Epigenetic Regulation of Organ Regeneration in Zebrafish
Abstract
1. Introduction
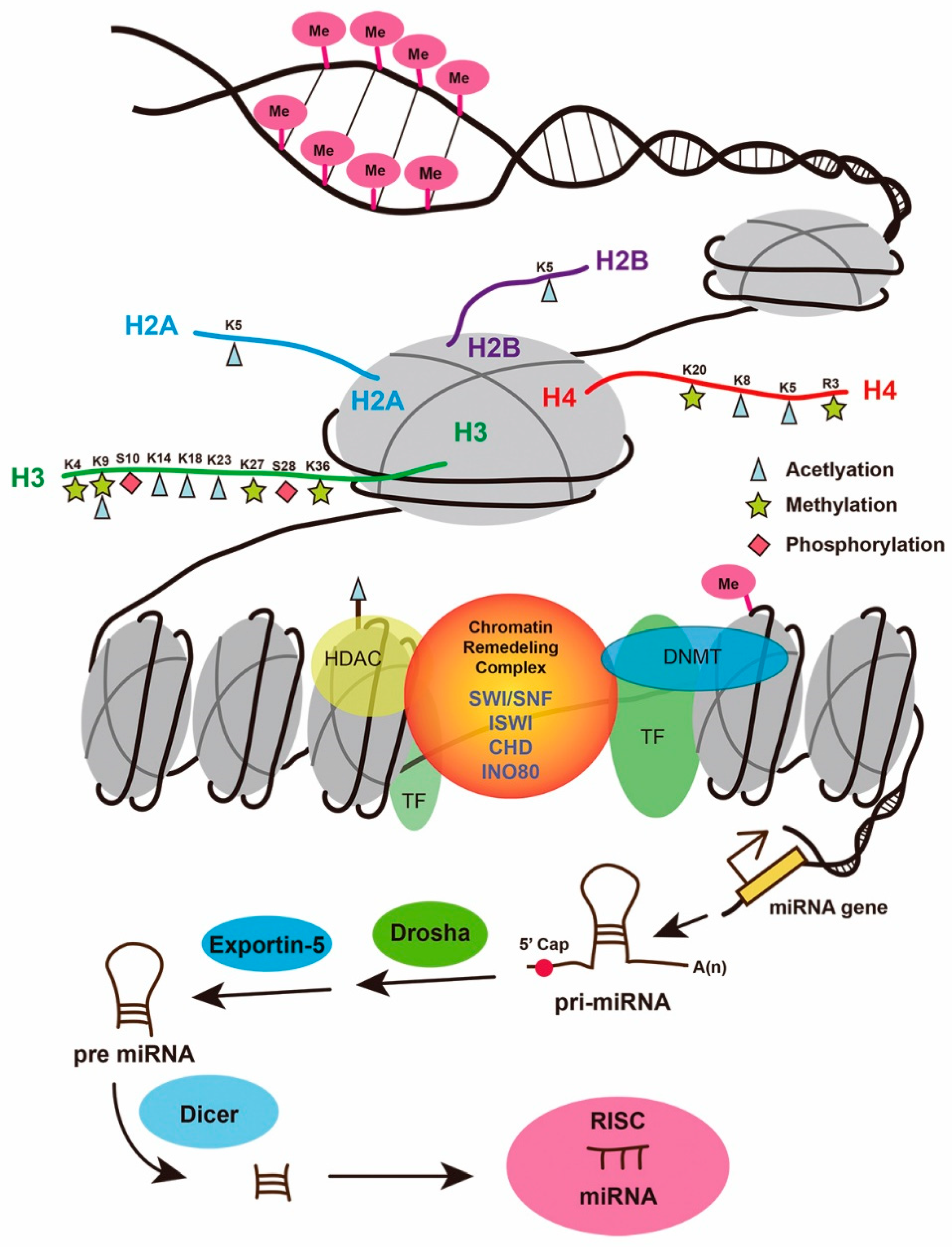
2. Role of DNA Methylation in Organ Regeneration
3. Histone Modification and Gene Regulation in Organ Regeneration
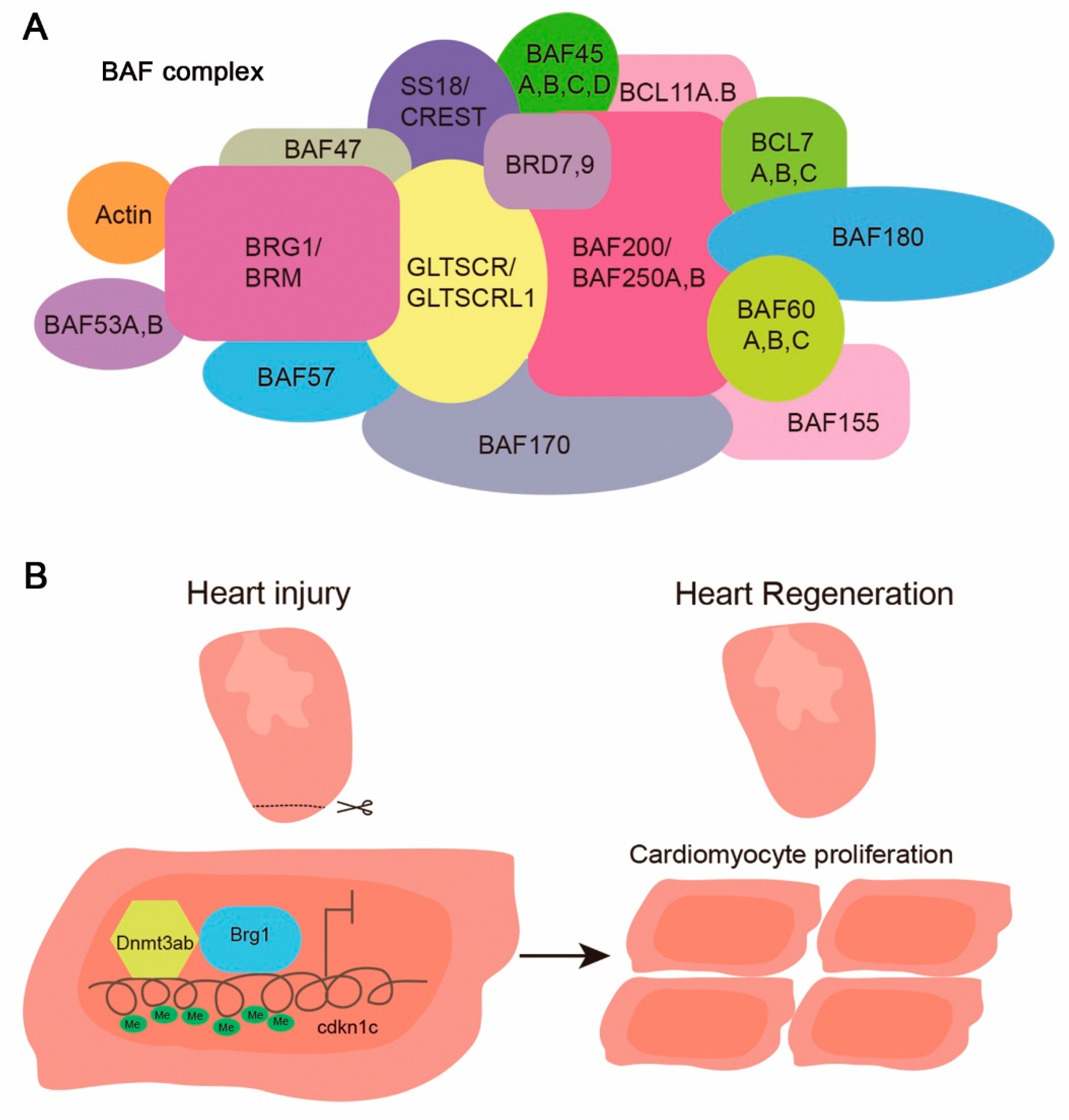
4. Role of the SWItch/Sucrose Non-Fermentable (SWI/SNF) Complex in Organ Regeneration
5. MicroRNA and Organ Regeneration
6. Perspective on Research Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosa-Garrido, M.; Chapski, D.J.; Vondriska, T.M. Epigenomes in Cardiovascular Disease. Circ. Res. 2018, 122, 1586–1607. [Google Scholar] [CrossRef] [PubMed]
- Gemberling, M.; Bailey, T.J.; Hyde, D.R.; Poss, K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013, 29, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Lenkowski, J.R.; Raymond, P.A. Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog. Retin. Eye Res. 2014, 40, 94–123. [Google Scholar] [CrossRef] [PubMed]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart Regeneration in Zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef] [PubMed]
- Chablais, F.; Veit, J.; Rainer, G.; Jazwinska, A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Devel. Biol. 2011, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Raya, A.; Koth, C.M.; Buscher, D.; Kawakami, Y.; Itoh, T.; Raya, R.M.; Sternik, G.; Tsai, H.-J.; Rodriguez-Esteban, C.; Izpisua-Belmonte, J.C. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2003, 100, 11889–11895. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; MacRae, C.A.; Stainier, D.Y.R.; Poss, K.D. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Akimenko, M.A.; Mari-Beffa, M.; Becerra, J.; Geraudie, J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 2003, 226, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Tornini, V.A.; Poss, K.D. Keeping at arm’s length during regeneration. Dev. Cell 2014, 29, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Pfefferli, C.; Jazwinska, A. The art of fin regeneration in zebrafish. Regeneration 2015, 2, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, E.J.; Paydar, I.; Anderson, K.K.; Patton, J.G. Regulation of zebrafish fin regeneration by microRNAs. Proc. Natl. Acad. Sci. USA 2008, 105, 18384–18389. [Google Scholar] [CrossRef] [PubMed]
