Identification and Characterization of a Predominant Hydrophobin in the Edible Mushroom Grifola frondosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture
2.2. High-Throughput Genome Sequencing and De Novo Assembly
2.3. Identification of Putative Hydrophobins
2.4. Bioinformatic Analysis of Hydrophobins
2.5. RNA Extraction and cDNA Synthesis
2.6. Quantification of Hydrophobin Gene Transcription
2.7. Heterologous Expression of Hydrophobin in Pichia Pastoris (Komagataella phaffii)
2.8. Water Contact Angle (WCA) Determination
2.9. X-ray Photoelectron Spectroscopy (XPS) Analysis
2.10. Thioflavin T (ThT) Fluorescence Measurement
2.11. Atomic Force Microscopy (AFM) Detection
2.12. Determination of Surface Hydrophobicity with 1-Anilino-8-naphthalenesulfonate (1,8-ANS)
2.13. Particle Size and Zeta Potential Measurements
2.14. Emulsification Assay
2.15. Statistical Analysis
3. Results and Discussion
3.1. Sequence Search and Annotation of Putative Hydrophobin Genes in Grifola frondosa
3.2. Bioinformatic Analysis of Hydrophobins
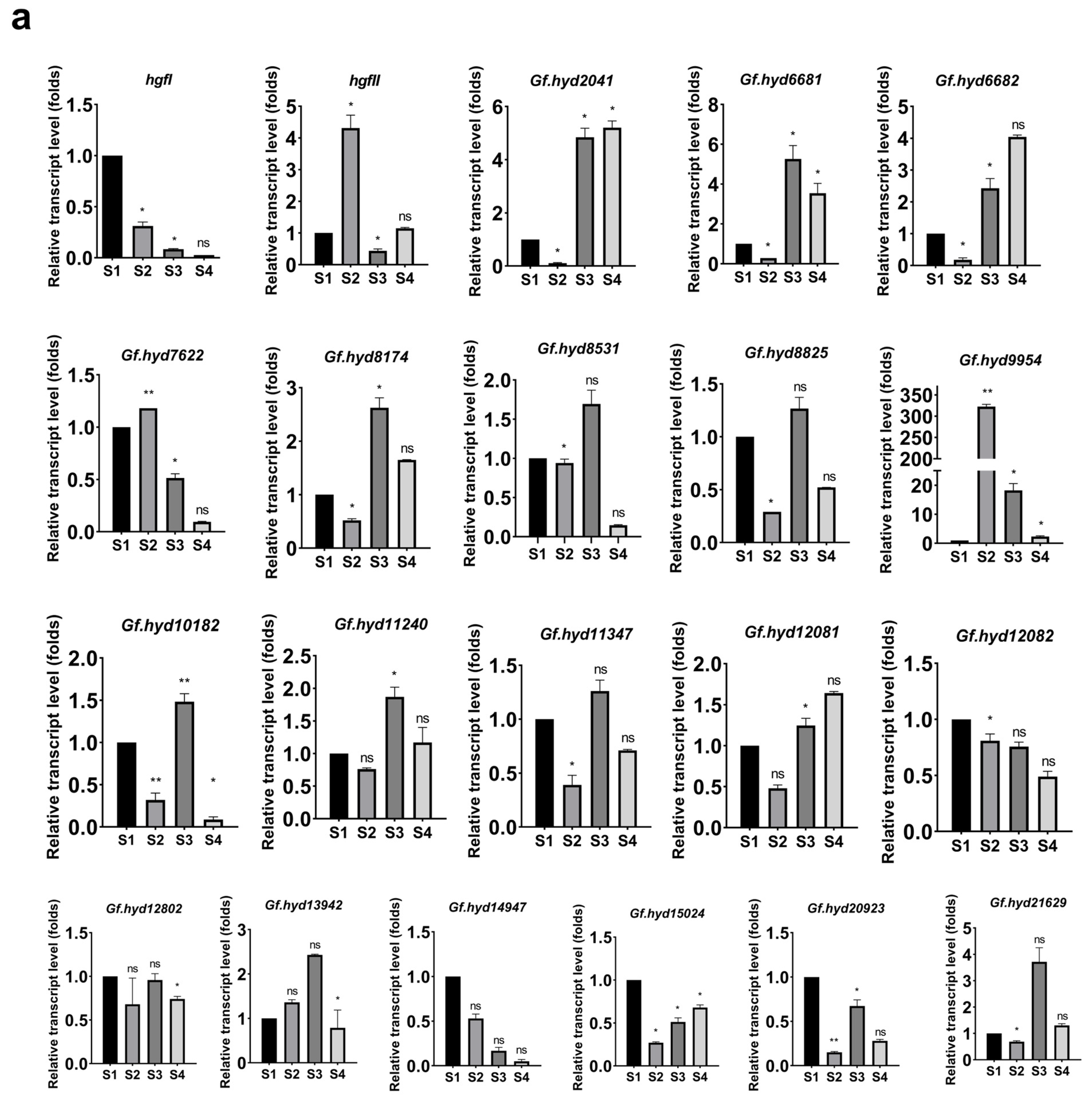
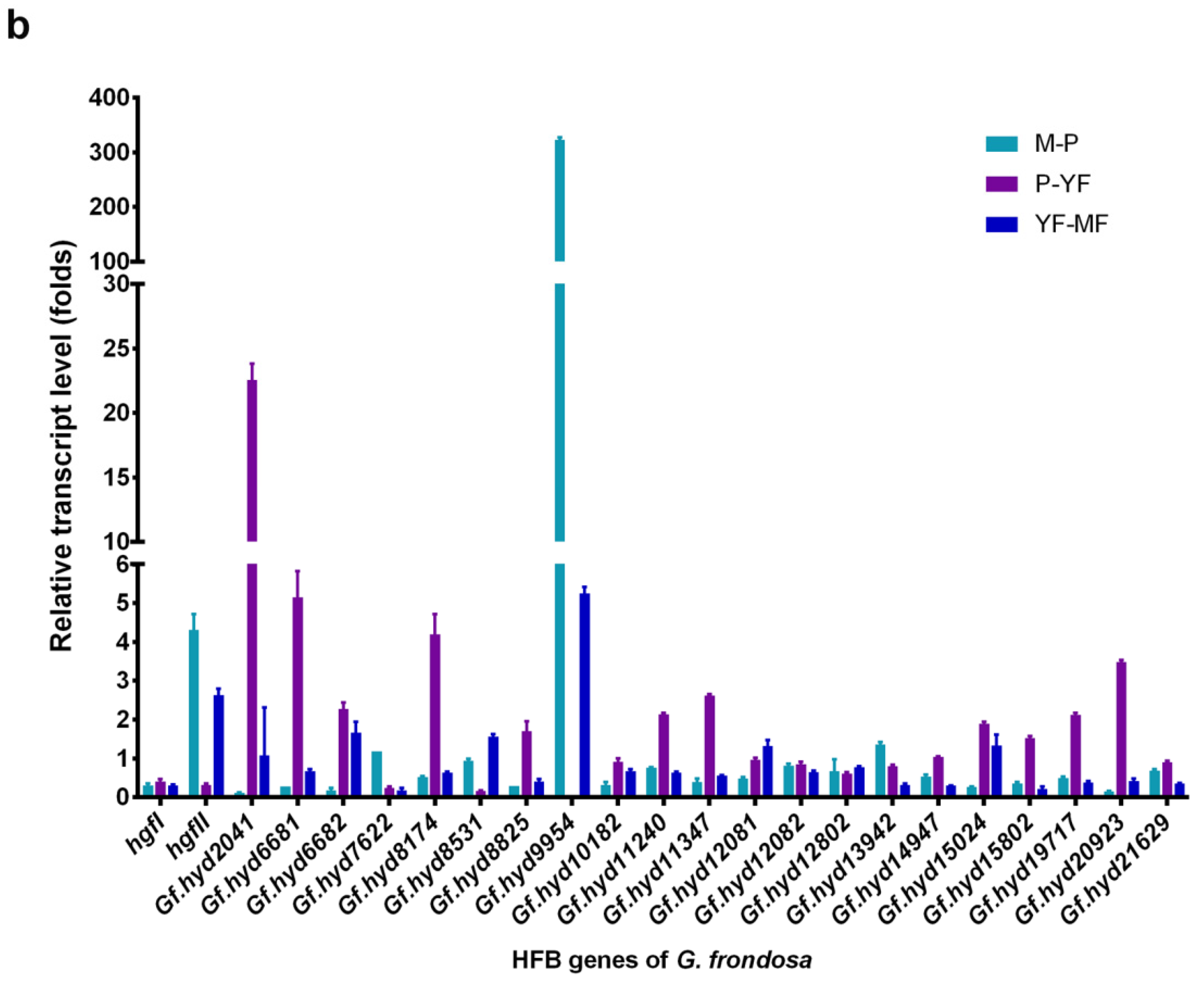
3.3. Expression Profile Analysis of Hydrophobin Genes during the Life Cycle of G. frondosa CICC®50075
3.4. rGf.hyd9954 Exhibits Wettability Alteration Ability Confirmed by Water Contact Angle
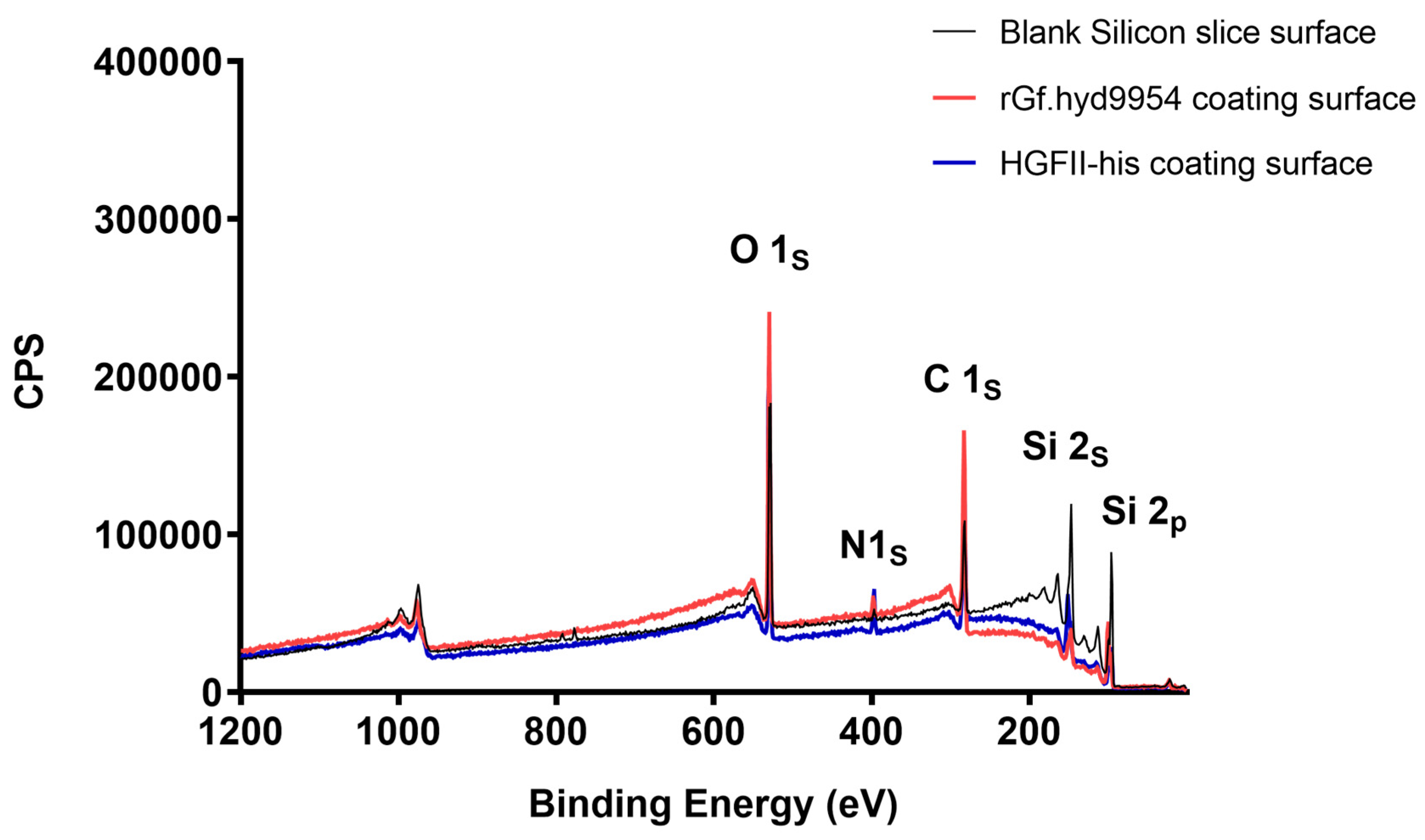
3.5. The Modification Ability of rGf.hyd9954 on a Silicon Slice
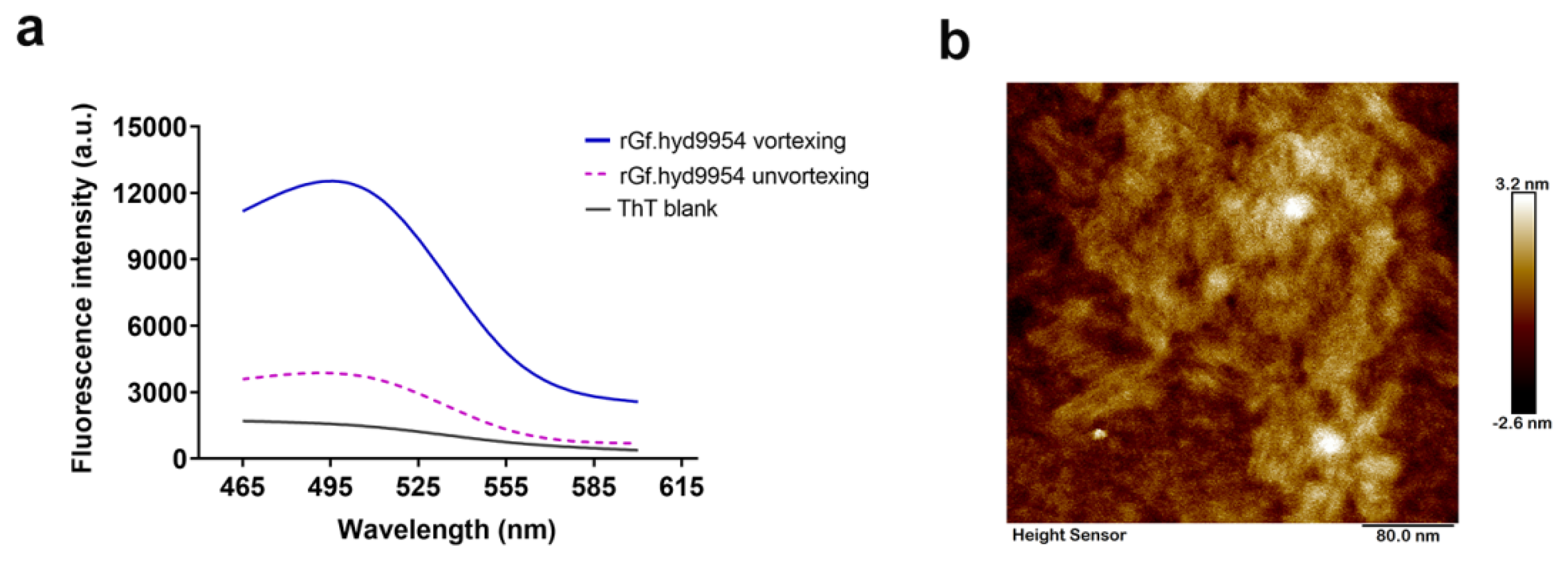
3.6. Monitoring of Self-Assembly by the Thioflavin T- (ThT-) Binding Assay
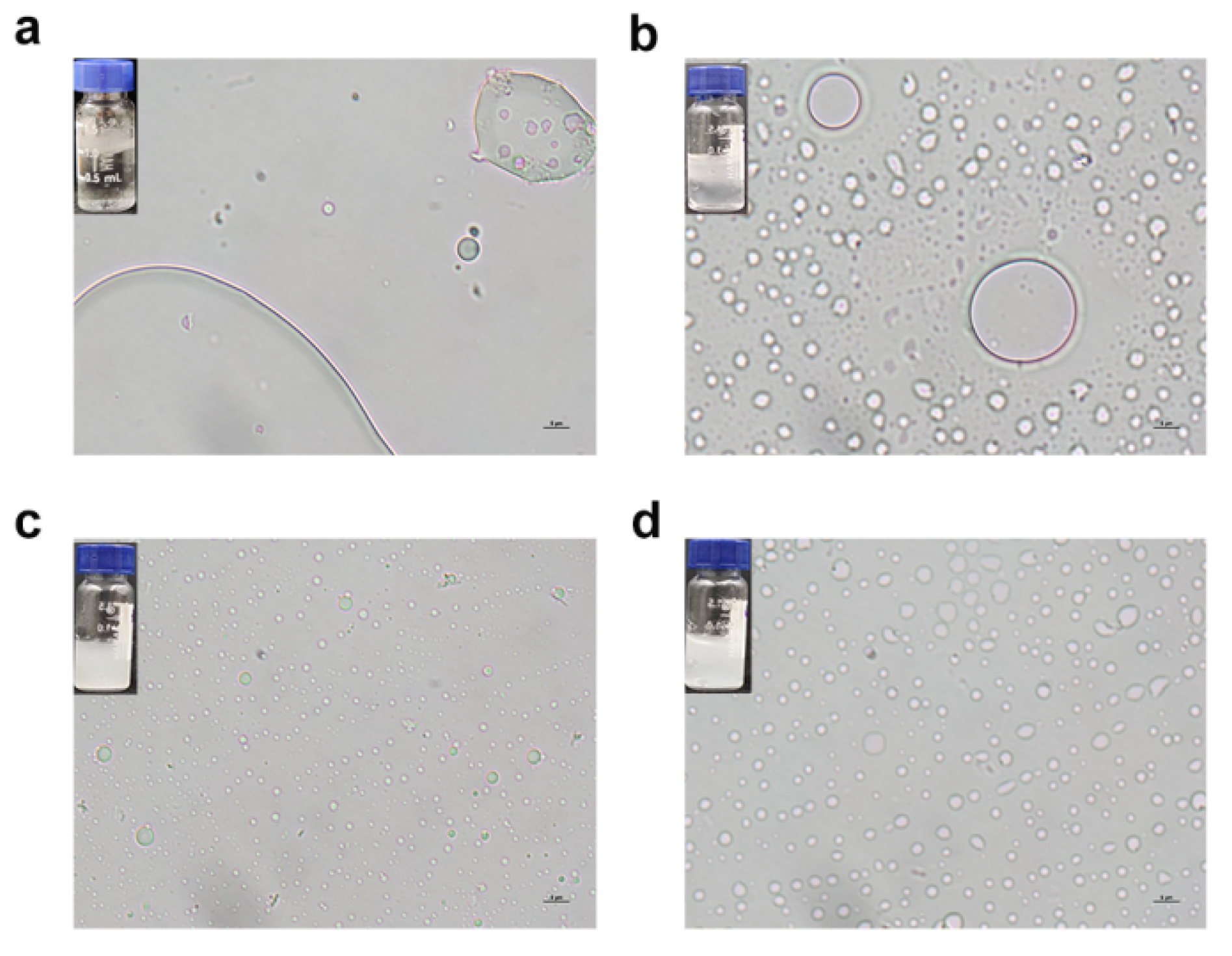
3.7. The rGf.hyd9954 Self-Assembly Behavior
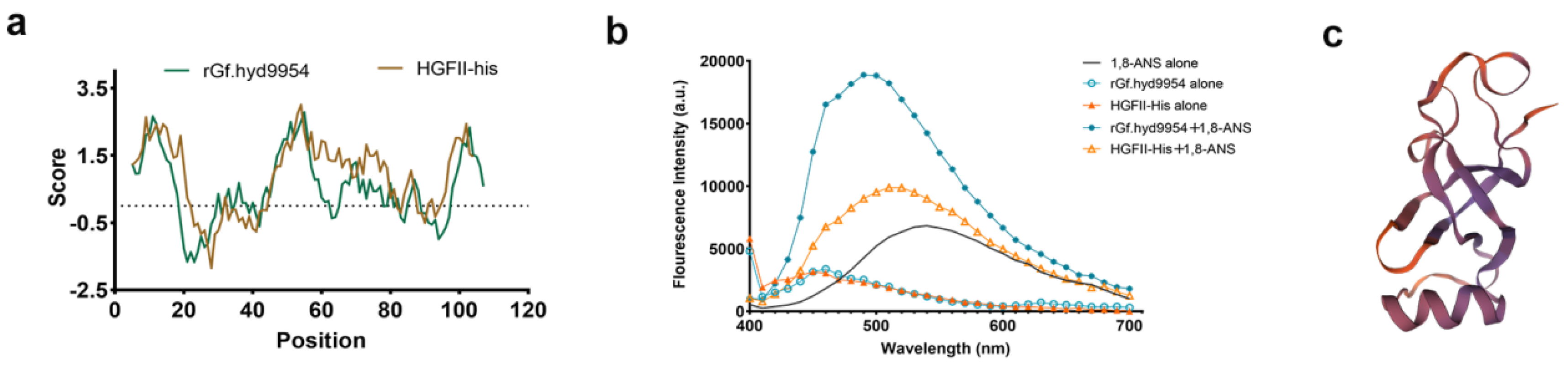
3.8. 1,8-ANS Fluorescence Measurements
3.9. The Putative Tertiary Structure of rGf.hyd9954
3.10. The Particle Size and Zeta Potential of rGf.hyd9954
3.11. The Emulsification Properties of rGf.hyd9954
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schor, M.; Reid, J.L.; MacPhee, C.E.; Stanley-Wall, N.R. The Diverse Structures and Functions of Surfactant Proteins. Trends Biochem. Sci. 2016, 41, 610–620. [Google Scholar] [CrossRef]
- De Vries, O.M.; Moore, S.; Arntz, C.; Wessels, J.G.; Tudzynski, P. Identification and characterization of a tri-partite hydrophobin from Claviceps fusiformis. A novel type of class II hydrophobin. Eur. J. Biochem. 1999, 262, 377–385. [Google Scholar] [CrossRef]
- Wessels, J.G.H. Fungal hydrophobins: Proteins that function at an interface. Trends Plant Sci 1996, 1, 9–15. [Google Scholar] [CrossRef]
- Wösten, H.A.; de Vocht, M.L. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 2000, 1469, 79–86. [Google Scholar] [CrossRef]
