Identification of Homeobox Transcription Factors in a Dimorphic Fungus Talaromyces marneffei and Protein-Protein Interaction Prediction of RfeB
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Conditions
2.2. Quantitative Real-Time PCR
2.3. Sequence Analysis
2.4. Heatmap Generation
2.5. Phylogenetic Analysis
2.6. Modeling Structure of RfeB in Complex with TmSwi5
2.7. Molecular Dynamics Simulation
2.8. Protein-Protein Binding Free Energy
3. Results
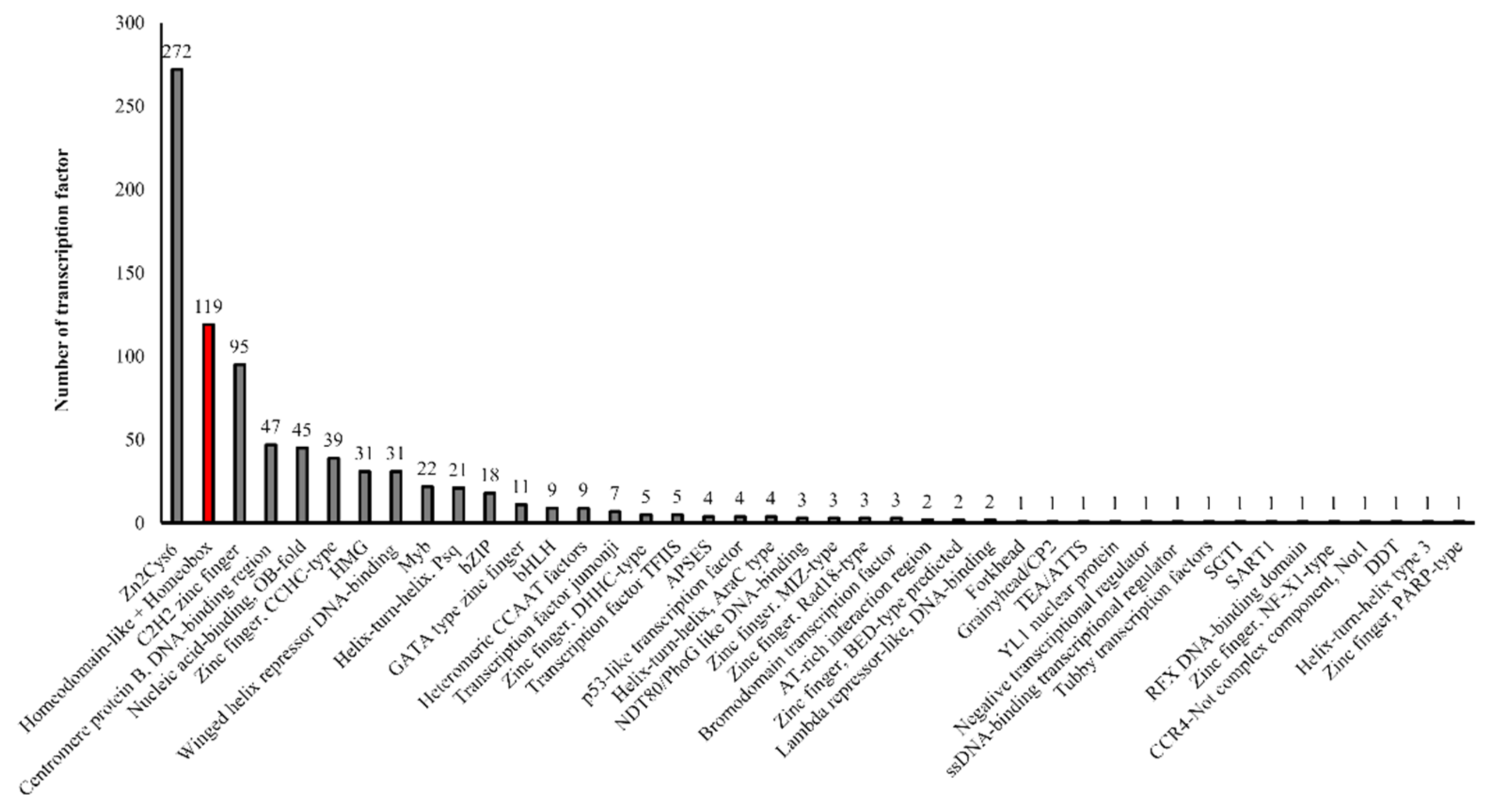
3.1. Homeobox Proteins in T. marneffei
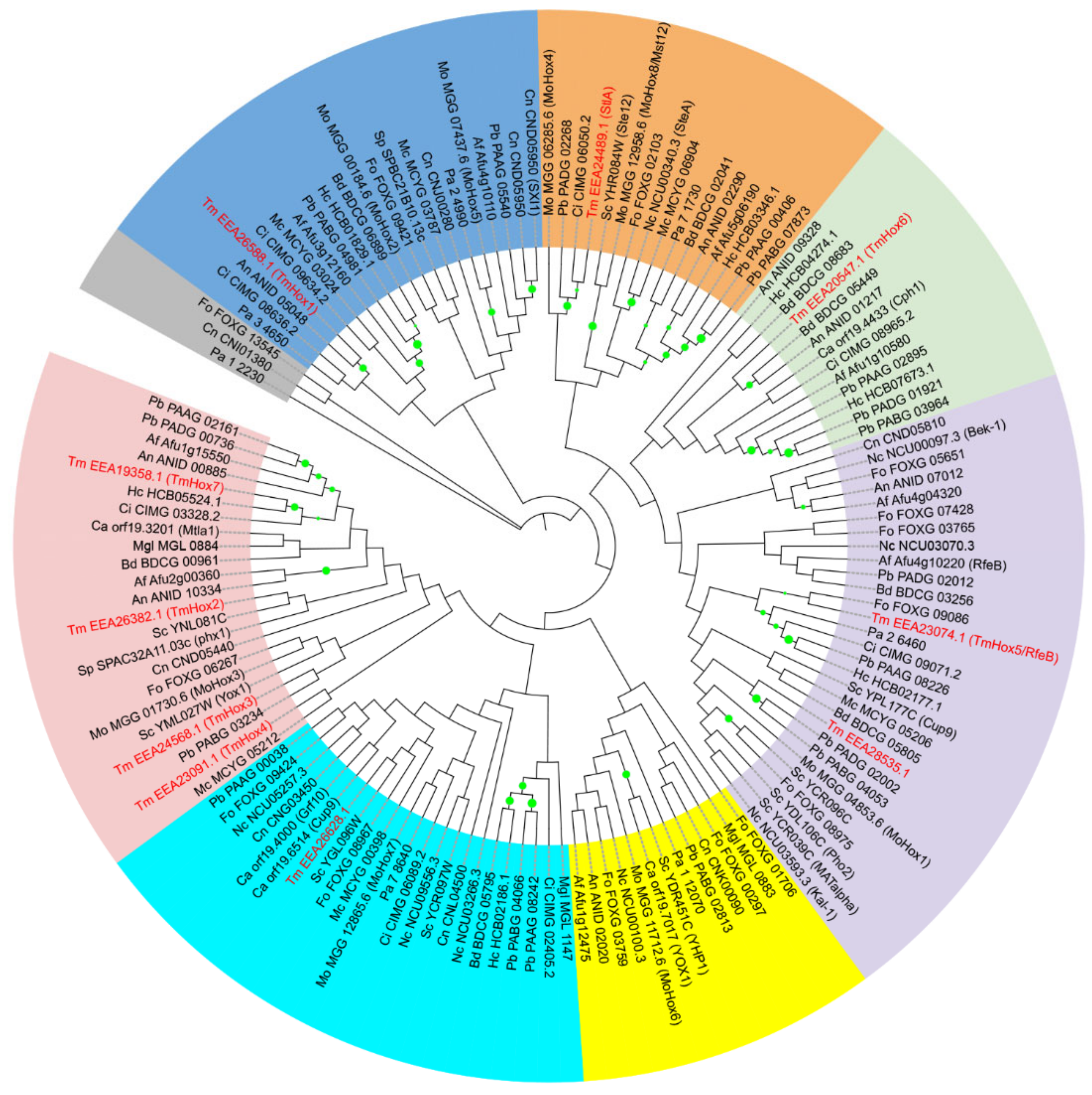
3.2. Phylogenetic Analysis of T. marneffei Homeodomain-Containing Proteins
3.3. Gene Expression Analysis of TmHox Genes
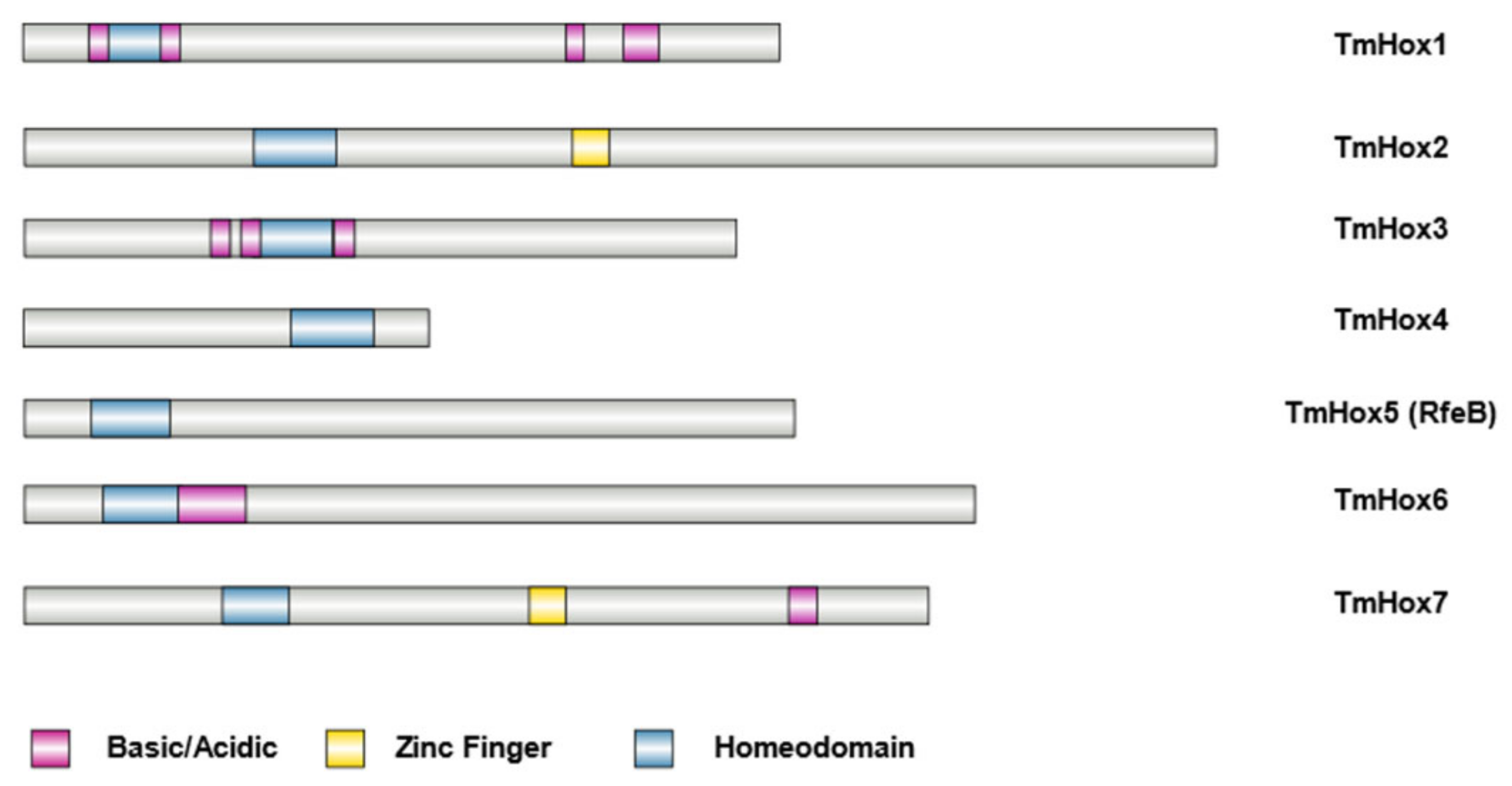
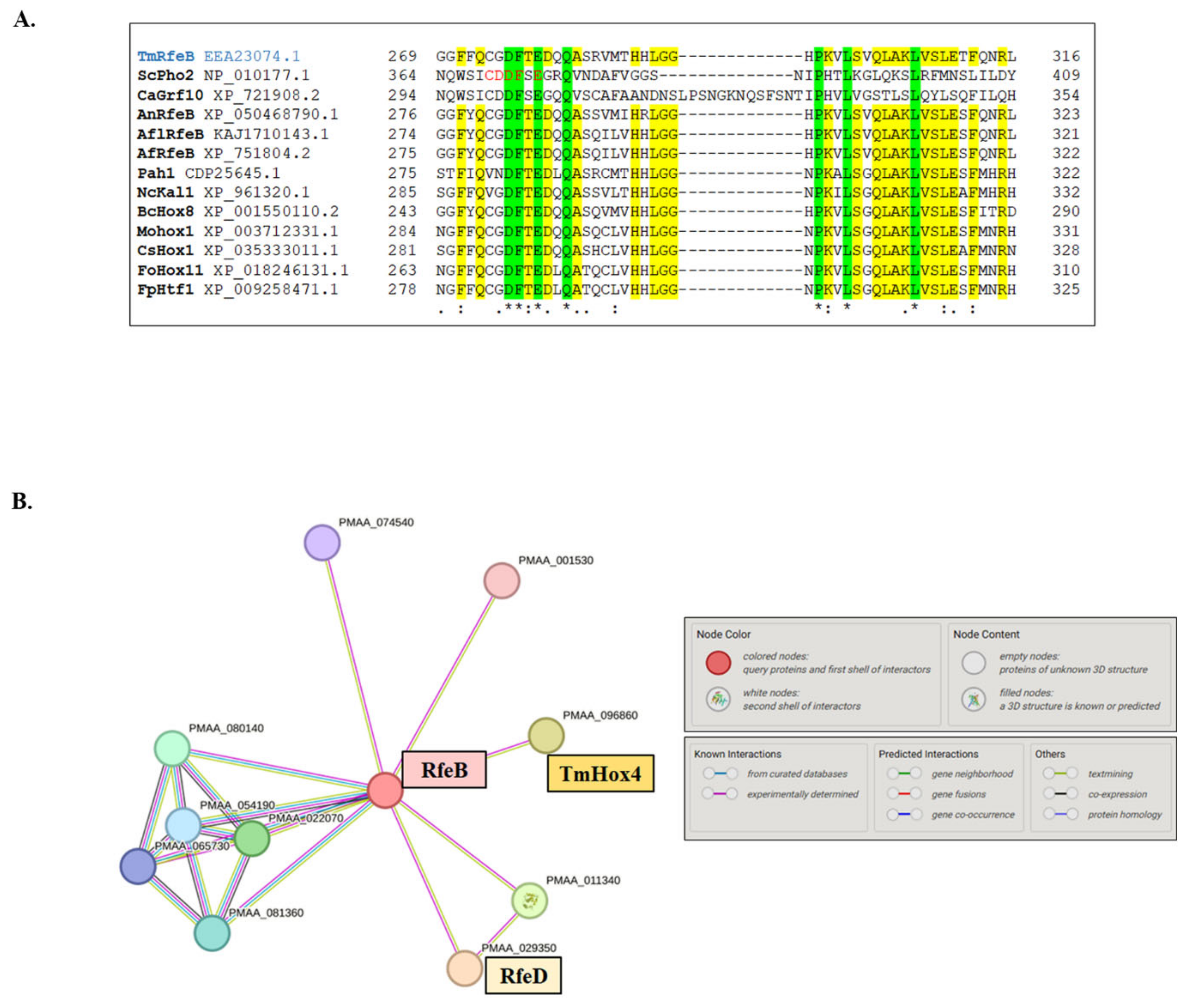
3.4. Sequence Analysis of RfeB Protein in T. marneffei
3.5. In Silico Structure Prediction of RfeB-TmSwi5-DNA Complex
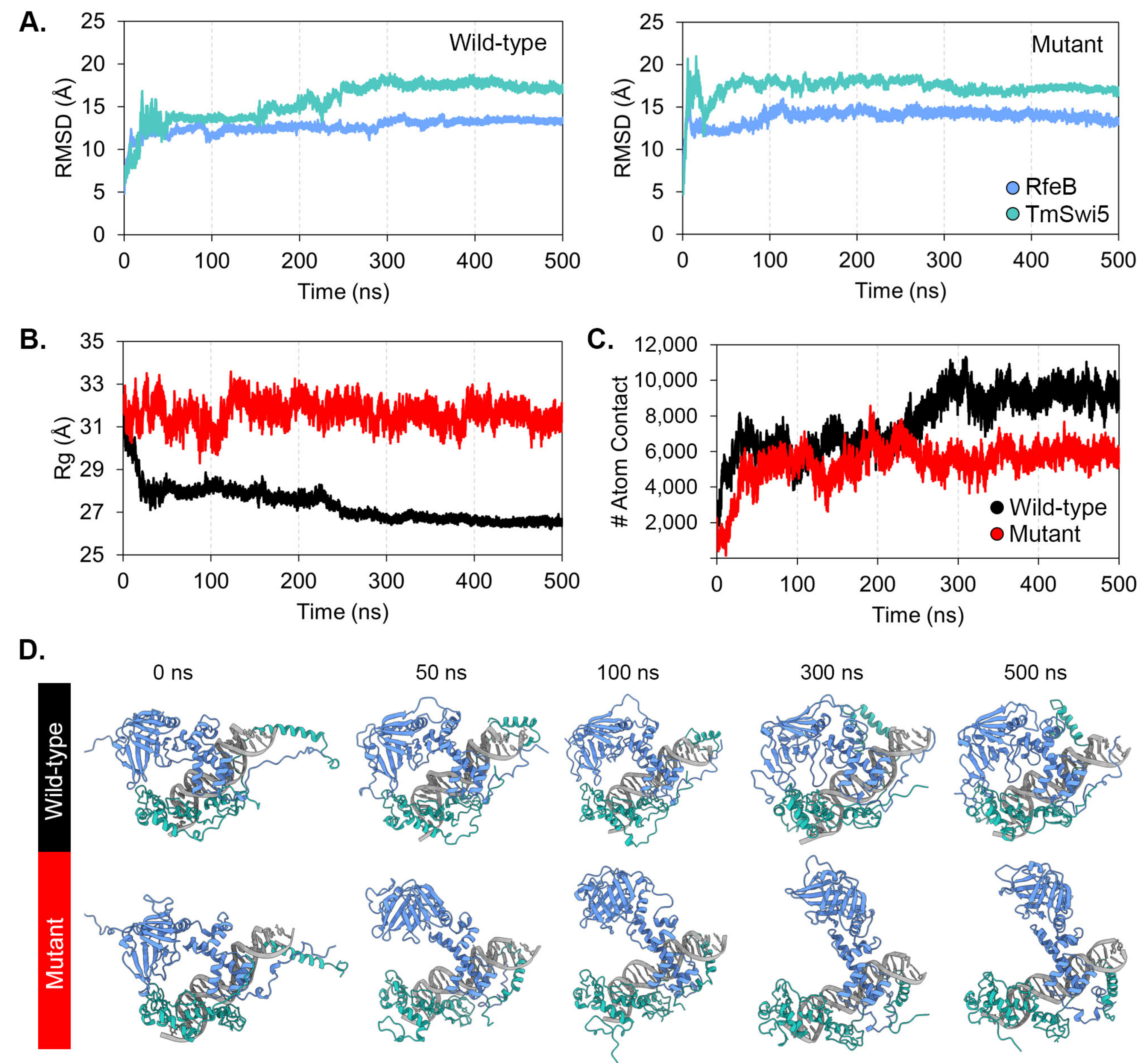
3.6. Molecular Dynamics Simulation
3.7. Protein-Protein Binding Free Energy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiedemann, B.; Weisner, J.; Rauh, D. Chemical modulation of transcription factors. Medchemcomm 2018, 9, 1249–1272. [Google Scholar] [CrossRef] [PubMed]
- Pais, P.; Costa, C.; Cavalheiro, M.; Romão, D.; Teixeira, M.C. Transcriptional control of drug resistance, virulence and immune system evasion in pathogenic fungi: A cross-species comparison. Front. Cell. Infect. Microbiol. 2016, 6, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.J.; Liu, Q.; Wu, D.L.; Tie, L. Current strategies and progress for targeting the “undruggable” transcription factors. Acta Pharmacol. Sin. 2022, 43, 2474–2481. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. Homeosis: The first 100 years. Trends Genet. 1994, 10, 341–343. [Google Scholar] [CrossRef]
- Pearson, J.C.; Lemons, D.; McGinnis, W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005, 6, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Frischer, L.E.; Hagen, F.S.; Garber, R.L. An inversion that disrupts the Antennapedia gene causes abnormal structure and localization of RNAs. Cell 1986, 47, 1017–1023. [Google Scholar] [CrossRef]
- Maeda, R.K.; Karch, F. The bithorax complex of Drosophila an exceptional Hox cluster. Curr. Top. Dev. Biol. 2009, 88, 1–33. [Google Scholar] [CrossRef]
- Quinonez, S.C.; Innis, J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014, 111, 4–15. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Lincoln, C.; Long, J.; Yamaguchi, J.; Serikawa, K.; Hake, S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 1994, 6, 1859–1876. [Google Scholar] [CrossRef] [PubMed]
- Derelle, R.; Lopez, P.; Le Guyader, H.; Manuel, M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol. Dev. 2007, 9, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Coppin, E.; Berteaux-Lecellier, V.; Bidard, F.; Brun, S.; Ruprich-Robert, G.; Espagne, E.; Aït-Benkhali, J.; Goarin, A.; Nesseir, A.; Planamente, S.; et al. Systematic deletion of homeobox genes in Podospora anserina uncovers their roles in shaping the fruiting body. PLoS ONE 2012, 7, e37488. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Chen, B.; Mukhtar, I.; Xie, B.; Li, Z.; Meng, L. Characterization and expression pattern of homeobox transcription factors in fruiting body development of straw mushroom Volvariella volvacea. Fungal Biol. 2019, 123, 95–102. [Google Scholar] [CrossRef]
- Muraguchi, H.; Umezawa, K.; Niikura, M.; Yoshida, M.; Kozaki, T.; Ishii, K.; Sakai, K.; Shimizu, M.; Nakahori, K.; Sakamoto, Y.; et al. Strand-specific RNA-Seq analyses of fruiting body development in Coprinopsis cinerea. PLoS ONE 2015, 10, e0141586. [Google Scholar] [CrossRef]
- Pelkmans, J.F.; Patil, M.B.; Gehrmann, T.; Reinders, M.J.; Wösten, H.A.; Lugones, L.G. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 2017, 7, 310. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.E. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 2012, 191, 33–64. [Google Scholar] [CrossRef]
- Wong Sak Hoi, J.; Dumas, B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 2010, 9, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Mead, M.E.; Stanton, B.C.; Kruzel, E.K.; Hull, C.M. Targets of the Sex Inducer homeodomain proteins are required for fungal development and virulence in Cryptococcus neoformans. Mol. Microbiol. 2015, 95, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.; Scharfenstein, L.; Mack, B.M.; Chang, P.K.; Wei, Q.; Lebar, M.; Carter-Wientjes, C.; Majumdar, R.; Mitra, C.; et al. The Aspergillus flavus Homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 2017, 9, 315. [Google Scholar] [CrossRef]