- Sehring, I.M.; Jahn, C.; Weidinger, G. Zebrafish fin and heart: What’s special about regeneration? Curr. Opin. Genet. Dev. 2016, 40, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Kragl, M.; Knapp, D.; Nacu, E.; Khattak, S.; Maden, M.; Epperlein, H.H.; Tanaka, E.M. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 2009, 460, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Knopf, F.; Hammond, C.; Chekuru, A.; Kurth, T.; Hans, S.; Weber, C.W.; Mahatma, G.; Fisher, S.; Brand, M.; Schulte-Merker, S.; et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 2011, 20, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Holdway, J.E.; Poss, K.D. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 2012, 22, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Wehner, D.; Weidinger, G. Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. 2015, 31, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Mokalled, M.H.; Patra, C.; Dickson, A.L.; Endo, T.; Stainier, D.Y.; Poss, K.D. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 2016, 354, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Kroehne, V.; Freudenreich, D.; Hans, S.; Kaslin, J.; Brand, M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 2011, 138, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Kyritsis, N.; Kizil, C.; Zocher, S.; Kroehne, V.; Kaslin, J.; Freudenreich, D.; Iltzsche, A.; Brand, M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 2012, 338, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Kizil, C.; Kyritsis, N.; Dudczig, S.; Kroehne, V.; Freudenreich, D.; Kaslin, J.; Brand, M. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev. Cell 2012, 23, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Ceci, M.; Mariano, V.; Romano, N. Zebrafish as a translational regeneration model to study the activation of neural stem cells and role of their environment. Rev. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Goldman, D. Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 2016, 40, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Uygur, A.; Lee, R.T. Mechanisms of Cardiac Regeneration. Dev. Cell 2016, 36, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Hui, S.P. Regeneration of Zebrafish CNS: Adult Neurogenesis. Neural Plast. 2016, 2016, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ganz, J.; Brand, M. Adult Neurogenesis in Fish. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Poss, K.D. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 2010, 11, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2017, 8, 5629–5637. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Traube, F.R.; Carell, T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017, 14, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, G.; Kreutz, M.R. The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders. Front. Mol. Neurosci. 