- Shin, Y.K.; Kim, D.W.; Lee, S.W.; Lee, M.J.; Gi Baek, S.; Lee, T.; Yun, S.H. Functional roles of all five putative hydrophobin genes in growth, development, and secondary metabolism in Fusarium graminearum. Fungal Genet. Biol. 2022, 160, 103683. [Google Scholar] [CrossRef]
- van Wetter, M.A.; Wösten, H.A.; Sietsma, J.H.; Wessels, J.G. Hydrophobin gene expression affects hyphal wall composition in Schizophyllum commune. Fungal Genet. Biol. 2000, 31, 99–104. [Google Scholar] [CrossRef]
- Linder, M.B.; Szilvay, G.R.; Nakari-Setälä, T.; Penttilä, M.E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef]
- Seidl-Seiboth, V.; Gruber, S.; Sezerman, U.; Schwecke, T.; Albayrak, A.; Neuhof, T.; von Döhren, H.; Baker, S.E.; Kubicek, C.P. Novel hydrophobins from Trichoderma define a new hydrophobin subclass: Protein properties, evolution, regulation and processing. J. Mol. Evol. 2011, 72, 339–351. [Google Scholar] [CrossRef]
- Wessels, J.G.H. Developmental regulation of fungal cell-wall formation. Annu. Rev. Phytopathol. 1994, 32, 413–437. [Google Scholar] [CrossRef]
- Lo, V.; Lai, J.-I.C.; Sunde, M. Fungal hydrophobins and their self-assembly into functional nanomaterials. Adv. Exp. Med. Biol. 2019, 1174, 161–185. [Google Scholar]
- van Wetter, M.A.; Wösten, H.A.; Wessels, J.G. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol. Microbiol. 2000, 36, 201–210. [Google Scholar] [CrossRef]
- Liu, D.; Sun, X.; Diao, W.; Qi, X.; Bai, Y.; Yu, X.; Li, L.; Fang, H.; Chen, Z.; Liu, Q.; et al. Comparative transcriptome analysis revealed candidate genes involved in fruiting body development and sporulation in Ganoderma lucidum. Arch. Microbiol. 2022, 204, 514. [Google Scholar] [CrossRef]
- Tao, Y.; Chen, R.; Yan, J.; Long, Y.; Tong, Z.; Song, H.; Xie, B. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef]
- Kazmierczak, P.; Kim, D.H.; Turina, M.; Van Alfen, N.K. A Hydrophobin of the chestnut blight fungus, Cryphonectria parasitica, is required for stromal pustule eruption. Eukaryot. Cell 2005, 4, 931–936. [Google Scholar] [CrossRef]
- Song, H.Y.; Kim, D.H.; Kim, J.M. Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes. Sci. Rep. 2018, 8, 8983. [Google Scholar] [CrossRef]
- Ma, H.; Snook, L.A.; Tian, C.; Kaminskyj, S.G.; Dahms, T.E. Fungal surface remodelling visualized by atomic force microscopy. Mycol. Res. 2006, 110 Pt 8, 879–886. [Google Scholar] [CrossRef]
- Scholtmeijer, K.; de Vocht, M.L.; Rink, R.; Robillard, G.T.; Wösten, H.A. Assembly of the fungal SC3 hydrophobin into functional amyloid fibrils depends on its concentration and is promoted by cell wall polysaccharides. J. Biol. Chem. 2009, 284, 26309–26314. [Google Scholar] [CrossRef]