- Cary, J.W.; Entwistle, S.; Satterlee, T.; Mack, B.M.; Gilbert, M.K.; Chang, P.K.; Scharfenstein, L.; Yin, Y.; Calvo, A.M. The transcriptional regulator Hbx1 affects the expression of thousands of genes in the aflatoxin-producing fungus Aspergillus flavus. G3 2019, 9, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Satterlee, T.; Nepal, B.; Lorber, S.; Puel, O.; Calvo, A.M. The Transcriptional regulator HbxA governs development, secondary metabolism, and virulence in Aspergillus fumigatus. Appl. Environ. Microbiol. 2020, 86, e01779-19. [Google Scholar] [CrossRef] [PubMed]
- Son, S.H.; Son, Y.E.; Cho, H.J.; Chen, W.; Lee, M.K.; Kim, L.H.; Han, D.M.; Park, H.S. Homeobox proteins are essential for fungal differentiation and secondary metabolism in Aspergillus nidulans. Sci. Rep. 2020, 10, 6094. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.S.; Zheng, J.; Yin, Y.; Lorber, S.; Puel, O.; Dhingra, S.; Espeso, E.A.; Calvo, A.M. Homeobox transcription factor HbxA influences expression of over one thousand genes in the model fungus Aspergillus nidulans. PLoS ONE 2023, 18, e0286271. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Wangsanut, T.; Fonzi, W.A.; Rolfes, R.J. The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans. FEMS Yeast Res. 2015, 15, fov093. [Google Scholar] [CrossRef]
- Wangsanut, T.; Ghosh, A.K.; Metzger, P.G.; Fonzi, W.A.; Rolfes, R.J. Grf10 and Bas1 regulate transcription of adenylate and one-carbon biosynthesis genes and affect virulence in the human fungal pathogen Candida albicans. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Wangsanut, T.; Arnold, S.J.Y.; Jilani, S.Z.; Marzec, S.; Monsour, R.C.; Rolfes, R.J. Grf10 regulates the response to copper, iron, and phosphate in Candida albicans. G3 2023, 13, jkad070. [Google Scholar] [CrossRef]
- Antal, Z.; Rascle, C.; Cimerman, A.; Viaud, M.; Billon-Grand, G.; Choquer, M.; Bruel, C. The homeobox BcHOX8 gene in Botrytis cinerea regulates vegetative growth and morphology. PLoS ONE 2012, 7, e48134. [Google Scholar] [CrossRef]
- Fu, T.; Han, J.H.; Shin, J.H.; Song, H.; Ko, J.; Lee, Y.H.; Kim, K.T.; Kim, K.S. Homeobox transcription factors are required for fungal development and the suppression of host defense mechanisms in the Colletotrichum scovillei-pepper pathosystem. mBio 2021, 12, e0162021. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Izumitsu, K.; Irie, T.; Suzuki, K. The homeobox transcription factor CoHox1 is required for the morphogenesis of infection hyphae in host plants and pathogenicity in Colletotrichum orbiculare. Mycoscience 2018, 60, 110–115. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, X.; Xie, Q.; Huang, Q.; Zhang, C.; Zhai, H.; Xu, L.; Lu, G.; Shim, W.B.; Wang, Z. A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS ONE 2012, 7, e45432. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, S.Y.; Kim, K.S.; Rho, H.S.; Chi, M.H.; Choi, J.; Park, J.; Kong, S.; Park, J.; Goh, J.; et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009, 5, e1000757. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, S.; Zhao, X.; Chen, X.; Zheng, W.; Lu, G.; Xu, J.R.; Wang, Z. A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 2010, 23, 366–375. [Google Scholar] [CrossRef]
- Arnaise, S.; Zickler, D.; Poisier, C.; Debuchy, R. pah1: A homeobox gene involved in hyphal morphology and microconidiogenesis in the filamentous ascomycete Podospora anserina. Mol. Microbiol. 2001, 39, 54–64. [Google Scholar] [CrossRef]
- Bhoite, L.T.; Allen, J.M.; Garcia, E.; Thomas, L.R.; Gregory, I.D.; Voth, W.P.; Whelihan, K.; Rolfes, R.J.; Stillman, D.J. Mutations in the pho2 (bas2) transcription factor that differentially affect activation with its partner proteins bas1, pho4, and swi5. J. Biol. Chem. 2002, 277, 37612–37618. [Google Scholar] [CrossRef]
- Berben, G.; Legrain, M.; Hilger, F. Studies on the structure, expression and function of the yeast regulatory gene PHO2. Gene 1988, 66, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Watanabe, D.; Nogami, S.; Morishita, S.; Ohya, Y. Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr. Genet. 2009, 55, 365–380. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, E.J.; Lopez-Maury, L.; Bähler, J.; Roe, J.H. A metabolic strategy to enhance long-term survival by Phx1 through stationary phase-specific pyruvate decarboxylases in fission yeast. Aging 2014, 6, 587–601. [Google Scholar] [CrossRef]