2018, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Seritrakul, P.; Gross, J.M. Tet-mediated DNA hydroxymethylation regulates retinal neurogenesis by modulating cell-extrinsic signaling pathways. PLoS Genet. 2017, 13, e1006987. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Bosch, J.A.; Goll, M.G.; Hesselson, D.; Dong, P.D.; Shin, D.; Chi, N.C.; Shin, C.H.; Schlegel, A.; Halpern, M.; et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev. Biol. 2009, 334, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jia, X.; Yuan, H.; Ma, K.; Chen, Y.; Jin, Y.; Deng, M.; Pan, W.; Chen, S.; Chen, Z.; et al. DNA methyltransferase 1 functions through C/ebpa to maintain hematopoietic stem and progenitor cells in zebrafish. J. Hematol. Oncol. 2015, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Tittle, R.K.; Sze, R.; Ng, A.; Nuckels, R.J.; Swartz, M.E.; Anderson, R.M.; Bosch, J.; Stainier, D.Y.; Eberhart, J.K.; Gross, J.M. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev. Biol. 2011, 350, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Chidester, S.; Zavala, C.V.; Manos, E.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007, 21, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Jafri, I.F.; Chidester, S.; James, S.R.; Karpf, A.R.; Cairns, B.R.; Jones, D.A. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 2010, 285, 4110–4121. [Google Scholar] [CrossRef] [PubMed]
- Thummel, R.; Burket, C.T.; Hyde, D.R. Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. ScientificWorldJournal 2006, 6 (Suppl. 1), 65–81. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.; Grant, A.R.; Cornblath, E.; Goldman, D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 19814–19819. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.; Elsaeidi, F.; Goldman, D. Injury-dependent Muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J. Neurosci. 2012, 32, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Hirose, K.; Shimoda, N.; Kikuchi, Y. Transient reduction of 5-methylcytosine and 5-hydroxymethylcytosine is associated with active DNA demethylation during regeneration of zebrafish fin. Epigenetics 2013, 8, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Shimoda, N.; Takanaga, S.; Hozumi, S.; Kikuchi, Y. Expression patterns of dnmt3aa, dnmt3ab, and dnmt4 during development and fin regeneration in zebrafish. Gene Expr. Patterns 2014, 14, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Purushothaman, S.; Meghah, V.; Bhatti, B.; Poruri, A.; Meena Lakshmi, M.G.; Sarath Babu, N.; Narasimha Murthy, C.L.; Mandal, K.K.; Kumar, A.; et al. Role of annexin gene and its regulation during zebrafish caudal fin regeneration. Wound Repair Regen. 