- Wang, L.; Lu, C.; Fan, M.; Liao, B. Coriolopsis trogii hydrophobin genes favor a clustering distribution and are widely involved in mycelial growth and primordia formation. Gene 2021, 802, 145863. [Google Scholar] [CrossRef]
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V.; et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef]
- Klimes, A.; Dobinson, K.F. A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 2006, 43, 283–294. [Google Scholar] [CrossRef]
- Cai, F.; Zhao, Z.; Gao, R.; Chen, P.; Ding, M.; Jiang, S.; Fu, Z.; Xu, P.; Chenthamara, K.; Shen, Q.; et al. The pleiotropic functions of intracellular hydrophobins in aerial hyphae and fungal spores. PLoS Genet. 2021, 17, e1009924. [Google Scholar] [CrossRef]
- Piombo, E.; Guaschino, M.; Jensen, D.F.; Karlsson, M.; Dubey, M. Insights into the ecological generalist lifestyle of Clonostachys fungi through analysis of their predicted secretomes. Front. Microbiol. 2023, 14, 1112673. [Google Scholar] [CrossRef]
- Hou, S.; Yang, K.; Qin, M.; Feng, X.Z.; Guan, L.; Yang, Y.; Wang, C. Patterning of cells on functionalized poly (dimethylsiloxane) surface prepared by hydrophobin and collagen modification. Biosens. Bioelectron. 2008, 24, 918–922. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, Y.; Li, S.; Xu, H.; Linder, M.B.; Qiao, M. Hydrophilic modification of polystyrene with hydrophobin for time-resolved immunofluorometric assay. Biosens. Bioelectron. 2010, 26, 1074–1079. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, L.; Zhu, X.; Wang, Y.; Yang, H.; Wang, Z. A rapid and ultrasensitive thrombin biosensor based on a rationally designed trifunctional protein. Adv. Healthc. Mater. 2020, 9, e2000364. [Google Scholar] [CrossRef]
- Puspitasari, N.; Tsai, S.L.; Lee, C.K. Fungal hydrophobin RolA enhanced PETase hydrolysis of polyethylene terephthalate. Appl. Biochem. Biotechnol. 2021, 193, 1284–1295. [Google Scholar] [CrossRef]
- Hähl, H.; Vargas, J.N.; Griffo, A.; Laaksonen, P.; Szilvay, G.; Lienemann, M.; Jacobs, K.; Seemann, R.; Fleury, J.B. Pure protein bilayers and vesicles from native fungal hydrophobins. Adv. Mater 2017, 29, 1602888. [Google Scholar] [CrossRef]
- Fang, G.; Tang, B.; Liu, Z.; Gou, J.; Zhang, Y.; Xu, H.; Tang, X. Novel hydrophobin-coated docetaxel nanoparticles for intravenous delivery: In vitro characteristics and in vivo performance. Eur. J. Pharm. Sci. 2014, 60, 1–9. [Google Scholar] [CrossRef]