- Yu, J.; Yu, M.; Song, T.; Cao, H.; Pan, X.; Yong, M.; Qi, Z.; Du, Y.; Zhang, R.; Yin, X.; et al. A homeobox transcription factor UvHOX2 regulates chlamydospore formation, conidiogenesis, and pathogenicity in Ustilaginoidea virens. Front. Microbiol. 2019, 10, 1071. [Google Scholar] [CrossRef] [PubMed]
- Torres-Guzmán, J.C.; Domínguez, A. HOY1, a homeo gene required for hyphal formation in Yarrowia lipolytica. Mol. Cell. Biol. 1997, 17, 6283–6293. [Google Scholar] [CrossRef]
- Wahl, R.; Zahiri, A.; Kämper, J. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol. Microbiol. 2010, 75, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.A.; Wallen, R.M.; Zhao, C.; Shi, T.; Perlin, M.H. Coordinate regulation of Ustilago maydis ammonium transporters and genes involved in mating and pathogenicity. Fungal Biol. 2018, 122, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Wallen, R.M.; Richardson, K.; Furnish, M.; Mendoza, H.; Dentinger, A.; Khanal, S.; Perlin, M.H. Hungry for Sex: Differential roles for Ustilago maydis b locus omponents in haploid cells vis à vis nutritional availability. J. Fungi 2021, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Ohm, R.A.; de Jong, J.F.; de Bekker, C.; Wösten, H.A.; Lugones, L.G. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011, 81, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Köhler, J.; Fink, G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef]
- Lo, H.J.; Köhler, J.R.; DiDomenico, B.; Loebenberg, D.; Cacciapuoti, A.; Fink, G.R. Nonfilamentous C. albicans mutants are avirulent. Cell 1997, 90, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.M.; Bignell, E.; Warn, P.; Jones, M.D.; Denning, D.W.; Mühlschlegel, F.A.; Rogers, T.R.; Haynes, K. Candida glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol. Microbiol. 2003, 50, 1309–1318. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, C.; Liu, X.; Ma, Z. A transcription factor FgSte12 is required for pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Vallim, M.A.; Miller, K.Y.; Miller, B.L. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 2000, 36, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Borneman, A.R.; Hynes, M.J.; Andrianopoulos, A. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 2001, 157, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Hatamoto, O.; Masuda, T.; Sato, T.; Takeuchi, M. Function analysis of steA homolog in Aspergillus oryzae. Fungal Genet. Biol. 2007, 44, 330–338. [Google Scholar] [CrossRef]
- Katayama, T.; Bayram, Ö.; Mo, T.; Karahoda, B.; Valerius, O.; Takemoto, D.; Braus, G.H.; Kitamoto, K.; Maruyama, J.I. Novel Fus3- and Ste12-interacting protein FsiA activates cell fusion-related genes in both Ste12-dependent and -independent manners in Ascomycete filamentous fungi. Mol. Microbiol. 2021, 115, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhang, B.; Gai, Y.; Sun, X.; Chung, K.R.; Li, H. Cell-wall-degrading enzymes required for virulence in the host selective toxin-producing necrotroph Alternaria alternata of citrus. Front. Microbiol. 2019, 10, 2514. [Google Scholar] [CrossRef]
- Li, D.; Bobrowicz, P.; Wilkinson, H.H.; Ebbole, D.J. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 2005, 170, 1091–1104. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, W.; Chen, Y.; Xiang, M.; Liu, X. DdaSTE12 is involved in trap formation, ring inflation, conidiation, and vegetative growth in the nematode-trapping fungus Drechslerella dactyloides. Appl. Microbiol. Biotechnol. 2021, 105, 7379–7393. [Google Scholar] [CrossRef] [PubMed]
- Bai, N.; Xie, M.; Liu, Q.; Wang, W.; Liu, Y.; Yang, J. AoSte12 is required for mycelial development, conidiation, trap morphogenesis, and secondary metabolism by regulating hyphal fusion in nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Spectr. 2023, 11, e0395722. [Google Scholar] [CrossRef]
- Gu, S.Q.; Li, P.; Wu, M.; Hao, Z.M.; Gong, X.D.; Zhang, X.Y.; Tian, L.; Zhang, P.; Wang, Y.; Cao, Z.Y.; et al. StSTE12 is required for the pathogenicity of Setosphaeria turcica by regulating appressorium development and penetration. Microbiol. Res. 2014, 169, 817–823. [Google Scholar] [CrossRef]
- Park, G.; Xue, C.; Zheng, L.; Lam, S.; Xu, J.R. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 2002, 15, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Schamber, A.; Leroch, M.; Diwo, J.; Mendgen, K.; Hahn, M. The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 2010, 11, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, G.; Fujii, S.; Tsuge, S.; Shiraishi, T.; Kubo, Y. The Colletotrichum lagenariu Ste12-like gene CST1 is essential for appressorium penetration. Mol. Plant Microbe Interact. 2003, 16, 315–325. [Google Scholar] [CrossRef][Green Version]
