindrical to oblong, aseptate, rounded at ends, straight or slightly curved, hyaline.
Culture characteristics: Colonies on PDA reaching 30–32 mm diameter after a week at 27 °C, flat with entire edge, circular, fluffy, pale grey in the center, white at the edge; reverse pale brown in the center.
Material examined: Thailand, Chiang Rai Province, Phan District, on stem of Oryza sativa, 28 June 2022, Sahar Absalan, PA156, (MFLU 24-0318); (living culture MFLUCC 24-0034).
GenBank numbers: MFLUCC 24-0033: ITS = PQ376618, LSU = PQ376621, rpb2 = N/A, tef1 = PQ412474; MFLUCC 24-0034: ITS = PQ376619, LSU = PQ376622, rpb2 = PQ412500, tef1 = PQ412475.
Notes: The ex-type strain of S. poaceicola (MFLUCC 16–0880) was originally isolated from a dead culm of grass as a saprobic fungus in Thailand [74]. Two strains isolated from Chiang Rai (Wiang Chai and Phan) were identified as S. poaceicola with 100% ML and 1.00 BYPP support (Figure 8). Recently, this species was isolated from rice in Indonesia, causing sheath rot [75]. However, our strains were isolated as saprobes from the stem (MFLUCC 24-0034) and panicle (MFLUCC 24-0033) (Table 2). Even though this species was found as a saprobe from Thailand, it may be an emerging pathogen. All studies reported S. poaceicola by its sexual morph, and this is the first report of this species in the asexual morph.
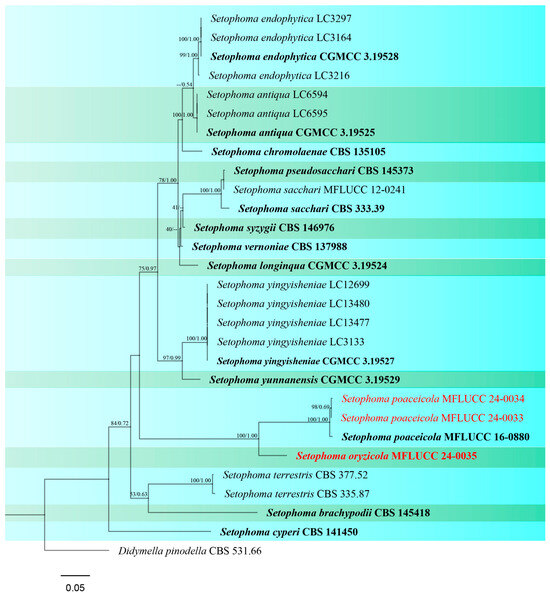
Figure 8.
Phylogenetic tree of Setophoma generated from a maximum likelihood analysis based on concatenated ITS, LSU, rpb2, and tef1 sequence data. A total of 29 strains were included in the combined analyses, which comprised 2509 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for RAxML and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Didymella pinodella (CBS 531.66), and the scale bar represents the expected number of nucleotide substitutions per site.
Setophoma oryzicola S. Absalan, S. Lumyong & K. D. Hyde, sp. nov. (Figure 9).
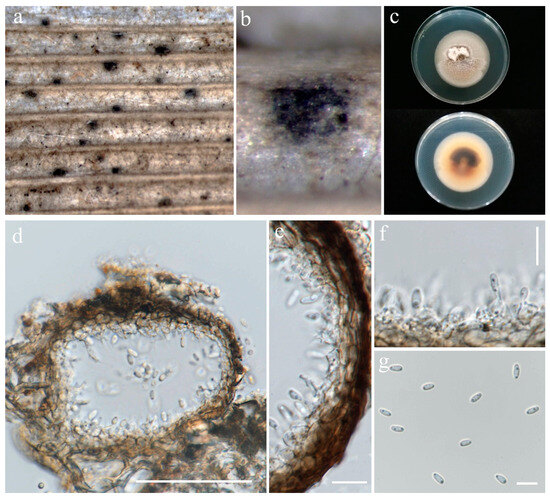
Figure 9.
Setophoma oryzicola (MFLU 24-0097, holotype). (a,b) Appearance of conidiomata on the stem of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Section through pycnidium; (e) Pycnidial wall; (f) Conidiogenous cells; (g) Conidia. Scale bars: (d) = 50 µm; (e,f) = 10 µm; (g) = 5 µm.
Index Fungorum number: IF902536; Facesoffungi number: FoF 14289
Etymology: Name refers to the host genus Oryza from which it was isolated.
Saprobic on stem of Oryza sativa. Sexual morph: Not observed. Asexual morph: Conidiomata 71–127 μm diameter, 45–76 μm height, pycnidial, scattered, immersed to semi-immersed, globose to subglobose, dark brown to black. Pycnidial wall 5–11 μm wide, brown, composed of 3–5 layers of cell walls arranged in a textura angularis. Conidiophores reduced to conidiogenous cells lining inner cavity. Conidiogenous cells 3–5 × 3–5.5 μm (= 3.5 × 4.5 μm, n = 10), phialidic, hyaline, aseptate, smooth, urceolate to ovoid or sometimes ampulliform. Conidia 3.5–6 × 2–3 μm (= 5 × 2.5 μm, n = 30), smooth, aseptate, ellipsoid to oblong, rounded at ends, with two guttulates, hyaline.
Culture characteristics: Colonies on PDA reaching 16–17 mm diameter after a week at 27 °C, medium dense, irregular, with rhizoid edge, slightly raised, fluffy, white; reverse yellowish-brown.
Material examined: Thailand, Chiang Rai Province, Phan District, on stem of Oryza sativa, 28 June 2022, Sahar Absalan, PA157, (MFLU 24-0097, holotype); (ex-type living culture MFLUCC 24-0035).
GenBank numbers: ITS = PQ376620, LSU = PQ376623, rpb2 = PQ412501, tef1 = PQ412476
Notes: Setophoma oryzicola was isolated from a stem of rice as a saprobic fungus (Table 2). In our phylogenetic analyses (Figure 8), S. oryzicola is located on a separate branch, forming a well-supported lineage (100% ML and 1.00 BYPP) basal to S. poaceicola clade. In a pairwise comparison, S. oryzicola and S. poaceicola (MFLUCC 16–0880) differ in ITS (15.2%), LSU (1.7%), and RPB2 (24.4%). It is challenging to differentiate between these two species based on morphology. However, conidiogenous cells in S. poaceicola are mainly ampulliform, while they are usually urceolate in S. oryzicola. Hence, we introduce S. oryzicola as a new species based on morphological examination and phylogenetic analysis.
Ophiosphaerella agrostidis Dern., M.P.S. Câmara, N.R. O’Neill, Berkum & M.E. Palm, Mycologia 92(2): 320 (2000). (Figure S5).
Index Fungorum number: IF464614; Facesoffungi number: FoF00258
For description see Câmara et al. [76].
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, Huai Sak subdistrict, on stem of Oryza sativa, 17 December 2021, Sahar Absalan, HS58-1, (MFLU 24-0094); (living culture MFLUCC 24-0029).
GenBank numbers: ITS = PQ374211, LSU = PQ374216, SSU = PQ394981, tef1 = PQ412454.
Notes: Phylogenetic analyses of concatenated ITS, LSU, SSU, and tef1 sequence data showed that our strain, MFLUCC 24-0029, is grouped with Ophiosphaerella agrostidis (Figure 10) which was first reported from Agrostis palustris [76]. Most of the species within Ophiosphaerella are pathogens on various hosts; however, studies in Thailand revealed that O. agrostidis can thrive as a saprobe [71,74]. In this study, MFLUCC 24-0029 was isolated from a dead rice plant stem (Table 2), representing a new host record from Oryza sativa.
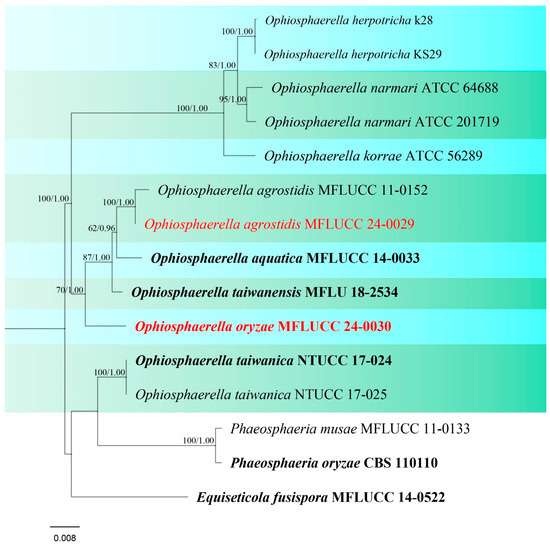
Figure 10.
Phylogenetic tree of Ophiosphaerella generated from a maximum likelihood analysis based on a concatenated ITS, LSU, SSU, and tef1 sequence data. A total of 15 strains were included in the combined analyses, which comprised 3262 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for IQ-Tree and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Phaeosphaeria musae (MFLUCC 11-0133), P. oryzae (CBS 110110), and Equiseticola fusispora (MFLUCC 14-0522), and the scale bar represents the expected number of nucleotide substitutions per site.
Ophiosphaerella oryzae S. Absalan, S. Lumyong & K. D. Hyde, sp. nov. (Figure 11).

Figure 11.
Ophiosphaerella oryzae (MFLU 24-0095, holotype). (a,b) Appearance of ascomata on the stem of Oryza sativa; (c) Section through ascoma; (d) Ascoma with ostiole; (e) Peridium; (f) Ascogenous cells; (g–j) Asci; (k,l) Ascospores. Scale bars: (c) = 100 µm; (d) = 50 µm; (e) = 10 µm; (f–l) = 20 µm.