- Han, Z.; Song, B.; Yang, J.; Wang, B.; Ma, Z.; Yu, L.; Li, Y.; Xu, H.; Qiao, M. Curcumin-encapsulated fusion protein-based nanocarrier demonstrated highly efficient epidermal growth factor receptor-targeted treatment of colorectal cancer. J. Agric. Food Chem. 2022, 70, 15464–15473. [Google Scholar] [CrossRef]
- Niu, B.; Huang, Y.; Zhang, S.; Wang, D.; Xu, H.; Kong, D.; Qiao, M. Expression and characterization of hydrophobin HGFI fused with the cell-specific peptide TPS in Pichia pastoris. Protein Expr. Purif. 2012, 83, 92–97. [Google Scholar] [CrossRef]
- Zhao, L.; Ma, S.; Pan, Y.; Zhang, Q.; Wang, K.; Song, D.; Wang, X.; Feng, G.; Liu, R.; Xu, H.; et al. Functional modification of fibrous PCL scaffolds with fusion protein VEGF-HGFI enhanced cellularization and vascularization. Adv. Healthc. Mater. 2016, 5, 2376–2385. [Google Scholar] [CrossRef] [PubMed]
- Oude Vrielink, A.S.; Bomans, P.H.; Vredenbregt, E.J.; Wirix, M.J.; Sommerdijk, N.A.; Luiten, O.J.; Voets, I.K. Suspended crystalline films of protein hydrophobin I (HFBI). J. Colloid Interface Sci. 2015, 447, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, B.; Zhang, Y.; Zhu, Y.; Song, B.; Xu, H.; Zhai, Y.; Qiao, M.; Sun, F. A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI. Nat. Commun. 2021, 12, 7257. [Google Scholar] [CrossRef]
- Wösten, H.A.; Scholtmeijer, K. Applications of hydrophobins: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, K.A.; Hooley, P.; Cox, P.W. Bioinformatics predicts diverse Aspergillus hydrophobins with novel properties. Food Hydrocoll. 2012, 27, 503–516. [Google Scholar] [CrossRef]
- Dynesen, J.; Nielsen, J. Surface hydrophobicity of Aspergillus nidulans conidiospores and its role in pellet formation. Biotechnol. Prog. 2003, 19, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Maeda, H.; Yoneda, S.; Ohtaki, S.; Yamagata, Y.; Hasegawa, F.; Gomi, K.; Nakajima, T.; Abe, K. The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol. Microbiol. 2005, 57, 1780–1796. [Google Scholar] [CrossRef]
- Dubey, M.K.; Jensen, D.F.; Karlsson, M. Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol. 2014, 14, 18. [Google Scholar] [CrossRef]
- Xu, D.; Wang, Y.; Keerio, A.A.; Ma, A. Identification of hydrophobin genes and their physiological functions related to growth and development in Pleurotus ostreatus. Microbiol. Res. 2021, 247, 126723. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Nene, S.N.; Joshi, K.S. Production of hydrophobins from fungi. Process. Biochem. 2017, 61, 1–11. [Google Scholar] [CrossRef]