- Deng, F.; Allen, T.D.; Nuss, D.L. Ste12 transcription factor homologue CpST12 is down-regulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 2007, 6, 235–244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, Y.; Wang, J.; Yang, K.; Fan, L.; Wang, Z.; Yin, Y. Regulation of conidiation, polarity growth, and pathogenicity by MrSte12 transcription factor in entomopathogenic fungus, Metarhizium rileyi. Fungal Genet. Biol. 2021, 155, 103612. [Google Scholar] [CrossRef]
- Gruber, S.; Zeilinger, S. The transcription factor Ste12 mediates the regulatory role of the Tmk1 MAP kinase in mycoparasitism and vegetative hyphal fusion in the filamentous fungus Trichoderma atroviride. PLoS ONE 2014, 9, e111636. [Google Scholar] [CrossRef] [PubMed]
- Schalamun, M.; Hinterdobler, W.; Schinnerl, J.; Brecker, L.; Schmoll, M. The transcription factor STE12 influences growth on several carbon sources and production of dehydroacetic acid (DHAA) in Trichoderma reesei. Sci. Rep. 2024, 14, 9625. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Kamiya, S.; Ozaki, Y.; Kameoka, S.; Kayano, Y.; Saikia, S.; Akano, F.; Uemura, A.; Takagi, H.; Terauchi, R.; et al. A nuclear protein NsiA from Epichloë festucae interacts with a MAP kinase MpkB and regulates the expression of genes required for symbiotic infection and hyphal cell fusion. Mol. Microbiol. 2020, 114, 626–640. [Google Scholar] [CrossRef] [PubMed]
- Rispail, N.; Di Pietro, A. Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant Microbe Interact. 2009, 22, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Asunción García-Sánchez, M.; Martín-Rodrigues, N.; Ramos, B.; de Vega-Bartol, J.J.; Perlin, M.H.; Díaz-Mínguez, J.M. fost12, the Fusarium oxysporum homolog of the transcription factor Ste12, is upregulated during plant infection and required for virulence. Fungal Genet. Biol. 2010, 47, 216–225. [Google Scholar] [CrossRef]
- Wei, Q.; Du, Y.; Jin, K.; Xia, Y. The Ste12-like transcription factor MaSte12 is involved in pathogenicity by regulating the appressorium formation in the entomopathogenic fungus, Metarhizium acridum. Appl. Microbiol. Biotechnol. 2017, 101, 8571–8584. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Springer, D.J.; Behr, M.J.; Samsonoff, W.A.; Chaturvedi, S.; Chaturvedi, V. Transcription factor STE12alpha has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot. Cell 2006, 5, 1065–1080. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, X.; Liu, W.; Chu, X.; Sun, Q.; Tan, C.; Yang, Q.; Jiao, M.; Guo, J.; Kang, Z. The transcription factor PstSTE12 is required for virulence of Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2018, 19, 961–974. [Google Scholar] [CrossRef]
- Lyu, X.; Wang, Q.; Liu, A.; Liu, F.; Meng, L.; Wang, P.; Zhang, Y.; Wang, L.; Li, Z.; Wang, W. The transcription factor Ste12-like increases the mycelial abiotic stress tolerance and regulates the fruiting body development of Flammulina filiformis. Front. Microbiol. 2023, 14, 1139679. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.M. Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog. 2015, 11, e1004608. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.S.; Tebbets, B. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 2007, 10, 314–319. [Google Scholar] [CrossRef]
- Boyce, K.J.; Andrianopoulos, A. Fungal dimorphism: The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol. Rev. 2015, 39, 797–811. [Google Scholar] [CrossRef]
- Kummasook, A.; Tzarphmaag, A.; Thirach, S.; Pongpom, M.; Cooper, C.R., Jr.; Vanittanakom, N. Penicillium marneffei actin expression during phase transition, oxidative stress, and macrophage infection. Mol. Biol. Rep. 2011, 38, 2813–2819. [Google Scholar] [CrossRef]
- Amsri, A.; Jeenkeawpieam, J.; Sukantamala, P.; Pongpom, M. Role of acuK in control of iron acquisition and gluconeogenesis in Talaromyces marneffei. J. Fungi 2021, 7, 798. [Google Scholar] [CrossRef]