2016, 24, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; Tsun, Z.Y.; Izpisua Belmonte, J.C. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 19889–19894. [Google Scholar] [CrossRef] [PubMed]
- Dupret, B.; Volkel, P.; Vennin, C.; Toillon, R.A.; Le Bourhis, X.; Angrand, P.O. The histone lysine methyltransferase Ezh2 is required for maintenance of the intestine integrity and for caudal fin regeneration in zebrafish. Biochim. Biophys. Acta 2017, 1860, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cai, C.; Tang, D.; Sun, S.; Li, H. Effect of histone deacetylase inhibitors trichostatin A and valproic acid on hair cell regeneration in zebrafish lateral line neuromasts. Front. Cell. Neurosci. 2014, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Russell, J.O.; Tian, J.; Gao, C.; Kobayashi, M.; Feng, R.; Yuan, X.; Shao, C.; Ding, H.; Poddar, M.; et al. Hdac1 Regulates Differentiation of Bipotent Liver Progenitor Cells During Regeneration via Sox9b and Cdk8. Gastroenterology 2018. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Sharma, P.; Kaur, S.; Khursheed, M.A.; Gupta, S.; Ahuja, R.; Kurup, A.J.; Chaudhary, M.; Ramachandran, R. Histone Deacetylase-Mediated Muller Glia Reprogramming through Her4.1-Lin28a Axis Is Essential for Retina Regeneration in Zebrafish. iScience 2018, 7, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.I.; Lessard, J.; Crabtree, G.R. Understanding the words of chromatin regulation. Cell 2009, 136, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Hota, S.K.; Bruneau, B.G. ATP-dependent chromatin remodeling during mammalian development. Development 2016, 143, 2882–2897. [Google Scholar] [CrossRef] [PubMed]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G.; et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef]
- Stankunas, K.; Hang, C.T.; Tsun, Z.-Y.; Chen, H.; Lee, N.V.; Wu, J.I.; Shang, C.; Bayle, J.H.; Shou, W.; Iruela-Arispe, M.L.; et al. Endocardial Brg1 Represses ADAMTS1 to Maintain the Microenvironment for Myocardial Morphogenesis. Dev. Cell 2008, 14, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Lou, X.; Alexander, J.M.; Sugizaki, H.; Delgado-Olguin, P.; Holloway, A.K.; Mori, A.D.; Wylie, J.N.; Munson, C.; Zhu, Y.; et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2011, 2, 187. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.M.; Howard, S.; Villa Del Campo, C.; Bollini, S.; Dube, K.N.; Masters, M.; Barnette, D.N.; Rohling, M.; Sun, X.; Hankins, L.E.; et al. BRG1-SWI/SNF-dependent regulation of the Wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat. Commun. 2017, 8, 16034. [Google Scholar] [CrossRef] [PubMed]