Index Fungorum number: IF902534; Facesoffungi number: FoF 16748
Etymology –Refers to the host genus Oryza from which it was isolated.
Saprobic on dead stem of Oryza sativa. Sexual morph: Ascomata 169–255 μm diameter, solitary, globose to subglobose, semi-immersed with erumpent neck, ostiolate, dark brown to black. Ostiole 30–101 × 90–115 μm (= 83 × 103 μm, n = 5) diameter, papillate. Peridium 20–35 μm wide, comprising 4–6 layers of dark brown pseudoparenchymatous cells arranged in a textura angulari forming a fairly thick wall. Asci 41.5–123 × 6–11.5 μm (= 83 × 7.5 μm, n = 20), 8-spored, arising from simple aseptate ascogenous cells, bitunicate, cylindrical and sometimes slightly curved, pedunculated, narrower toward the base. Ascospores 90–145 × 3–4 μm (= 118 × 3.5 μm, n = 30), filiform, smooth-walled, with 13 transverse septa, lying parallel or partly spirally twisted in ascus, yellow to pale brown. Asexual morph: Not observed.
Culture characteristics: Colonies on PDA reaching 17–18 mm diameter after a week at 27 °C, slightly raised, floccose to cottony, brownish grey with irregular fluffy white edge; reverse brown with irregular white edge.
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, Huai Sak subdistrict, on stem of Oryza sativa, 17 December 2021, Sahar Absalan, HS68-1, (MFLU 24-0095, holotype); (ex-type living culture MFLUCC 24-0030).
GenBank numbers: ITS = PQ374212, LSU = PQ374217, SSU = PQ394982, tef1 = PQ412455.
Notes: Ophiosphaerella oryzae is proposed as a new species based on the multi-gene phylogenetic analyses and morphology. Our strain (MFLUCC 24-0030), clustered in a clade with other species, comprising O. agrostidis, O. aquatica, and O. taiwanensis, in a separate branch with 70% ML, and 1.00 BYPP support (Figure 10). A comparison of the nucleotide differences between O. oryzae and the closely related species, O. taiwanensis, revealed 0.46% (across 428 nucleotides) and 6.3% (across 620 nucleotides) base pair differences in ITS and tef1 gene regions, respectively. Morphological characters of Ophiosphaerella oryzae has smaller ascomata, shorter ostiole, and lacks periphyses in the ostiole when compared to O. taiwanensis [77].
Pyrenochaetopsidaceae Valenz.-Lopez, Crous, Cano, Guarro & Stchigel
Valenzuela-Lopez et al. [78] introduced Pyrenochaetopsidaceae to accommodate Pyrenochaetopsis as the type genus. Based on their phylogenetic studies, Neopyrenochaetopsis and Xenopyrenochaetopsis were also included in this family. The family was initially characterized by its asexual morph until Mapook et al. [20] reported the sexual morph of Pyrenochaetopsis for the first time. The family contains species that have been found in different habitats such as plants, water, soil, cysts of plant-parasitic nematodes, and as opportunistic infections in humans [78].
Pyrenochaetopsis indica (T.S. Viswan.) de Gruyter, Aveskamp & Verkley, Mycologia 102(5): 1077 (2010). (Figure S6).
Index Fungorum number: IF514656; Facesoffungi number: FoF 16503
For description see de Gruyter et al. [79].
Material examined: Thailand, Chiang Rai Province, Phan District, on panicle of Oryza sativa, 9 November 2021, Sahar Absalan, NS22-1, (MFLU 24-0197); (MFLUCC 24-0036).
GenBank numbers: MFLUCC 24-0036: ITS = PQ376610, LSU = PQ376624, rpb2 = N/A, tub2 = PQ412483; MFLUCC 24-0037: ITS = PQ376611, LSU = PQ376625, rpb2 = PQ412493, tub2 = PQ412484.
Notes: In the combined ITS, LSU, rpb2, and tub2 phylogeny, our two isolates, MFLUCC 24-0036 and MFLUCC 24-0037, clustered with the ex-type strain of Pyrenochaetopsis indica (CBS 124454) with 95% ML, and 0.98 BYPP support (Figure 12). Pyrenochaetopsis indica was originally introduced as Pyrenochaeta indica by de Gruyter et al. [80], isolated from Saccharum officinarum leaves in India. Valenzuela-Lopez et al. [78] presented new genomic sequence data for the ex-type strain of Pyrenochaeta indica and introduced a new genus, Pyrenochaetopsis, typified by Pyrenochaetopsis indica. However, adequate micro-morphological characteristics of this species were not available prior to the present study. We illustrate P. indica on rice as a new host record isolated from panicle and stem, collected from two locations in Chiang Rai Province (Table 2).
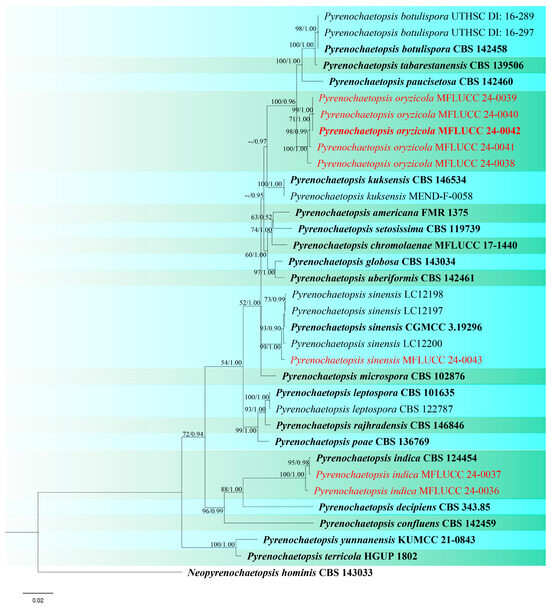
Figure 12.
Phylogenetic tree of Pyrenochaetopsis generated from a maximum likelihood analysis based on concatenated ITS, LSU, rpb2, and tub2 sequence data. A total of 35 strains were included in the combined analyses, which comprised 2344 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for RAxML and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Neopyrenochaetopsis hominis (CBS 143033), and the scale bar represents the expected number of nucleotide substitutions per site.

Figure 13.
Sexual morph of Pyrenochaetopsis oryzicola (MFLU 24-0098, holotype). (a,b) Appearance of ascomata on the panicle of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Ascoma; (e) Peridium; (f) Ascogenous cells bearing asci; (g–j) Asci; (k–m) Ascospores. Scale bars: (d) = 20 µm; (e–m) = 10 µm.
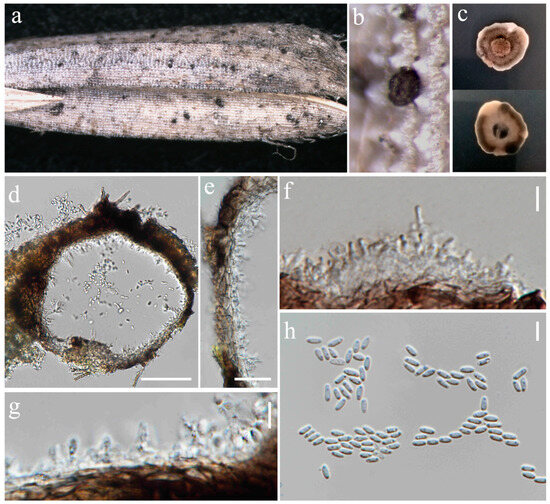
Figure 14.
Asexual morph of Pyrenochaetopsis oryzicola (MFLU 24-0319, epitype). (a,b) Appearance of conidiomata on the panicle of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Section through pycnidium; (e) Pycnidial wall; (f,g) Conidiogenous cells; (h) Conidia. Scale bars: (d) = 50 µm; (e) = 10 µm; (f–h) = 5 µm.
Index Fungorum number: IF902535; Facesoffungi number: FoF 16749
Etymology: Refers to the host genus Oryza from which it was isolated.
Saprobic on panicle of Oryza sativa. Sexual morph: Ascomata 76–113 μm diameter, 60–98 μm high, gregarious, scattered to clustered, globose with a short cylindrical opening at apex, mostly visible superficially on the host, sometimes semi-immersed, dark brown to black. Peridium 5–11 μm wide, having equal thickness, greyish-brown to brown with several layers of flattened, pseudoparenchymatous cells. Asci 42–84 × 7–10 μm (= 53.5 × 8.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical to slightly clavate, with short pedicel, apically rounded. Ascospores 16–19 × 3.5–5.5 μm (= 18 × 4 μm, n = 30), fusiform, slightly curved, 1–3-septate, slightly constricted at central septum, not constricted at other septa, smooth-walled, enlarged at second cell from above, hyaline as immature, becoming shiney yellowish-brown when mature, with small guttules. Asexual morph: Conidiomata 136–141 μm diameter, 98–116 μm high, visible as raised and superficial on the host, solitary, scattered, sometimes clustered, globosoe to slightly ovoid, dark brown. Pycnidial wall 6.5–22 µm wide, composed of several layers of pseudoparenchymatous cells, outer layers pigmented with brown, and becoming paler towards the inner layers. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 0.5–2.5 × 1–2 μm (= 1.5 × 1 μm, n = 15), phialidic, hyaline, aseptate, emerging from the cavity inside of the pycnidial wall, difficult to distinguish from the pycnidial wall. Conidia 3.5–5 × 1.5–2 μm (= 4.5 × 2 μm, n = 30), aseptate, cylindrical to ellipsoid, with two guttules, hyaline.