- Dankai, W.; Pongpom, M.; Vanittanakom, N. Validation of reference genes for real-time quantitative RT-PCR studies in Talaromyces marneffei. J. Microbiol. Methods 2015, 118, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.; Jang, S.; Kim, S.; Kong, S.; Choi, J.; Ahn, K.; Kim, J.; Lee, S.; Kim, S.; et al. FTFD: An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 2008, 24, 1024–1025. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, H.; Luo, X.; Li, H.; Gao, Q.; Zhang, L.; Teng, Y.; Zhao, Q.; Zuo, Z.; Ren, J. IBS 2.0: An upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. 2022, 50, W420–W426. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Kissinger, C.R.; Liu, B.; Martin-Blanco, E.; Kornberg, T.B.; Pabo, C.O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: A framework for understanding homeodomain-DNA interactions. Cell 1990, 63, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.B.; Yang, Y.; Wang, B.; Füsti-Molnár, L.; Weaver, M.N.; Merz, K.M., Jr. Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF). J. Chem. Theory Comput. 2010, 6, 2935–2947. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Wangsanut, T.; Tobin, J.M.; Rolfes, R.J. Functional mapping of transcription factor Grf10 that regulates adenine-responsive and filamentation genes in Candida albicans. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Yang, E.; Chow, W.N.; Wang, G.; Woo, P.C.; Lau, S.K.; Yuen, K.Y.; Lin, X.; Cai, J.J. Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei. PLoS Genet. 2014, 10, e1004662. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, S.; Payne, M.; Canovas, D.; Pase, L.; Ngaosuwankul, N.; Beard, S.; Oshlack, A.; Smyth, G.K.; Chaiyaroj, S.C.; Boyce, K.J.; et al. Cell-type-specific transcriptional profiles of the dimorphic pathogen Penicillium marneffei reflect distinct reproductive, morphological, and environmental demands. G3 2013, 3, 1997–2014. [Google Scholar] [CrossRef]
- Lin, X.; Ran, Y.; Gou, L.; He, F.; Zhang, R.; Wang, P.; Dai, Y. Comprehensive transcription analysis of human pathogenic fungus Penicillium marneffei in mycelial and yeast cells. Med. Mycol. 2012, 50, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Vonk, P.J.; Ohm, R.A. The role of homeodomain transcription factors in fungal development. Fungal Biol. Rev. 2018, 32, 219–230. [Google Scholar] [CrossRef]
- Brazas, R.M.; Bhoite, L.T.; Murphy, M.D.; Yu, Y.; Chen, Y.; Neklason, D.W.; Stillman, D.J. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J. Biol. Chem. 1995, 270, 29151–29161. [Google Scholar] [CrossRef] [PubMed]
- Justice, M.C.; Hogan, B.P.; Vershon, A.K. Homeodomain-DNA interactions of the Pho2 protein are promoter-dependent. Nucleic Acids Res. 1997, 25, 4730–4739. [Google Scholar] [CrossRef]
- Oladejo, D.O.; Duselu, G.O.; Dokunmu, T.M.; Isewon, I.; Oyelade, J.; Okafor, E.; Iweala, E.E.; Adebiyi, E. In silico structure prediction, molecular docking, and dynamic simulation of Plasmodium falciparum AP2-I transcription factor. Bioinform. Biol. Insights 2023, 17, 11779322221149616. [Google Scholar] [CrossRef] [PubMed]
- Curry, L.; Soleimani, M. Belzutifan: A novel therapeutic for the management of von Hippel-Lindau disease and beyond. Future Oncol. 2024, 20, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.S. Exploiting fungal virulence-regulating transcription factors as novel antifungal drug targets. PLoS Pathog. 2015, 11, e1004936. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Dai, L.; Ma, Y.; Wang, J.; Yan, H.; Jin, Y.; Zhu, X.; Liu, Z. Homeobox proteins are potential biomarkers and therapeutic targets in gastric cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 866. [Google Scholar] [CrossRef] [PubMed]
- Chin, F.W.; Chan, S.C.; Veerakumarasivam, A. Homeobox gene expression dysregulation as potential diagnostic and prognostic biomarkers in bladder cancer. Diagnostics 2023, 13, 2641. [Google Scholar] [CrossRef]
- King, N. The unicellular ancestry of animal development. Dev. Cell 2004, 7, 313–325. [Google Scholar] [CrossRef]
- Pruksaphon, K.; Nosanchuk, J.D.; Ratanabanangkoon, K.; Youngchim, S. Talaromyces marneffei Infection: Virulence, intracellular lifestyle and host defense mechanisms. J. Fungi 2022, 8, 200. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, J.; Chen, W.; Geng, X.; He, T.; Ding, S.; Sun, B.; Li, H.; Chen, L. Identification and characterization of the homeobox gene family in Fusarium pseudograminearum reveal their roles in pathogenicity. Biochem. Genet. 2022, 60, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Feng, B.; Chen, D.; Yao, Z.; Yang, B. Genome-wide identification and expression analysis of homeobox genes in Fusarium oxysporum f. sp. lycopersici. J. Plant Pathol. 2023, 105, 137–145. [Google Scholar] [CrossRef]
- Barbaric, S.; Münsterkötter, M.; Goding, C.; Hörz, W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 1998, 18, 2629–2639. [Google Scholar] [CrossRef]