- Yang, J.; Ge, L.; Song, B.; Ma, Z.; Yang, X.; Wang, B.; Dai, Y.; Xu, H.; Qiao, M. A novel hydrophobin encoded by hgfII from Grifola frondosa exhibiting excellent self-assembly ability. Front. Microbiol. 2022, 13, 990231. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Song, B.; Yu, L.; Yang, J.; Han, Z.; Yang, J.; Wang, B.; Song, D.; Xu, H.; Qiao, M. Efficient expression of hydrophobin HGFII-his via POT1-mediated δ integration strategy and its potential in curcumin nanoformulation. Colloids Surf. A Physicochem. Eng. Asp. 2023, 656, 130344. [Google Scholar] [CrossRef]
- Montoya, S.; Orrego, C.E.; Levin, L. Growth, fruiting and lignocellulolytic enzyme production by the edible mushroom Grifola frondosa (maitake). World J. Microbiol. Biotechnol. 2012, 28, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, J.; Wang, B.; Ge, L.; Yang, X.; Wang, X.; Dai, Y.; Niu, B.; Xu, H.; Qiao, M. The enhancement of surface activity and nanoparticle stability through the alteration of charged amino acids of HGFI. Colloids Surf. B Biointerfaces 2019, 175, 703–712. [Google Scholar] [CrossRef]
- Li, Y.; Li, K.; Wang, X.; Cui, M.; Ge, P.; Zhang, J.; Qiu, F.; Zhong, C. Conformable self-assembling amyloid protein coatings with genetically programmable functionality. Sci. Adv. 2020, 6, eaba1425. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, S.; Huang, Y.; Li, S.; Xu, H.; Zhang, X.; Bai, Y.; Qiao, M. Expression and characterization of a Grifola frondosa hydrophobin in Pichia pastoris. Protein Expr. Purif. 2010, 72, 19–25. [Google Scholar] [CrossRef]
- Petrlova, J.; Petruk, G.; Huber, R.G.; McBurnie, E.W.; van der Plas, M.J.A.; Bond, P.J.; Puthia, M.; Schmidtchen, A. Thrombin-derived C-terminal fragments aggregate and scavenge bacteria and their proinflammatory products. J. Biol. Chem. 2020, 295, 3417–3430. [Google Scholar] [CrossRef] [PubMed]
- Lo, V.C.; Ren, Q.; Pham, C.L.; Morris, V.K.; Kwan, A.H.; Sunde, M. Fungal Hydrophobin Proteins Produce Self-Assembling Protein Films with Diverse Structure and Chemical Stability. Nanomaterials 2014, 4, 827–843. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, A.; Emide, D.; Saitta, F.; Fessas, D.; Iametti, S.; Barbiroli, A.; Marti, A. Impact of Thermal Treatment on the Starch-Protein Interplay in Red Lentils: Connecting Molecular Features and Rheological Properties. Molecules 2022, 27, 1266. [Google Scholar] [CrossRef]