Culture characteristics: Colonies on PDA reaching 31–33 mm diameter after a week at 27 °C, circular, medium dense, cottony, olivaceous grey with pale grey margin; reverse grey.
Material examined: Thailand, Chiang Rai Province, Phan District, on dead panicle of Oryza sativa, 9 November 2021, Sahar Absalan, NS28-2a, (MFLU 24-0098, holotype); (ex-type living culture MFLUCC 24-0038). Thailand, Chiang Rai Province, Mueang Chiang Rai District, Tha Sut subdistrict, on dead panicle of O. sativa, 17 December 2021, Sahar Absalan, TS110, (MFLU 24-0319, epitype, designated here); (ex-epitype living culture MFLUCC 24-0042).
GenBank numbers: MFLUCC 24-0038: ITS = PQ376612, LSU = PQ376626, rpb2 = PQ412494, tub2 = PQ412478; MFLUCC 24-0039: ITS = PQ376613, LSU = PQ376627, rpb2 = PQ412495, tub2 = PQ412479; MFLUCC 24-0040: ITS = PQ376614, LSU = PQ376628, rpb2 = PQ412496, tub2 = PQ412480; MFLUCC 24-0041: ITS = PQ376615, LSU = PQ376629, rpb2 = PQ412497, tub2 = PQ412481; MFLUCC 24-0042: ITS = PQ376616, LSU = PQ376630, rpb2 = PQ412498, tub2 = PQ412482
Notes: According to the phylogenetic analyses, our five strains grouped in a single clade, distinct from closely related species (Figure 12). Three strains (MFLUCC 24-0038, MFLUCC 24-0041, MFLUCC 24-0042) were isolated from the panicle samples, while two strains (MFLUCC 24-0039, MFLUCC 24-0040) were obtained from the stem. Among the isolates, MFLUCC 24-0039, MFLUCC 24-0040, MFLUCC 24-0041, and MFLUCC 24-0042, collected from the Mueang Chiang Rai district, all displayed asexual morphs. In contrast, strain MFLUCC 24-0038, collected from the Phan district, developed a sexual morph (Table 2). Pyrenochaetopsis oryzicola is described as a new species based on morphological characteristics of sexual and asexual morphs, as well as phylogenetic analysis of multigene sequence data. We also established the connection between the sexual and asexual stages of this species. Strain MFLU 24-0319 is designated an epitype as it is important to also have authentic material of the asexual morph.
Pyrenochaetopsis sinensis G.S. Li, J.M. Liang & L. Cai, Fungal Diversity 96: 65 (2019). (Figure 15).
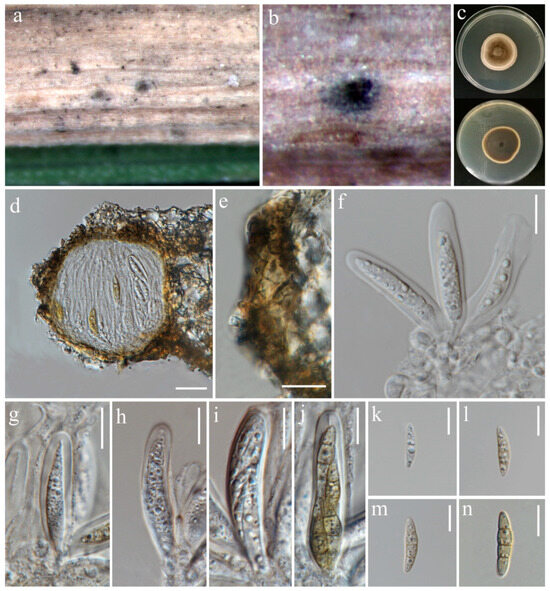
Figure 15.
Pyrenochaetopsis sinensis (MFLU 24-0099, new host record). (a,b) Appearance of ascomata on the leaf of Oryza sativa; (c) Top and reverse of colony on PDA; (d) Section through ascoma; (e) Peridium; (f) Ascogenous cells bearing asci; (g–j) Asci; (k–n) Ascospores. Scale bars: (d) = 20 µm; (e–n) = 10 µm.
Index Fungorum number: IF556011; Facesoffungi number: FoF 05965
On leaf spots of Oryza sativa. Sexual morph: Ascomata 75–90 μm diameter, 63–70 μm high, solitary, immersed to semi-immersed, globose to sub-globose, dark grey to black, with a dull appearance Peridium 13–30 μm wide, thin at the apex, thick at both sides, composed of several layers of pseudoparenchymatous cells, yellowish-brown to hyaline toward the inner layers Asci 25–45 × 7–9 μm (= 36 × 7.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical to clavate, rounded at the apex. Ascospores 13–23 × 3–4.5 μm (= 21 × 4 μm, n = 30), fusiform to oblong, 4-septate, second cell from top enlarged, narrower toward the end cells, slightly curved, smooth-walled, hyaline when immature, turning yellowish-grey when mature, with small guttules. Asexual morph: Not observed.
Culture characteristics: Colonies on PDA reaching 18–21 mm diameter after a week at 27 °C, circular, cottony, olivaceous grey with yellowish white margin; reverse dark grey with yellowish white margin.
Material examined: Thailand, Chiang Rai Province, Mae Sai District, on leaf spot of Oryza sativa, 11 April 2022, Sahar Absalan, M2S120, (MFLU 24-0099); (living culture MFLUCC 24-0043).
GenBank numbers: ITS = PQ376617, LSU = PQ376631, rpb2 = PQ412499, tub2 = PQ412485
Notes: Pyrenochaetopsis sinensis was introduced as a new species by its asexual morph, isolated from soil [81]. In this study, the sexual morph of P. sinensis is reported and illustrated for the first time, isolated from leaf spots of Oryza sativa (variety: RD-MAEJO2) (Table 2). In the phylogenetic analysis our strain grouped with the ex-type (CGMCC 3.19296) and three strains of P. sinensis (Figure 12). The sexual morph of closely related species is unavailable for morphological comparison. Herein, P. sinensis is reported as a new host record and its sexual morph is illustrated for the first time.
Didymellaceae Gruyter, Aveskamp & Verkley
de Gruyter et al. [82] established the Didymellaceae to include the three genera Ascochyta, Didymella (type genus), and Phoma as well as other related phoma-like species that were classified in this family. Didymellaceae contains 44 genera [22], found in a wide range of habitats worldwide. Most members are plant pathogens that affect a variety of hosts and mostly cause lesions on leaves and stems [83,84].
Epicoccum catenisporum Valenz.-Lopez, Stchigel, Crous, Guarro & J.F. Cano, Studies in Mycology 90: 30 (2017). (Figure S7).
Index Fungorum number: IF819762; Facesoffungi number: FoF 16706
For description see Valenzuela-Lopez et al. [78].
Material examined: Thailand, Chiang Rai Province, Wiang Chiang Rung District, on leaf spot of seedling of Oryza sativa, 4 July 2022, Sahar Absalan, WCR164, (MFLU 24-0093); (living culture MFLUCC 24-0028).
GenBank numbers: MFLUCC 24-0025: ITS = PQ358494, LSU = PQ358538, rpb2 = PQ412489; MFLUCC 24-0027: ITS = PQ358495, LSU = PQ358539, rpb2 = PQ412490; MFLUCC 24-0028: ITS = PQ358496, LSU = PQ358540, rpb2 = PQ412491
Notes: Epicoccum catenisporum (CBS 181.80, ex-type) was formerly identified as Phoma sorghina, isolated from leaf spots of rice in Guinea-Bissau [78,85]. Three isolates (MFLUCC 24-0025, MFLUCC 24-0027, MFLUCC 24-0028) collected from three locations in Chiang Rai are phylogenetically related to E. catenisporum. Of these, two strains (MFLUCC 24-0025 and MFLUCC 24-0028) were isolated from leaf spots on seedlings, including two different rice varieties (Pathum Thani 60 and RD6). MFLUCC 24-0027 was obtained from a dead stem collected from Phan district as a saprobic fungus (Table 2). This is a new geographical record of Epicoccum catenisporum from Thailand.
Epicoccum latusicollum Q. Chen, Crous & L. Cai, Studies in Mycology 87: 144. (Figure S8).
Index Fungorum number: IF818960; Facesoffungi number: FoF 06580
For description see Chen et al. [86].
GenBank numbers: ITS = PQ358497, LSU = PQ358541, rpb2 = PQ412492
Notes: Our isolate (MFLUCC 24-0026) clustered with Epicoccum latusicollum (CGMCC 3.18346, ex-type) with 93% ML, and 1.00 BYPP support in the combined ITS, LSU, and rpb2 phylogenetic analysis (Figure 16). The type of E. latusicollum was reported from leaves of Sorghum bicolor in China. Other hosts associated with this species include leaves of Vitex negundo and Camellia sinensis (as an endophyte) in China, and Podocarpus macrophyllus in Japan [86]. In our study, this species was isolated from rice leaves collected in Mae Chan district of Chiang Rai, Thailand, and is reported here as a new host record based on phylogenetic analysis and morphological assessment.