- Brazas, R.M.; Stillman, D.J. The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc. Natl. Acad. Sci. USA 1993, 90, 11237–11241. [Google Scholar] [CrossRef] [PubMed]
- Ejzykowicz, D.E.; Cunha, M.M.; Rozental, S.; Solis, N.V.; Gravelat, F.N.; Sheppard, D.C.; Filler, S.G. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol. Microbiol. 2009, 72, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.T.; MacCallum, D.M.; Clancy, S.D.; Odds, F.C.; Brown, A.J.; Butler, G. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 2004, 53, 969–983. [Google Scholar] [CrossRef]
- Mulhern, S.M.; Logue, M.E.; Butler, G. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 2006, 5, 2001–2013. [Google Scholar] [CrossRef]
- Wakade, R.S.; Krysan, D.J. The Cbk1-Ace2 axis guides Candida albicans from yeast to hyphae and back again. Curr. Genet. 2021, 67, 461–469. [Google Scholar] [CrossRef]





| Division | Species | Genes | Key Findings | Reference |
|---|---|---|---|---|
| Ascomycota | A. flavus (Mycotoxigenic mold) | hbx1-8 | The hbx1 gene is required for production of conidia, sclerotia, and aflatoxin. | [21,22] |
| A. fumigatus (Human pathogen) | hbxA | The hbxA gene regulates asexual development, secondary metabolism, and virulence. | [23] | |
| A. nidulans (Mold model) | hbxA-H | - The hbxA and hbxB genes regulate asexual and sexual development. - The hbxB gene regulates trehalose biosynthesis and stress tolerance. - The hbxD gene is important for normal conidial production. - The hbxA gene regulates the expression of other key regulators involved in developmental processes, secondary metabolism, and virulence. | [24,25] | |
| Botrytis cinerea (Plant pathogen) | BcHOX1-8 | The BcHOX8 gene regulates morphology and pathogenicity. | [30] | |
| C. albicans (Human pathogen) | GRF10 | - The GRF10 gene is required for morphogenesis and pathogenicity. - The GRF10 gene regulates the response to purine, phosphate, iron and copper. | [27,28,29] | |
| Colletotrichum scovillei (Plant pathogen) | CsHOX1-10 | - The CsHOX1 gene is required for host defense suppression. - The CsHOX2 gene is important for conidiation and appressorium development. | [31] | |
| Colletotrichum orbiculare (Plant pathogen) | CoHOX1-9 | The CoHOX3 gene is essential for appressorium formation and pathogenicity. | [32] | |
| Fusarium spp. (Plant pathogen) | FgHTF1 FvHTF1 FoHTF1 | The Fusarium HTF1 gene is required for phialide development and conidiogenesis. | [33] | |
| Magnaporthe oryzae (Plant pathogen) | MoHOX1-8 | The MoHOX2 (HTF1) and MoHOX7 genes are required for conidiation, appressorium development, and pathogenicity. | [34,35] | |
| Neurospora crassa (Mold model) | kal-1 | The kal-1 gene is necessary for basal hyphal growth, conidiation, and nutrient sensing. | [26] | |
| Podospora anserina (Mold model) | pah1-7 | - The pah1 gene is involved in hyphal morphogenesis and production of microconidiation. The pah1 has no role in sexual development after fertilization. - The pah2 and pah5 genes play important role in shaping fruiting body and sexual asci spore formation. | [14,36] | |
| S. cerevisiae (Yeast model) | PHO2 | - PHO2 controls sporulation and bud morphogenesis. PHO2 regulates the cellular response to phosphate and adenine starvation. - PHO2 activates the gene involved in mating type switching during vegetative cell division. | [37,38,39] | |
| S. pombe (Yeast model) | phx1 | The phx1 gene is required for sporulation and regulates thiamine responsive genes. | [40] | |
| Ustilaginoidea virens (Plant pathogen) | UvHOX2 | The UvHOX2 gene regulates chlamydospore formation, conidiation, and pathogenicity. | [41] | |
| Yarrowia lipolytica (Dimorphic yeast model) | HOY1 | The HOY1 gene regulates filamentation. | [42] | |
| Basidiomycota | Volvariella volvacea (Mushroom) | VvHox1-8 | Gene expression analyses shown that VvHox genes are differentially expressed during each step of fruiting body development. | [15] |
| Ustilago maydis (Plant pathogen) | bE and bW | - During sexual reproduction, the genes for bE/bW complex is required for stable dikaryon formation, hyphal proliferation, and pathogenicity. - In haploid cells, the genes for bE/bW complex is necessary for filamentous growth in response to nitrogen starvation. | [43,44,45] | |
| C. neoformans (Human pathogen) | SXI2a and SXI1α | - The SXI2a and SXI1α genes are required for sexual development. - Sxi2a and Sxi1α proteins regulate genes involved in sexual development, nutrient starvation, and virulence. | [20] | |