- Niu, B.; Li, M.; Jia, J.; Zhang, C.; Fan, Y.-Y.; Li, W. Hydrophobin-enhanced stability, dispersions and release of curcumin nanoparticles in water. J. Biomater. Sci. Polym. Ed. 2020, 31, 1793–1805. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.G.; Andersen, M.R.; Pedersen, M.H.; Frisvad, J.C.; Søndergaard, I. Hydrophobins from Aspergillus species cannot be clearly divided into two classes. BMC Res. Notes 2010, 3, 344. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, F.; Xu, Y.; Liu, G.; Dong, C. Cysteine-rich hydrophobin gene family: Genome wide analysis, phylogeny and transcript profiling in Cordyceps militaris. Int. J. Mol. Sci. 2021, 22, 643. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Gibon, J.; Kohler, A.; Duffy, K.; Hoegger, P.J.; Velagapudi, R.; Han, J.; Kües, U.; Grigoriev, I.V.; Martin, F. Phylogenetic, genomic organization and expression analysis of hydrophobin genes in the ectomycorrhizal basidiomycete Laccaria bicolor. Fungal Genet. Biol. 2012, 49, 199–209. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, K.; Liu, T.; Zhang, Y.; Tang, Z.; Dong, J.; Wang, F. The characterization and expression analysis under stress conditions of PCST1 in Arabidopsis. Plant Signal. Behav. 2022, 17, 2134675. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Huang, J.; Jiang, S.; Chen, Y.; Yin, Y.; Pan, Y.; Yu, G.; Li, Y.; Wong, B.H.; et al. Transcriptome and proteome exploration to provide a resource for the study of Agrocybe aegerita. PLoS ONE 2013, 8, e56686. [Google Scholar] [CrossRef]
- Kim, H.-I.; Lee, C.-S.; Park, Y.-J. Further characterization of hydrophobin genes in genome of Flammulina velutipes. Mycoscience 2016, 57, 320–325. [Google Scholar] [CrossRef]
- Yamada, M.; Sakuraba, S.; Shibata, K.; Inatomi, S.; Okazaki, M.; Shimosaka, M. Cloning and characterization of a gene coding for a hydrophobin, Fv-hyd1, specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes. Appl. Microbiol. Biotechnol. 2005, 67, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Winandy, L.; Hilpert, F.; Schlebusch, O.; Fischer, R. Comparative analysis of surface coating properties of five hydrophobins from Aspergillus nidulans and Trichoderma reseei. Sci. Rep. 2018, 8, 12033. [Google Scholar] [CrossRef] [PubMed]
- de Vocht, M.L.; Scholtmeijer, K.; van der Vegte, E.W.; de Vries, O.M.; Sonveaux, N.; Wösten, H.A.; Ruysschaert, J.M.; Hadziloannou, G.; Wessels, J.G.; Robillard, G.T. Structural characterization of the hydrophobin SC3, as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys. J. 1998, 74, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Pothiratana, C.; Fuangsawat, W.; Jintapattanakit, A.; Teerapatsakul, C.; Thachepan, S. Putative hydrophobins of black poplar mushroom (Agrocybe cylindracea). Mycology 2020, 12, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, P.; Zhou, F.; Gao, L.; Sun, D.; Meng, Y.; Wang, X. Zinc-Guided 3D Graphene for Thermally Chargeable Supercapacitors to Harvest Low-Grade Heat. Molecules 2022, 27, 1239. [Google Scholar] [CrossRef] [PubMed]