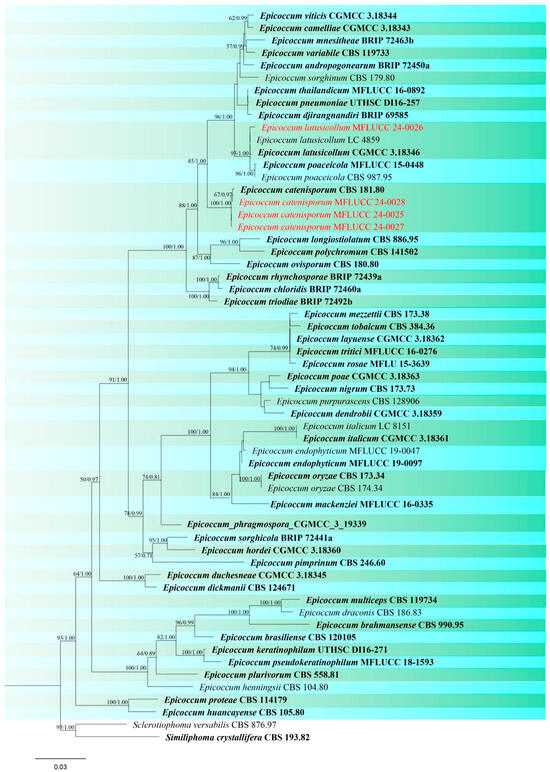
Figure 16.
Phylogenetic tree of Epicoccum generated from a maximum likelihood analysis based on concatenated ITS, LSU, and rpb2 sequence data. A total of 58 strains were included in the combined analyses, which comprised 1993 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for RAxML and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Sclerotiophoma versabilis (CBS 876.97) and Similiphoma crystallifera (CBS 193.82), and the scale bar represents the expected number of nucleotide substitutions per site.
Remotididymella capsici (Bond.-Mont.) L.W. Hou, L. Cai & Crous, Studies in Mycology 96: 337 (2020). (Figure S9).
Index Fungorum number: IF834117; Facesoffungi number: FoF 16497
Material examined: Thailand, Chiang Rai Province, Wiang Chai District, endophytic from seedling of Oryza sativa, 4 July 2022, Sahar Absalan, WC167a, (living culture MFLUCC 24-0031).
GenBank numbers: ITS = PQ358526, LSU = PQ373118, rpb2 = PQ412488
Notes: Remotididymella capsici was introduced by Hou et al. [87] isolated from leaf spots on Capsicum annuum in Fiji, which was originally identified as Ascochyta capsici. However, phylogenetic studies revealed that the ex-type strain (CBS 679.77) was clustered in Remotididymella resulting in a separate lineage from other species (Figure 17). Morphological characteristics do not exist since their culture remained sterile. In this study, we obtained one strain identified as R. capsici, which was isolated as an endophytic fungus from a symptomless seedling a new host record of rice and new geographical record from Thailand. The isolate also did not produce any fruiting body in culture.
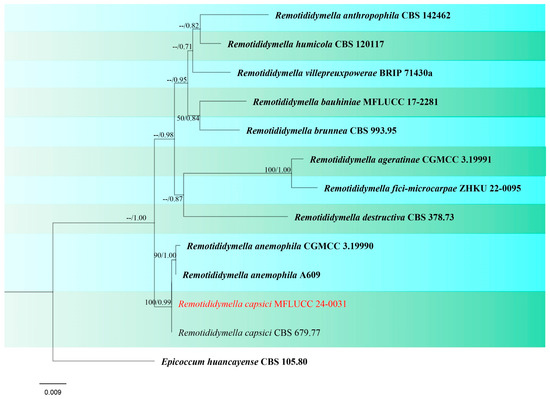
Figure 17.
Phylogenetic tree of Remotididymella generated from a maximum likelihood analysis based on concatenated ITS, LSU, and rpb2 sequence data. A total of 13 strains were included in the combined analyses, which comprised 1570 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for RAxML and ≥0.7 for Bayesian posterior probabilities (BYPP). The isolate from the current study is in red, while ex-type strains are in bold black. The tree was rooted with Epicoccum huancayense (CBS 105.80), and the scale bar represents the expected number of nucleotide substitutions per site.
Didymosphaeriaceae Munk
Didymosphaeriaceae was introduced by Munk [88] and encompasses 33 genera [22]. Didymosphaeria Fuckel is considered the type genus of Didymosphaeriaceae. The family includes species that are saprotrophic, endophytic, and pathogenic occurring on a wide range of substrates such as wood and herbaceous stems, pods, leaves, and soil [54,89,90]. The sexual morph of Didymosphaeriaceae is characterized by globose to subglobose ascomata with central ostiole; cellular or trabeculate pseudoparaphyses; 2–4-spored or 8-spored, bitunicate, pedicellate asci; 1–3-septate or muriform ascospores which are pigmented and in ellipsoid or oblong shapes. The asexual morph can be fusicladium-like and phoma-like [18,21].
Pseudopithomyces chartarum (Berk. & M.A. Curtis) Jun F. Li, Ariyaw. & K. D. Hyde. (Figure S10).
Index Fungorum number: IF551393; Facesoffungi number: FoF 00938
For description see Ariyawansa et al. [91].
Material examined: Thailand, Chiang Rai Province, Wiang Chiang Rung District, on leaf spot of Oryza sativa, 4 July 2022, Sahar Absalan, WCR162, (MFLU 24-0101); (living culture MFLUCC 24-0048).
GenBank numbers: MFLUCC 24-0047: ITS = PQ358511, LSU = PQ380128, SSU = PQ380126, tef1 = N/A; MFLUCC 24-0048: ITS = PQ358512, LSU = PQ380129, SSU = PQ380127, tef1 = PQ412473.
Notes: The first occurrence of Pseudopithomyces chartarum (formerly known as Pithomyces chartarum) on rice was reported in India, causing glume blotch of rice [92]. Phylogenetic analyses of concatenated ITS, LSU, SSU, and tef1 sequence datasets indicated that our two strains (MFLUCC 24-0047 and MFLUCC 24-0048) have a close affinity with Pseudopithomyces chartarum with 82% ML, 0.96 BYPP support (Figure 18). Therefore, based on similarity in the morphological characteristics and phylogenetic analysis, we identified the strains as P. chartarum. The two strains were isolated from two different parts of the rice plant, panicle (rice cultivar: RD15) and leaf (rice cultivar: RD6), collected from two districts (Phan and Wiang Chiang Rung) in Chiang Rai Province (Table 2).
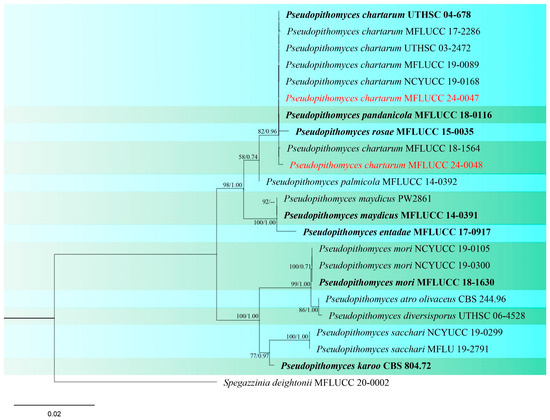
Figure 18.
Phylogenetic tree of Pseudopithomyces generated from a maximum likelihood analysis based on concatenated ITS, LSU, SSU, and tef1 sequence data. A total of 23 strains were included in the combined analyses, which comprised 3009 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for IQ-Tree and ≥0.7 for Bayesian posterior probabilities (BYPP). Isolates from the current study are in red, while ex-type strains are in bold black. The tree was rooted with Spegazzinia deightonii (MFLUCC 20-0002), and the scale bar represents the expected number of nucleotide substitutions per site.
Paraphaeosphaeria oryzae S. Absalan, S. Lumyong & K. D. Hyde, sp. nov. (Figure 19).
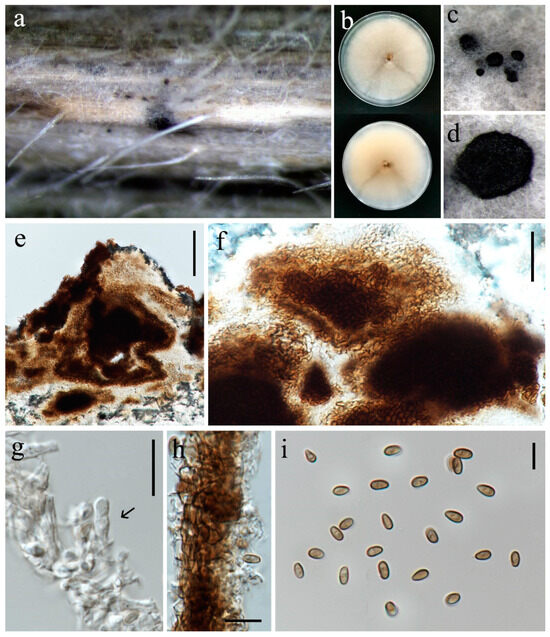
Figure 19.
Paraphaeosphaeria oryzae (MFLU 24-0096, holotype). (a) Appearance of conidiomata on leaf of Oryza sativa; (b) Top and reverse of colony on PDA; (c,d) Conidiomata superficial on PDA after three weeks; (e,f) Cut through the pycnidium; (g) Conidiogenous cell (arrow); (h) Pycnidial wall; (i) Conidia. Scale bars: (e) = 50 μm; (f–h) = 20 μm; (g) = 10 μm; (i) = 5 μm.
Index Fungorum number: IF902539; Facesoffungi number: FoF 16750
Etymology: Name refers to the genus Oryza from which it was isolated
On a leaf spot of Oryza sativa. Sexual morph: Not observed. Asexual morph: Conidiomata 100–120 μm diameter, pycnidial, solitary, scattered, immersed on the host, and superficial in agar, globose to subglobose. Pycnidial wall 14–20 μm wide, comprising several layers of irregular shaped, relatively thick-walled cells of textura angularis, pale brown to brown. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 5–9 × 3–5.5 µm (= 6 × 4 μm, n = 10), holoblastic, hyaline, doliiform, discrete or lining on mass of cells that are clumped together and protrude into the conidiomatal cavity. Conidia 4.5–6 × 2.5–3.5 µm (= 5 × 3 μm, n = 30) ellipsoid to ovoid, aseptate, smooth, thin-walled, initially hyaline, becoming greyish brown.