| Schizophyllum commune (Mushroom) | hom1 and hom2 | - The hom1 and hom2 genes regulate fructification process. - The hom1 gene stimulates biomass production while the hom2 gene represses vegetative growth. | [17,46] | |
| Coprinopsis cinerea (Mushroom) | RNA-seq analyses | Many homeobox genes show changes in expression levels during fruitification. | [16] |
| Division | Species | Genes | Key Findings | Reference |
|---|---|---|---|---|
| Ascomycota | S. cerevisiae | STE12 | Role in mating and pseudohyphal/invasive growth. | [19] |
| C. albicans | CPH1 | Role in mating, filamentation, biofilm formation, and virulence. | [47,48] | |
| C. glabrata | STE12 | Required for virulence and nitrogen starvation-induced filamentation. | [49] | |
| Fusarium graminearum | FgSte12 | - Required for virulence and secretion of cellulase and protease. - Not involved in mycelial growth. | [50] | |
| A. nidulans | steA | - Required for sexual reproduction. - Not involved in mycelial growth. | [51] | |
| T. marneffei | stlA | - The stlA gene can complement the sexual defect of an A. nidulans steA mutant. - No role on vegetative growth, asexual development, and dimorphic switching. | [52] | |
| A. oryzae | steA | - Involved in regulating the expression of cell wall-degrading enzymes. - Role in cell fusion | [53,54] | |
| Alternaria alternata (Plant fungal pathogen) | STE12 | - Role in pathogenicity. -No role in growth, ACT toxin production, and response to oxidative and osmotic stress. | [55] | |
| N. crassa | STE12/pp-1 | Role in aerial hyphae/colony growth, conidiophore development and fertility. | [56] | |
| Drechslerella dactyloides (Nematophagus fungus) | DdaSTE12 | Role in nematode trap formation, ring cell inflation, conidiation and vegetative growth. | [57] | |
| Arthrobotrys oligospora (Nematophagus fungus) | AoSte12 | - Role in nematode trap morphogenesis, conidiation, hyphal fusion and mycelial growth. - Involved in stress tolerance response and production of secondary metabolites. | [58] | |
| Setosphaeria turcica (Plant pathogen) | StSte12 | Role in pathogenicity by regulating vegetative growth, conidiation, appressorial development, and penetration. | [59] | |
| M. oryzae | MST12 | - Regulating infectious growth and pathogenicity. - No role in mycelial growth. | [60] | |
| B. cinerea | STE12 | Role in radial growth, pigment production, sclerotia formation, and pathogenicity. | [61] | |
| Colletotrichum lagenariu (Plant pathogen) | CST1 | Role in pathogenicity by controlling the production of infectious hyphae from appressoria. | [62] | |
| Cryphonectria parasitica (Plant pathogen) | CpST12 | Role in virulence and female fertility. | [63] | |
| Metarhizium rileyi (Entomopathogenic fungus) | MrSte12 | - Role in virulence, appressorium formation, hyphal morphogenesis, and conidiogenesis. - Role in stress tolerance. | [64] | |
| Trichoderma atroviride (Plant fungal pathogen) | STE12 | Role in carbon source-dependent growth, hyphal fusion, lytic enzyme expression, and mycoparasitic activity. | [65] | |
| Trichoderma reesei (Industrial fungus) | STE12 | - Regulation of cellulase gene expression, carbon source utilization, and secondary metabolites. - Involvement in iron homeostasis via the regulation of iron transport genes and siderophore-associated genes. | [66] | |
| Epichloë festucae (Endophytic fungus) | STE12 | Role in pathogenicity, but not hyphal fusion. | [67] | |
| F. oxysporum | STE12 | Required for invasive growth and virulence, but not hyphal fusion. | [68,69] | |
| Metarhizium acridum (Entomopathogenic fungus) | MaSte12 | Role in pathogenicity by regulating the appressorium formation. | [70] | |
| Basidiomycota | C. gattii (Human fungal pathogen) | Ste12alpha | Role in mating, melanin production, and ecological fitness. | [71] |
| Puccinia striiformis f. sp. tritici (Plant fungal pathogen) | PstSTE12 | - Role in pathogenicity, haustorium formation, fungal colonization and hyphal development. - The PstSTE12 gene can complement the mating defect of a S. cerevisiae ste12 mutant. | [72] | |
| Flammulina filiformis (Mushroom) | - Role in fruiting body development. - Role in tolerance to salt stress, cold stress and oxidative stress. | [73] |
| Primer Name | Type | Sequences | Gene ID | Name |
|---|---|---|---|---|
| q-HOX1-F q-HOX1-R | Forward Reverse | GTCGAAAATCGAAGCCGCTC TCTCAGTGTTGTTCGACGGG | PMAA_076490 | EEA26588.1 (Hox1) |
| q-HOX2-F q-HOX2-R | Forward Reverse | AGTCAGTGCACTTGGCTCTC AGGGGCTCTACAAAAGGCAC | PMAA_074540 | EEA26382.1 (Hox2) |