- Sunde, M.; Kwan, A.H.; Templeton, M.D.; Beever, R.E.; Mackay, J.P. Structural analysis of hydrophobins. Micron 2008, 39, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Stringer, M.A.; Dean, R.A.; Sewall, T.C.; Timberlake, W.E. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 1991, 5, 1161–1171. [Google Scholar] [CrossRef]
- Kwan, A.H.; Winefield, R.D.; Sunde, M.; Matthews, J.M.; Haverkamp, R.G.; Templeton, M.D.; Mackay, J.P. Structural basis for rodlet assembly in fungal hydrophobins. Proc. Natl. Acad. Sci. USA 2006, 103, 3621–3626. [Google Scholar] [CrossRef]
- Gebbink, M.F.; Claessen, D.; Bouma, B.; Dijkhuizen, L.; Wösten, H.A. Amyloids—A functional coat for microorganisms. Nat. Rev. Microbiol. 2005, 3, 333–341. [Google Scholar] [CrossRef]
- Lim, H.J.; Lee, E.H.; Yoon, Y.; Chua, B.; Son, A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Cittadino, G.M.; Andrews, J.; Purewal, H.; Estanislao Acuña Avila, P.; Arnone, J.T. Functional Clustering of Metabolically Related Genes Is Conserved across Dikarya. J. Fungi. 2023, 9, 523. [Google Scholar] [CrossRef]


| Name | CDS(bp) | DNA (bp) | Number of Introns | Number of Exons | Number of Amino Acids | Molecular Weight (Da) | Class of Hydrophobin |
|---|---|---|---|---|---|---|---|
| Gf.hyd2041 | 429 | 2613 | 3 | 4 | 142 | 14,361.64 | I |
| Gf.hyd6681 | 357 | 1227 | 2 | 3 | 118 | 11,782.75 | I |
| Gf.hyd6682 | 342 | 1692 | 4 | 5 | 113 | 11,372.04 | I |
| Gf.hyd7622 | 324 | 2225 | 2 | 3 | 107 | 10,470.30 | I |
| Gf.hyd8174 | 429 | 3784 | 7 | 7 | 142 | 14,106.29 | I |
| Gf.hyd8531 | 324 | 2445 | 2 | 3 | 107 | 10,456.27 | I |
| Gf.hyd8825 | 384 | 1881 | 2 | 3 | 127 | 12,549.59 | I |
| rGf.hyd9954 | 336 | 1789 | 2 | 3 | 111 | 10,917.37 | I |
| Gf.hyd10182 | 303 | 1601 | 2 | 3 | 100 | 9723.19 | I |
| Gf.hyd11240 | 1179 | 2562 | 2 | 3 | 392 | 36,493.27 | I |
| Gf.hyd11347 | 1131 | 2510 | 3 | 4 | 376 | 35,047.55 | I |
| Gf.hyd12081 | 417 | 1040 | 1 | 1 | 138 | 13,564.40 | I |
| Gf.hyd12082 | 699 | 1976 | 3 | 4 | 232 | 23,671.27 | I |
| Gf.hyd12802 | 429 | 2157 | 2 | 3 | 142 | 13,949.09 | I |
| Gf.hyd13942 | 339 | 2051 | 2 | 3 | 112 | 11,241.08 | I |
| Gf.hyd14947 | 312 | 1594 | 2 | 3 | 103 | 10,078.58 | I |
| Gf.hyd15024 | 420 | 1817 | 4 | 4 | 139 | 13,846.22 | I |
| Gf.hyd20923 | 321 | 596 | 3 | 3 | 106 | 10,708.47 | I |
| Gf.hyd21629 | 312 | 548 | 2 | 3 | 103 | 9782.11 | I |
| Name | Sub-Location * | Theoretical PI | GRAVY ** | Signal Peptide | Number of Negative Charged Residues | Number of Positive Charged Residues | GO *** Terms |