Culture characteristics: Colonies on PDA reaching 86–89 mm diameter after a week at 27 °C, circular, slightly fluffy to floccose aerial mycelia, dull white; reverse white.
Material examined: Thailand, Chiang Rai Province, Wiang Chiang Rung District, on leaf spot of Oryza sativa, 4 July 2022, Sahar Absalan, WCR161, (MFLU 24-0096, holotype); (ex-type living culture MFLUCC 24-0032).
GenBank numbers: ITS = PQ374213, LSU = PQ374215, tub2 = PQ412477
Notes: Based on combined multi-gene phylogenetic analysis, isolate (MFLUCC 24-0032) clustered as a separate lineage, basal to Paraphaeosphaeria traversiae and P. viridescens with 64% ML and 0.91 BYPP support (Figure 20). The ex-type of P. viridescens (CBS 854.73) was isolated from fresh water in Montenegro [89], while the ex-type of P. traversiae (BRIP 75020a) was isolated from a leaf spot of Cyperus aromaticus in Australia [93]. Morphologically, P. oryzae differs from P. viridescens in the color and size of the pycnidial wall (brown, 14–20 μm in P. oryzae vs. pale orange, 5–12 µm in P. viridescens) and conidia (greyish brown, 5 × 3 μm in P. oryzae vs. greenish yellow, 4–4.5 × 1.8–2.2 in P. viridescens). Furthermore, P. viridescens produces a green pigment in the culture media, while this pigment production is absent in P. oryzae. Thus, according to macro- and micro-morphology differences and pairwise sequence comparison (8.7% nucleotide differences across 509 nucleotides in ITS, 1.4% across 844 nucleotides in LSU, 11.7% across 314 nucleotides in tub2) we propose this taxon as a new species.
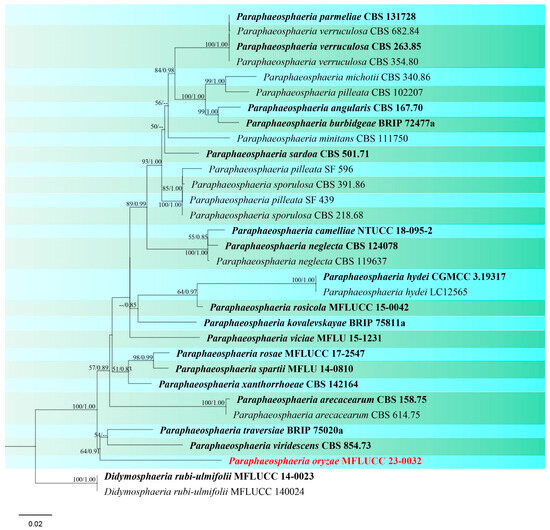
Figure 20.
Phylogenetic tree of Paraphaeosphaeria generated from a maximum likelihood analysis based on concatenated ITS, LSU, and tub2 sequence data. A total of 32 strains were included in the combined analyses, which comprised 1885 characters after alignment. Nodes on the tree were supported by bootstrap values, with values ≥50% for IQ-Tree and ≥0.7 for Bayesian posterior probabilities (BYPP). The isolate from the current study is in red, while ex-type strains are in bold black. The tree was rooted with Didymosphaeria rubi-ulmifolii (MFLUCC 14-0023 and MFLUCC 14-0024), and the scale bar represents the expected number of nucleotide substitutions per site.
4. Discussion
Pleosporales is an order with two suborders, Pleosporineae and Massarineae, containing a quarter of all dothideomycetous species [18,94]. In the current study, all five discovered fungal families belong to the suborder Pleosporineae, which includes many economically important plant pathogen genera with huge species diversity [95]. There are several genera in Pleosporales known as graminicolous fungi such as Curvularia, Bipolaris, and phoma-like playing important roles in plant diseases. However, many studies have shown that these fungal pathogens can also exist in different nutrient modes as endophytes and saprobes [58,96]. This study also showed that the species belonging to Pleosporales have diverse life modes, thriving as pathogenic, saprobic and endophytic in Oryza sativa (Table 2).
Pleosporaceae, recognized as the representative family of Pleosporales, is the largest family in the order with 23 accepted genera [21,22]. Bipolaris and Curvularia are two significant hyphomycetous genera in the family known by their asexual morph [58,97,98,99,100]. Species of Bipolaris and Curvularia are distributed worldwide and are associated with a wide range of hosts (more than 60 host genera) especially grasses and cereals [26,101,102]. Many species of Bipolaris and Curvularia have been reported from Thailand. Marin-Felix et al. [60] reported one new species and five new host records of Bipolaris during their investigation in Chiang Mai Province. Morphologically, strains of B. oryzae isolated in our study have the highest resemblance to strains reported by Marin-Felix et al. [60], in contrast to strains reported from other countries. Bipolaris oryzae was the dominant species in our study, with ten strains isolated from different parts of rice. Curvularia is another important genus in this family. There are many reports of Curvularia species from Poaceae, including rice in the world [44,45,58,63,68,103,104]. We report three species of Curvularia including C. geniculata and C. plantarum as saprobes, and C. chiangmaiensis associated with leaf spots. Marin-Felix et al. [60] introduced five new species of Curvularia from different monocotyledon plants in Chiang Mai, but they did not report any strains from rice. Ferdinandez et al. [44,45] showed a high diversity of Curvularia species in cereal crops including rice in Sri Lanka. Curvularia plantarum, was first reported from Saccharum officinarum in China [68], and was also isolated from rice as a new host record in Sri Lanka [45]. In the current study, we recorded C. plantarum as the first geographical report from Thailand.
Phaeosphaeriaceae is another important and large family within Pleosporales, playing a crucial role as plant pathogens and causing infections in significant agricultural crops [105,106]. In addition, numerous species are saprophytes on monocotyledons, particularly on poaceous hosts. Some species may exist as saprobes on herbaceous dicotyledonous hosts, while others can act as endophytes [18,70]. While Phaeosphaeria musae has been documented on various plant families including Arecaceae, Asparagaceae, Marantaceae, and Musaceae [71,107,108,109], its presence in rice was not previously reported. Moreover, phylogenetic analyses revealed that two sexual strains and one asexual strain isolated in this study clustered together with P. musae, which was previously known only from its sexual morph. Hence, we report the asexual form of this species for the first time. Another genus within Phaeosphaeriaceae, Setophoma, was initially described by de Gruyter et al. [80]. Phookamsak et al. [110] illustrated the sexual form of Setophoma sacchari and S. poaceicola [74] for the first time from Thailand. In the current study, two strains of S. poaceicola demonstrated its asexual morph for the first time. Setophoma species have been reported from numerous host plants as well as members of the family Poaceae (e.g., sugarcane, corn, grasses) [74,111,112,113,114,115,116]. However, an association between Setophoma and rice had not been documented before. Ophiosphaerella species are globally reported as pathogens or saprobes on Poaceae, Cyperaceae, Asparagaceae, and Zingiberaceae [23,71,76,77,117,118]. Wetzel et al. [119], Hutchens et al. [120], and Tomioka et al. [121] have recorded a variety of Ophiosphaerella species isolated from grasses and cereal crops. The present study reports the first association between Ophiosphaerella and rice.
Pyrenochaetopsidaceae contains three genera Neopyrenochaetopsis, Pyrenochaetopsis, and Xenopyrenochaetopsis. Despite broad world distribution, this study reveals the first record of association between rice and Pyrenochaetopsis. Species of Pyrenochaetopsis are frequently characterized by their asexual morph but two studies have documented the sexual morph of Pyrenochaetopsis [20,43]. In the present study, the sexual morphs of P. sinensis were isolated as a new host record, and P. oryzicola was described as a new species.
Didymellaceae encompasses cosmopolitan species with a broad distribution. More than 50% of its members are plant pathogens causing great losses to a wide range of economic crops [83,122]. Epicoccum, a significant genus within this family, can be found in the air, soil, human toenails, diverse plant material, and aquatic environments [86,123,124,125]. Plant-associated species have different lifestyles including endophytes [126]. Epicoccum nigrum and E. sorghinum have been reported on rice causing brown blotches on glumes and leaf spot disease, respectively [127,128,129]. We isolated three strains of E. catenisporum and one of E. latusicollum. Epicoccum catenisporum was isolated from leaf spots of rice in 1978 and deposited as an ex-holotype living culture, which was later discovered as a novel species by Valenzuela-Lopez et al. [78]. Epicoccum latusicollum was isolated as a pathogen associated with rice sheath rot in Ethiopia [130]. However, scant and insufficient information was provided on both morphology and phylogeny to verify the species identification. Herein, E. latusicollum is reported as a new geographical record from Thailand, and identified as saprobe on rice.
Species of Didymosphaeriaceae are often saprobic [21]. Two genera, Pseudopithomyces and Paraphaeosphaeria, were isolated in our study. Pseudopithomyces chartarum is recognized as the causal agent of leaf spot diseases affecting a variety of plants, including medicinal plants, grasses, cereals, and legumes [131,132,133,134,135,136]. Instances of P. chartarum as a pathogen from rice have been documented in India, New Zealand, and Brazil [92,137,138]. Paraphaeosphaeria species have been isolated from different hosts and substrates such as tea, sugarcane, coconut, rotten wood, dead leaves, soil, and fresh water [89,139,140]. Herein, a new species of Paraphaeosphaeria was isolated from rice.