| q-HOX3-F q-HOX3-R | Forward Reverse | CGAGCACTCCCTGACCATTT GGTACTCGTCACCAGGGTTG | PMAA_085630 | EEA24568.1 (Hox3) |
| q-HOX4-F q-HOX4-R | Forward Reverse | ACCCAGCATCTTCGCCTATG GCACGCAAGACGTCTGTAAC | PMAA_096860 | EEA23091.1 (Hox4) |
| qRfeB F qRfeB R | Forward Reverse | CCAGATTCAATGCGGCAATAC TACTTAGTCCACCGAGTCCAT | PMAA_096690 | EEA23074.1 (Hox5) |
| q-HOX6-F q-HOX6-R | Forward Reverse | ACAGCTCATCGCTATCACGG GGACGGGGATGTTTGTCTGT | PMAA_043800 | EEA20547.1 (Hox6) |
| q-HOX7-F q-HOX7-R | Forward Reverse | TCGTCCCTGGACTCTGACAT TTGGGGACGTTGTTGCTTCT | PMAA_001530 | EEA19358.1 (Hox7) |
| act1-F act1-R | Forward Reverse | TGATGAGGCACAGTCTAAGC CTTCTCTCTGTTGGACTTGG | PMAA_012310 | EEA18960.1 (Actin) |
| Designated Hox Name | Name | Protein ID |
|---|---|---|
| TmHox1 | YOX1 | EEA26588.1 |
| TmHox2 | - | EEA26382.1 |
| TmHox3 | - | EEA24568.1 |
| TmHox4 | Homeobox protein TGIF2LX | EEA23091.1 |
| TmHox5 | RfeB/Hoy1 | EEA23074.1 |
| TmHox6 | - | EEA20547.1 |
| TmHox7 | Homeobox protein AKR | EEA19358.1 |
| Protein | Strain ID | Alternative Name | C Pasricha et al., 2013 [96] | C Yang et al., 2014 [95] | C Lin et al., 2012 [97] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 18224 ID | PM1 ID | Fold Change (log2) | % of Total Expression | b Pattern | Fold Change 37 vs. 25 | Pattern 37 vs. 25 | 37-log2 | 25-log2 | ||||||
| a 37 > 25 vs. 37 | 25 > 37 vs. 25 | 37 vs. 25 | Hyphal | Asexual | Yeast | |||||||||
| EEA26588.1 (Hox1) | PMAA_076490 | GQ26_0112560 | Yox1 | na | na | na | na | na | na | 0001 | 2.57 | up | 16.53 | 15.17 |
| EEA26382.1 (Hox2) | PMAA_074540 | GQ26_0110650 | na | na | na | na | na | na | 0100 | na | na | na | na | |
| EEA24568.1 (Hox3) | PMAA_085630 | GQ26_0280750 | na | na | na | na | na | na | 1100 | 25.87 | down | 7.48 | 12.17 | |
| EEA23091.1 (Hox4) | PMAA_096860 | GQ26_0160760 | TGIF2LX | 2.2 | −0.42 | −0.58 | 17 | 71 | 12 | 0011 | 8.49 | up | 8.05 | 4.96 |
| EEA23074.1 (Hox5) | PMAA_096690 | GQ26_0161010 | RfeB/Hoy1 | na | na | na | na | na | na | 1011 | na | na | na | na |
| EEA20547.1 (Hox6) | PMAA_043800 | GQ26_0142040 | na | na | na | na | na | na | 0011 | 6.98 | down | 10.64 | 13.44 | |
| EEA19358.1 (Hox7) | PMAA_001530 | GQ26_0024010 | na | na | na | na | na | na | 0111 | 7.85 | down | 13.42 | 16.40 | |
| EEA26628.1 | PMAA_015520 | na | na | na | na | na | na | 93.57 | up | 10.3 | 3.75 | |||
| EEA28535.1 | PMAA_033420 | na | na | na | na | na | na | na | na | na | na | |||
| GQ26_0480540 | Homeobox protein Meis3-like 1 | na | na | na | na | na | na | 0100 | na | na | na | na | ||
| EEA24489.1 (SteA) | PMAA_084900 | GQ26_0530470 | Ste12, StlA | na | na | na | na | na | na | 1111 | na | na | na | na |
| Component | Free Energy (kcal/mol) | |||
|---|---|---|---|---|
| RfeB-TmSwi5 | Pho2-ScSwi5 | |||
| Wild-Type | Triple Mutant | Wild-Type | Triple Mutant | |
| Gas phase | ||||
| Van der Waals | −177.96 ± 10.13 | −104.77 ± 7.05 | −241.17 ± 17.45 | −175.65 ± 9.08 |
| Electrostatic | 1387.77 ± 100.32 | 1960.49 ± 121.79 | −1262.06 ± 138.54 | −124.02 ± 110.26 |
| Solvent phase | ||||
| Polar solvation | −1281.85 ± 94.0 | −1875.47 ± 116.85 | 1437.11 ± 134.07 | 259.44 ± 106.89 |
| Non-polar solvation | −24.94 ± 1.12 | −15.47 ± 0.99 | −35.05 ± 2.46 | −25.51 ± 1.22 |
| Total | −96.98 ± 11.86 | −35.22 ± 9.55 | −101.17 ± 22.05 | −65.74 ± 11.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pongpom, M.; Khamto, N.; Sukantamala, P.; Kalawil, T.; Wangsanut, T. Identification of Homeobox Transcription Factors in a Dimorphic Fungus Talaromyces marneffei and Protein-Protein Interaction Prediction of RfeB. J. Fungi 2024, 10, 687. https://doi.org/10.3390/jof10100687
Pongpom M, Khamto N, Sukantamala P, Kalawil T, Wangsanut T. Identification of Homeobox Transcription Factors in a Dimorphic Fungus Talaromyces marneffei and Protein-Protein Interaction Prediction of RfeB. Journal of Fungi. 2024; 10(10):687. https://doi.org/10.3390/jof10100687
Chicago/Turabian StylePongpom, Monsicha, Nopawit Khamto, Panwarit Sukantamala, Thitisuda Kalawil, and Tanaporn Wangsanut. 2024. "Identification of Homeobox Transcription Factors in a Dimorphic Fungus Talaromyces marneffei and Protein-Protein Interaction Prediction of RfeB" Journal of Fungi 10, no. 10: 687. https://doi.org/10.3390/jof10100687
APA StylePongpom, M., Khamto, N., Sukantamala, P., Kalawil, T., & Wangsanut, T. (2024). Identification of Homeobox Transcription Factors in a Dimorphic Fungus Talaromyces marneffei and Protein-Protein Interaction Prediction of RfeB. Journal of Fungi, 10(10), 687. https://doi.org/10.3390/jof10100687