|---|---|---|---|---|---|---|---|
| Gf.hyd2041 | plas | 8.50 | 0.270 | Yes | 7 | 10 | 9 |
| Gf.hyd6681 | extr | 3.57 | 0.858 | Yes | 11 | 1 | 9 |
| Gf.hyd6682 | extr | 6.05 | 0.441 | Yes | 5 | 5 | null **** |
| Gf.hyd7622 | extr | 4.04 | 0.951 | Yes | 5 | 2 | 9 |
| Gf.hyd8174 | extr | 7.52 | 0.336 | Yes | 8 | 9 | 9 |
| Gf.hyd8531 | extr | 4.04 | 0.950 | Yes | 5 | 2 | 9 |
| Gf.hyd8825 | extr | 4.00 | 0.876 | Yes | 4 | 1 | 9 |
| rGf.hyd9954 | extr | 3.62 | 0.495 | Yes | 7 | 1 | 9 |
| Gf.hyd10182 | extr | 5.30 | 0.723 | Yes | 2 | 1 | 9 |
| Gf.hyd11240 | extr | 4.87 | 0.368 | Yes | 14 | 8 | 9 |
| Gf.hyd11347 | extr | 5.31 | 0.343 | Yes | 13 | 8 | 9 |
| Gf.hyd12081 | extr | 4.01 | 0.675 | Yes | 9 | 3 | null |
| Gf.hyd12082 | extr | 4.31 | 0.524 | No | 17 | 12 | 9 |
| Gf.hyd12802 | extr | 4.46 | 0.777 | Yes | 8 | 5 | 9 |
| Gf.hyd13942 | extr | 3.57 | 0.789 | Yes | 7 | 1 | 9 |
| Gf.hyd14947 | extr | 5.30 | 0.682 | Yes | 2 | 1 | 9 |
| Gf.hyd15024 | extr | 3.98 | 0.878 | Yes | 8 | 3 | 9 |
| Gf.hyd20923 | extr | 4.99 | 0.644 | Yes | 6 | 5 | null |
| Gf.hyd21629 | extr | 3.19 | 0.818 | No | 8 | 0 | 9 |
| Hydrophobins | Processing | WCA |
|---|---|---|
| Blank Teflon film | 86.32 ± 3.26° | |
| Blank mica slice | 8.86 ± 0.63° | |
| rGf.hyd9954 | a | 59.48 ± 2.47° |
| a + c | 62.17 ± 4.15° | |
| b | 20.73 ± 0.98° | |
| b + c | 17.43 ± 2.19° | |
| HGFII-his | a | 58.40 ± 2.71° |
| a + c | 71.79 ± 2.56° | |
| b | 17.38 ± 0.57° | |
| b + c | 7.22 ± 0.45° |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, B.; Wang, W.; Jia, C.; Han, Z.; Yang, J.; Yang, J.; Wu, Z.; Xu, H.; Qiao, M. Identification and Characterization of a Predominant Hydrophobin in the Edible Mushroom Grifola frondosa. J. Fungi 2024, 10, 25. https://doi.org/10.3390/jof10010025
Song B, Wang W, Jia C, Han Z, Yang J, Yang J, Wu Z, Xu H, Qiao M. Identification and Characterization of a Predominant Hydrophobin in the Edible Mushroom Grifola frondosa. Journal of Fungi. 2024; 10(1):25. https://doi.org/10.3390/jof10010025
Chicago/Turabian StyleSong, Bo, Wenjun Wang, Chunhui Jia, Zhiqiang Han, Jiyuan Yang, Jiuxia Yang, Zhenzhou Wu, Haijin Xu, and Mingqiang Qiao. 2024. "Identification and Characterization of a Predominant Hydrophobin in the Edible Mushroom Grifola frondosa" Journal of Fungi 10, no. 1: 25. https://doi.org/10.3390/jof10010025
APA StyleSong, B., Wang, W., Jia, C., Han, Z., Yang, J., Yang, J., Wu, Z., Xu, H., & Qiao, M. (2024). Identification and Characterization of a Predominant Hydrophobin in the Edible Mushroom Grifola frondosa. Journal of Fungi, 10(1), 25. https://doi.org/10.3390/jof10010025