Studying fungal species diversity on rice will help to improve agricultural practices, protect ecosystems, enhance economic outcomes, and ensure global food security. Establishing a comprehensive understanding of current fungal diversity provides a baseline for monitoring changes over time.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10110763/s1, Figure S1: Bipolaris oryzae (MFLU 24-0089); Figure S2: Curvularia chiangmaiensis (MFLU 24-0196); Figure S3: Curvularia geniculata (MFLU 24-0092); Figure S4: Curvularia plantarum (MFLU 24-0091, new geographical record); Figure S5: Ophiosphaerella agrostidis (MFLU 24-0094, new host record); Figure S6: Pyrenochaetopsis indica (MFLU 24-0197, new host record); Figure S7: Epicoccum catenisporum (MFLU 24-0093, new geographical record); Figure S8: Epicoccum latusicollum (MFLU 24-0317, new host record); Figure S9: Remotididymella capsici (MFLUCC 24-0031, new host and geographical record); Figure S10: Pseudopithomyces chartarum (MFLU 24-0101).
Author Contributions
Conceptualization, S.A., R.S.J. and S.L.; methodology, S.A. and A.A.; software, S.A.; formal analysis, S.A., A.A. and R.S.J.; writing—original draft preparation, S.A.; writing—review and editing, A.A., R.S.J., E.H.C.M. and K.D.H.; supervision, S.L. and R.S.J.; funding acquisition, S.L. and R.S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Research Council of Thailand (NRCT) grant “Biodiversity, taxonomy, phylogeny and evolution of Colletotrichum in northern Thailand” (grant no. NRCT-TRG010).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Acknowledgments
Sahar Absalan is grateful for Chiang Mai University Presidential Scholarship. The authors express their gratitude to Shaun Pennycook for his correction of Latin names for novelties. Sahar Absalan thanks Nootjarin Jungkhun and Naruemon Huanraluek for their kind help with the name of rice cultivars. Sahar Absalan would also like to thank Kritsana Jatuwong for her valuable assistance in reviewing the manuscript. This research was partially supported by Chiang Mai University, Thailand.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sharif, M.K.; Butt, M.S.; Anjum, F.M.; Khan, S.H. Rice bran: A novel functional ingredient. Crit. Rev. Food Sci. Nutr. 2014, 54, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Nakamura, K.; Cui, T.; Kayahara, H. High-performance liquid chromatographic determination of phenolic compounds in rice. J. Chromatogr. A 2005, 1063, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Amnuaylojaroen, T.; Chanvichit, P.; Janta, R.; Surapipith, V. Projection of rice and maize productions in Northern Thailand under climate change scenario RCP8.5. Agriculture 2021, 11, 23. [Google Scholar] [CrossRef]
- Berno, T.; Dentice, G.; Wisansing, J.J. Kin kao laew reu young (“Have You Eaten Rice Yet”)?: A New Perspective on Food and Tourism in Thailand. In Food Tourism in Asia; Springer: Singapore, 2019; pp. 17–30. [Google Scholar] [CrossRef]
- Statista. Available online: https://www.statista.com/statistics/1447114/thailand-export-volume-of-rice/#statisticContainer (accessed on 9 October 2024).
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Yadav, A.N.; Kour, D.; Kaur, T.; Devi, R.; Yadav, N. Agriculturally important fungi for crop productivity: Current research and future challenges. In Agriculturally Important Fungi for Sustainable Agriculture: Volume 1: Perspective for Diversity and Crop Productivity; Springer: Cham, Switzerland, 2020; pp. 275–286. [Google Scholar] [CrossRef]
- Chaiharn, M.; Chunhaleuchanon, S.; Lumyong, S. Screening siderophore producing bacteria as potential biological control agent for fungal rice pathogens in Thailand. World J. Microbiol. Biotechnol. 2009, 25, 1919–1928. [Google Scholar] [CrossRef]
- Suprapta, D.N. Potential of microbial antagonists as biocontrol agents against plant fungal pathogens. J. ISSAAS 2012, 18, 1–8. [Google Scholar]
- Naik, B.S.; Shashikala, J.; Krishnamurthy, Y.L.A. Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiol. Res. 2009, 164, 290–296. [Google Scholar] [CrossRef]
- Pili, N.N.; França, S.C.; Kyndt, T.; Makumba, B.A.; Skilton, R.; Höfte, M.; Mibey, R.K.; Gheysen, G. Analysis of fungal endophytes associated with rice roots from irrigated and upland ecosystems in Kenya. Plant Soil 2016, 405, 371–380. [Google Scholar] [CrossRef]
- Nassimi, Z.; Taheri, P. Endophytic fungus Piriformospora indica induced systemic resistance against rice sheath blight via affecting hydrogen peroxide and antioxidants. Biocontrol Sci. Technol. 2017, 27, 252–267. [Google Scholar] [CrossRef]
- Luttrell, E.S. The Ascostromatic Ascomycetes. Mycologia 1955, 47, 511–532. [Google Scholar] [CrossRef]
- Barr, M.E. Prodromus to Class Loculoascomycetes; University of Massachusetts: Amherst, MA, USA, 1987. [Google Scholar]
- Barr, M.E. New taxa and combinations in the Loculoascomycetes. Mycotaxon 1987, 29, 501–505. [Google Scholar]
- Hyde, K.D.; Jones, E.G.; Liu, J.-K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.-Q. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Pem, D.; Bhat, J.D.; et al. Refned families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Fournier, J.; Voglmayr, H. Two unusual new species of Pleosporales: Anteaglonium rubescens and Atrocalyx asturiensis. Sydowia 2018, 70, 129–140. [Google Scholar]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.T.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal Diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Sivanesan, A. Graminicolous Species of Bipolaris, Curvularia, Drechslera, Exserohilum and Their Teleomorphs; CAB International: Wallingford, UK, 1987. [Google Scholar]
- Dugan, F.M.; Peever, T.L. Morphological and cultural differentiation of described species of Alternaria from Poaceae. Mycotaxon 2002, 83, 229–264. [Google Scholar]
- Rosa, L.H.; Vaz, A.B.M.; Caligiorne, R.B. Endophytic fungi associated with the Antarctic grass Deschampsia antarctica Desv. (Poaceae). Polar Biol. 2009, 32, 161–167. [Google Scholar] [CrossRef]
- Manamgoda, D.; Rossman, A.; Castlebury, L.; Crous, P.; Madrid, H.; Chukeatirote, E.; Hyde, K. The genus Bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef] [PubMed]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, I.; Rathnayaka, A.; Marasinghe, D.; Calabon, M.; Gentekaki, E.; Lee, H.; Hurdeal, V.; Pem, D.; Dissanayake, S.; Wijesinghe, S.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 18 August 2024).
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database, fungal names linked with morphology; phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S. Primers for Elongation Factor 1-Alpha (EF1-alpha). 2001. Available online: https://www.docin.com/p-1613748809.html (accessed on 23 December 2023).
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Myllys, L.; Stenroos, S.; Thell, A. New genes for phylogenetic studies of lichenized fungi: Glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist 2002, 34, 237–246. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Keirnan, E.C.; Tan, Y.P.; Laurence, M.H.; Mertin, A.A.; Liew, E.C.Y.; Summerell, B.A.; Shivas, R.G. Cryptic diversity found in Didymellaceae from Australian native legumes. MycoKeys 2021, 78, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Phookamsak, R.; Jiang, H.; Suwannarach, N.; Lumyong, S.; Xu, J.C.; Xu, S.; Liao, C.F.; Chomnunti, P. Bambusicolous fungi in Pleosporales: Introducing four novel taxa and a new habitat record for Anastomitrabeculia didymospora. J. Fungi 2022, 8, 630. [Google Scholar] [CrossRef]
- Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Deshappriya, N.; Munasinghe, M.S.; Castlebury, L.A. Molecular phylogeny and morphology reveal three novel species of Curvularia (Pleosporales, Pleosporaceae) associated with cereal crops and weedy grass hosts. Mycol. Prog. 2021, 20, 431–451. [Google Scholar] [CrossRef]
- Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Munasinghe, M.S.; Castlebury, L.A. Molecular phylogeny and morphology reveal two new graminicolous species, Curvularia aurantia sp. nov. and C. vidyodayana sp. nov. with new records of Curvularia spp. from Sri Lanka. Fung. Syst. Evol. 2023, 12, 219–246. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the raxml web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree: Tree Figure Drawing Tool, Version 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Nitschke, T.R.J. Grundlage eines Systems der Pyrenomyceten. Verh Naturhist Vereines Preuss. Rheinl 1869, 26, 70–77. [Google Scholar]
- Wijayawardene, N.N.; Crous, P.W.; Kirk, P.M.; Hawksworth, D.L.; Boonmee, S.; Braun, U.; Dai, D.-Q.; D’Souza, M.J.; Diederich, P.; Dissanayake, A.J.; et al. Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Divers. 2014, 69, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.; Boudreaux, C. Fatal cerebral phaeohyphomycosis due to Curvularia lunata in an immunocompetent patient. J. Clin. Microbiol. 2004, 42, 5419–5423. [Google Scholar] [CrossRef] [PubMed]
- Ariyawansa, H.A.; Tanaka, K.; Thambugala, K.M.; Phookamsak, R.; Tian, Q.; Camporesi, E.; Hongsanan, S.; Monkai, J.; Wanasinghe, D.N.; Mapook, A. A molecular phylogenetic reappraisal of the Didymosphaeriaceae (=Montagnulaceae). Fungal Divers. 2014, 68, 69–104. [Google Scholar] [CrossRef]
- Liu, S.L.; Wang, X.W.; Li, G.J.; Deng, C.Y.; Rossi, W.; Leonardi, M.; Liimatainen, K.; Kekki, T.; Niskanen, T.; Smith, M.E.; et al. Fungal diversity notes 1717–1817: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2024, 124, 1–216. [Google Scholar] [CrossRef]
- Hongsanan, S.; Phookamsak, R.; Goonasekara, I.D.; Thambugala, K.M.; Hyde, K.D.; Bhat, J.D.; Suwannarach, N.; Cheewangkoon, R. Introducing a new pleosporalean family Sublophiostomataceae fam. nov. to accommodate Sublophiostoma gen. nov. Sci. Rep. 2021, 11, 9496. [Google Scholar] [CrossRef]
- Lourenço, C.C.G.; Alves, J.L.; Guatimosim, E.; Colman, A.; Barreto, R.W. Bipolaris marantae sp. nov., a novel Helminthosporoid species causing foliage blight of the garden plant Maranta leuconeura in Brazil. Mycobiology 2017, 45, 123–128. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; McKenzie, E.H.C.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Shivas, R.G.; Tan, Y.P.; Hyde, K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012, 56, 131–144. [Google Scholar] [CrossRef]
- Fisher, P.J.; Petrini, O. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.). New Phytol. 1992, 120, 137–143. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Senwanna, C.; Cheewangkoon, R.; Crous, P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 2017, 8, 1556–1574. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Nemati, Z.; Mostowfizadeh-Ghalamfarsa, R. Identification of some grass-associated species of Bipolaris, Curvularia and Exserohilum in selected regions of Iran. Rostaniha 2016, 17, 40–50. [Google Scholar] [CrossRef]
- Kusai, N.A.; Mior Zakuan Azmi, M.; Zulkifly, S.; Yusof, M.T.; Mohd Zainudin, N.A.I. Morphological and molecular characterization of Curvularia and related species associated with leaf spot disease of rice in Peninsular Malaysia. Rend. Lincei 2016, 27, 205–214. [Google Scholar] [CrossRef]
- Matsushima, T. Icones Microfungorum a Matsushima Lectorum; Taylor & Francis Group: Oxfordshire, UK, 1976; p. 955. [Google Scholar] [CrossRef]
- Dade, H.A. A revised list of Gold Coast fungi and plant diseases. Bull. Misc. Inf. Kew 1940, 6, 205–247. [Google Scholar] [CrossRef]
- Tai, F.L. Sylloge Fungorum Sinicorum; Science Press: Beijing, China, 1979. [Google Scholar]
- Photita, W.; Lumyong, S.; Lumyong, P.; McKenzie, E.H.C.; Hyde, K.D. Saprobic fungi on dead wild banana. Mycotaxon 2003, 85, 345–356. [Google Scholar]
- Raza, M.; Zhang, Z.-F.; Hyde, K.D.; Diao, Y.-Z.; Cai, L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 2019, 99, 1–104. [Google Scholar] [CrossRef]
- Barr, M.E. A classification of Loculoascomycetes. Mycologia 1979, 71, 935–957. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Verkley, G.; Shin, H.-D.; Barreto, R.; Alfenas, A.; Swart, W.; Groenewald, J.; Crous, P. Sizing up Septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef]
- Phookamsak, R.; Liu, J.-K.; McKenzie, E.H.C.; Manamgoda, D.S.; Ariyawansa, H.; Thambugala, K.M.; Dai, D.-Q.; Camporesi, E.; Chukeatirote, E.; Wijayawardene, N.N.; et al. Revision of Phaeosphaeriaceae. Fungal Divers. 2014, 68, 159–238. [Google Scholar] [CrossRef]
- Bakhshi, M.; Arzanlou, M.; Groenewald, J.Z.; Quaedvlieg, W.; Crous, P.W. Parastagonosporella fallopiae gen. et sp. nov. (Phaeosphaeriaceae) on Fallopia convolvulus from Iran. Mycol. Prog. 2019, 18, 203–214. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Maharachchikumbura, S.S.N. Exploring the diversity and systematics of Phaeosphaeriaceae: Taxonomic novelties from ecologically diverse habitats and their phylogenetic resolution. J. Fungi 2023, 9, 853. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Ariyawansa, H.A.; Goonasekara, I.D.; Phookamsak, R.; Dissanayake, A.; et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Ivayani; Widiastuti, A.; Suryanti; Suharjo, R.; Priyatmojo, A. Fungi associated with rice sheath rot in Lampung, Indonesia. Arch. Phytopathol. Pflanzenschutz 2022, 55, 2075–2097. [Google Scholar] [CrossRef]
- Câmara, M.P.S.; O’Neill, N.R.; Berkum, P.V.; Dernoeden, P.H.; Palm, M.E. Ophiosphaerella agrostis sp. nov. and its relationship to other species of Ophiosphaerella. Mycologia 2000, 92, 317–325. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Thambugala, K.M.; Wanasinghe, D.N.; Gentekaki, E.; Promputtha, I.; Kuo, C.H.; Hyde, K.D. Additions to Phaeosphaeriaceae (Pleosporales): Elongaticollum gen. nov., Ophiosphaerella taiwanensis sp. nov., Phaeosphaeriopsis beaucarneae sp. nov. and a new host record of Neosetophoma poaceicola from Musaceae. MycoKeys 2020, 70, 59–88. [Google Scholar] [CrossRef]
- Valenzuela-Lopez, N.; Cano-Lira, J.F.; Guarro, J.; Sutton, D.A.; Wiederhold, N.; Crous, P.W.; Stchigel, A.M. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2018, 90, 1–69. [Google Scholar] [CrossRef]
- de Gruyter, J.; Boerema, G.H. Contributions towards a monograph of Phoma (Coelomycetes) VIII. Section Paraphoma: Taxa with setose pycnidia. Persoonia 2002, 17, 541–561. [Google Scholar]
- de Gruyter, J.; Woudenberg, J.H.; Aveskamp, M.M.; Gerard, J.M.V.; Groenewald, J.Z.; Crous, P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 2010, 102, 1066–1081. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.M.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- de Gruyter, J.; Aveskamp, M.M.; Woudenberg, J.H.C.; Verkley, G.J.M.; Groenewald, J.Z.; Crous, P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009, 113, 508–519. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; de Gruyter, J.; Crous, P.W. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Divers. 2008, 31, 1–18. [Google Scholar]
- Chen, Q.; Zhang, K.; Zhang, G.; Cai, L. A polyphasic approach to characterise two novel species of Phoma (Didymellaceae) from China. Phytotaxa 2015, 197, 267–281. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; Verkley, G.J.M.; de Gruyter, J.; Murace, M.A.; Perelló, A.; Woudenberg, J.H.C.; Groenewald, J.Z.; Crous, P.W. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 2009, 101, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hou, L.W.; Duan, W.J.; Crous, P.W.; Cai, L. Didymellaceae revisited. Stud. Mycol. 2017, 87, 105–159. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Groenewald, J.; Pfenning, L.; Yarden, O.; Crous, P.; Cai, L. The phoma-like dilemma. Stud. Mycol. 2020, 96, 309–396. [Google Scholar] [CrossRef]
- Munk, A. The system of the Pyrenomycetes. Dansk Bot. Ark. 1953, 15, 1–163. [Google Scholar]
- Verkley, G.J.M.; Dukik, K.; Renfurm, R.; Goker, M.; Stielow, J.B. Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 2014, 32, 25–51. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Neptunomyces aureus gen. et sp. nov. (Didymosphaeriaceae, Pleosporales) isolated from algae in Ria de Aveiro, Portugal. MycoKeys 2019, 60, 31–44. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Haware, M.P.; Sharma, N.D. A new glume blotch of rice (Oryza sativa). Plant Dis. Rep. 1973, 57, 436–437. [Google Scholar]
- Tan, Y.P.; Shivas, R.G. Nomenclatural novelties. Index Austral. Fungi 2022, 1, 1–18. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainthworth and Bisby’s Dictionary of the Fungi, 10th ed.; Kirk, P., Cannon, P., Minter, D., Stalpers, J., Eds.; CABI: Wallingford, UK, 2008; ISBN 978-0-85199-826-8. [Google Scholar]
- Manamgoda, D.S.; Cai, L.; Bahkali, A.H.; Chukeatirote, E.; Hyde, K.D. Cochliobolus: An overview and current status of species. Fungal Divers. 2011, 51, 3–42. [Google Scholar] [CrossRef]
- Rai, M.; Agarkar, G. Plant–fungal interactions: What triggers the fungi to switch among lifestyles? Crit. Rev. Microbiol. 2016, 42, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.A.; Hussain, S.T.; Hasan, S.; McEvoy, P.; Sarwari, A. Disseminated Bipolaris infection in an immunocompetent host: An atypical presentation. J. Pak. Med. Assoc. 2000, 50, 68–71. [Google Scholar] [PubMed]
- Shoemaker, R.A. Nomenclature of Drechslera and Bipolaris, grass parasites segregated from Helminthosporium. Can. J. Bot. 1959, 37, 879–887. [Google Scholar] [CrossRef]
- Dai, F.C.; Wang, X.M.; Zhu, Z.D.; Gao, W.D.; Huo, N.X. Curvularia leaf spot of maize: Pathogens and varietal resistance. Acta Phytopathol. Sin. 1998, 2, 123–129. [Google Scholar]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. 2024. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 10 July 2024).
- Tan, Y.P.; Crous, P.W.; Shivas, R.G. Eight novel Bipolaris species identified from John L. Alcorn’s collections at the Queensland Plant Pathology Herbarium (BRIP). Mycol. Prog. 2016, 15, 1203–1214. [Google Scholar] [CrossRef]
- Tan, Y.P.; Crous, P.W.; Shivas, R.G. Cryptic species of Curvularia in the culture collection of the Queensland Plant Pathology Herbarium. MycoKeys 2018, 35, 1–25. [Google Scholar] [CrossRef]
- Gao, B.D.; Huang, W.; Xia, H. A new rice disease, black sheath spot, caused by Curvularia fallax in China. Plant Dis. 2012, 96, 1224. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Rossman, A.; Castlebury, L.A.; Chukeatirote, E. Article PHYTOTAXA. Phytotaxa 2015, 212, 175–198. [Google Scholar] [CrossRef]
- Shoemaker, R.A.; Babcock, C.E. Phaeosphaeria . Can. J. Bot. 1989, 67, 1500–1599. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; De Cock, A.W.A.M.; Dissanayake, A.J.; Glockling, S.L.; Goonasekara, I.D.; et al. One stop shop: Backbones trees for important phytopathogenic genera: I. Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef]
- Sawada, K. Descriptive Catalogue of Taiwan (Formosan) Fungi. Part XI; College of Agriculture National Taiwan University: Taipei, Taiwan, 1959; pp. 1–268. [Google Scholar]
- Photita, W.; Lumyong, S.; Lumyong, P.; Hyde, K.D.; McKenzie, E.H.C. Index of fungi described from the Musaceae. Mycotaxon 2002, 81, 491–503. [Google Scholar]
- Zhang, Y.; Schoch, C.L.; Fournier, J.; Crous, P.W.; De Gruyter, J.; Woudenberg, J.H.C.; Hirayama, K.; Tanaka, K.; Pointing, B.; Spatafora, J.W.; et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009, 64, 85–102. [Google Scholar] [CrossRef]
- Phookamsak, R.; Liu, J.K.; Manamgoda, D.S.; Chukeatirote, E.; Mortimer, P.E.; McKenzie, E.H.C.; Hyde, K.D. The sexual state of Setophoma. Phytotaxa 2014, 176, 260–269. [Google Scholar] [CrossRef]
- Schwartz, H.F.; Mohan, S.K. Compendium of Onion and Garlic Diseases; APS Press: St. Paul, MN, USA, 1995. [Google Scholar]
- Howard, R.J.; Garland, J.A.; Seaman, W.L.; Grafius, E.J. Diseases and pests of vegetable crops in Canada. J. Econ. Entomol. 1996, 89, 1045. [Google Scholar]
- Zhu, X.; Reid, L.M.; Woldemariam, T.; Tenuta, A.; Lachance, P.; Pouleur, S. Survey of corn diseases and pests in Ontario and Québec in 2004. Can. Plant Dis. Surv. 2005, 85, 31–34. [Google Scholar]
- Ikeda, K.; Kuwabara, K.; Urushibara, T.; Soyai, P.; Miki, S.; Shibata, S. Pink root rot of squash caused by Setophoma terrestris in Japan. J. Gen. Plant Pathol. 2012, 78, 372–375. [Google Scholar] [CrossRef]
- Yang, Y.; Zuzak, K.; Harding, M.; Neilson, E.; Feindel, D.; Feng, J. First report of pink root rot caused by Setophoma (Pyrenochaeta) terrestris on canola. Can. J. Plant. Pathol. 2017, 39, 354–360. [Google Scholar] [CrossRef]
- Liu, F.; Wang, J.; Li, H.; Wang, W.; Cai, L. Setophoma spp. on Camellia sinensis. Fung. Syst. Evol. 2019, 4, 43–57. [Google Scholar] [CrossRef]
- Flores, F.J.; Marek, S.M.; Orquera, G.; Walker, N.R. Molecular identification and multilocus phylogeny of Ophiosphaerella species associated with spring dead spot of bermudagrass. Crop Sci. 2017, 57, S-249–S-261. [Google Scholar] [CrossRef]
- Oki, T.; Yano, K.; Komatsu, K.; Shimomoto, Y.; Arie, T.; Morita, Y. Ophiosphaerella agrostidis causes leaf-sheath rot of Zingiber mioga. J. Gen. Plant Pathol. 2022, 88, 173–177. [Google Scholar] [CrossRef]
- Wetzel, H.C., III; Skinner, D.Z.; Tisserat, N.A. Geographic distribution and genetic diversity of three Ophiosphaerella species that cause spring dead spot of bermudagrass. Plant Dis. 1999, 83, 1160–1166. [Google Scholar] [CrossRef]
- Hutchens, W.J.; Henderson, C.A.; Bush, E.A.; Kerns, J.P.; McCall, D.S. Geographic distribution of Ophiosphaerella species in the mid-Atlantic United States. Plant Health Prog. 2021, 23, 93–100. [Google Scholar] [CrossRef]
- Tomioka, K.; Sekiguchi, H.; Nomiyama, K.; Kawaguchi, A.; Kawakami, A.; Masunaka, A.; Kobayashi, H.; Chiba, M.; Nagata, K.; Ishikawa, N. Early dead ripe of bread wheat and durum wheat caused by Ophiosphaerella korrae and evaluation of the fungal influence on barley, rice, and soybean. J. Gen. Plant Pathol. 2021, 87, 273–280. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; de Gruyter, J.; Woudenberg, J.H.C.; Verkley, G.J.M.; Crous, P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 2010, 65, 1–60. [Google Scholar] [CrossRef] [PubMed]
- Melgarejo, P.; Carrillo, R.; Sagasta, E.M. Mycoflora of peach twigs and flowers and its possible significance in biological control of Monilinia laxa. Trans. Br. Mycol. Soc. 1985, 85, 313–317. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.-H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag: Eching, Germany, 2007; pp. 1–672. [Google Scholar]
- Arenal, F.; Platas, G.; Martin, J.; Salazar, O.; Peláez, F. Evaluation of different PCR-based DNA fingerprinting techniques for assessing the genetic variability of isolates of the fungus Epicoccum nigrum. J. Appl. Microbiol. 1999, 87, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Taguiam, J.D.; Evallo, E.; Balendres, M.A. Epicoccum species: Ubiquitous plant pathogens and effective biological control agents. Eur. J. Plant Pathol. 2021, 159, 713–725. [Google Scholar] [CrossRef]
- Kodama, F.; Tsuchiya, S. Brown blotch on glume of rice plant caused by Epicoccum purpurascens Ehrenberg ex Schlechtendahl. Annu. Rep. Soc. Plant Prot. North Jpn. 1981, 32, 107–109. [Google Scholar]
- Liu, L.M.; Zhao, Y.; Zhang, Y.L.; Wang, L.; Hou, Y.X.; Huang, S.W. First report of leaf spot disease on rice caused by Epicoccum sorghinum in China. Plant Dis. 2020, 104, 2735. [Google Scholar] [CrossRef]
- Imran, M.; Khanal, S.; Zhou, X.G.; Antony-Babu, S.; Atiq, M. First report of leaf spot of rice caused by Epicoccum sorghinum in the United States. Plant Dis. 2022, 106, 2758. [Google Scholar] [CrossRef] [PubMed]
- Gizaw, T.; Zegeye, W.A.; Hika, N.; Degwale, A.; Setargie, A.; Tsegaw, M. Genetic diversity and pathogenicity of fungi associated with rice sheath rot in the Fogera plain, Northwest Ethiopia. J. Phytopathol. 2023, 172, e13252. [Google Scholar] [CrossRef]
- Ponappa, K.M. New record of fungi associated with Cannabis sativa. J. Mycol. Pl. Pathol. 1977, 7, 139. [Google Scholar]
- Russomanno, O.M.R.; Amaral, R.E.M.; Malavolta, V.M.A.; Alcantara, V.B.G.; Schammass, E.A. Ocorrência do fungo Pithomyces chartarum (Berk. & Curt.) M. B. Ellis em forrageiras pastoreadas por bovinos. Rev. Agric. Piracicaba 1985, 60, 249–265. [Google Scholar]
- Jonardhanan, K.K. Diseases of Major Medicinal Plants; Daya Publ. House: Delhi, India, 2002. [Google Scholar]
- Eken, C.; Jochum, C.C.; Yuen, G.Y. First report of leaf spot of smooth bromegrass caused by Pithomyces chartarum in Nebraska. Plant Dis. 2006, 90, 108. [Google Scholar] [CrossRef]
- Varga, Z.; Fischl, G. Pathogenic fungal species isolated from leaves and seeds of smooth brome (Bromus inermis Leyss.). Commun. Agric. Appl. Biol. Sci. 2006, 71, 1103–1108. [Google Scholar]
- Tóth, B.; Csosz, M.; Dijksterhuis, J.; Frisvad, J.C.; Vagra, J. Pithomyces chartarum as a pathogen of wheat. Plant Pathol. J. 2007, 89, 405–408. [Google Scholar]
- Lau, K.H.; Sheridan, J.E. Mycoflora of rice (Oryza sativa L.) seed in New Zealand. N. Z. J. Agric. Res. 1975, 18, 237–250. [Google Scholar] [CrossRef]
- Malavolta, V.M.A.; Amaral, R.D.M.; Alexandre, J. Fungi recorded on rice crops in different regions of Brazil. Biologico 1979, 45, 159–164. [Google Scholar]
- Verkley, G.J.; da Silva, M.; Wicklow, D.T.; Crous, P.W. Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Stud. Mycol. 2004, 50, 323–335. [Google Scholar]
- Ariyawansa, H.A.; Tsai, I.; Thambugala, K.M.; Chuang, W.Y.; Lin, S.R.; Hozzein, W.N.; Cheewangkoon, R. Species diversity of Pleosporalean taxa associated with Camellia sinensis (L.) Kuntze in Taiwan. Sci. Rep. 2020, 10, 12762. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).