- Lickert, H.; Takeuchi, J.K.; Von Both, I.; Walls, J.R.; McAuliffe, F.; Adamson, S.L.; Henkelman, R.M.; Wrana, J.L.; Rossant, J.; Bruneau, B.G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 2004, 432, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hota, S.K.; Zhou, Y.Q.; Novak, S.; Miguel-Perez, D.; Christodoulou, D.; Seidman, C.E.; Seidman, J.G.; Gregorio, C.C.; Henkelman, R.M.; et al. Cardiac-enriched BAF chromatin-remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Biol. Open 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhai, W.; Richardson, J.A.; Olson, E.N.; Meneses, J.J.; Firpo, M.T.; Kang, C.; Skarnes, W.C.; Tjian, R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004, 18, 3106–3116. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Archer, T.K. Analysis of the SWI/SNF chromatin-remodeling complex during early heart development and BAF250a repression cardiac gene transcription during P19 cell differentiation. Nucleic Acids Res. 2014, 42, 2958–2975. [Google Scholar] [CrossRef] [PubMed]
- Toto, P.C.; Puri, P.L.; Albini, S. SWI/SNF-directed stem cell lineage specification: Dynamic composition regulates specific stages of skeletal myogenesis. Cell. Mol. Life Sci. 2016, 73, 3887–3896. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Gao, L.; Hou, Y.; Xu, C.; Chang, N.; Wang, F.; Hu, K.; He, A.; Luo, Y.; Wang, J.; et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat. Commun. 2016, 7, 13787. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Kong, M.; Zeng, S.; Hao, C.; Li, M.; Li, L.; Xu, Z.; Zhu, M.; Xu, Y. Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/beta-catenin pathway in mice. FASEB J. 2018. [Google Scholar] [CrossRef]
- Wu, Q.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Nickerson, J.A.; Imbalzano, A.N. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017, 9, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Fu, Y.; Hu, F.; Lan, J.; Xu, F.; Yang, X.; Luo, X.; Wang, J.; Hu, J. Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway. Cell Death Dis. 2017, 8, e2607. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Lee, Y.C.; Yeh, S.H.; Chen, C.H.; Wu, C.C.; Wang, T.Y.; Chen, Y.N.; Hung, L.Y.; Liu, Y.W.; Chen, H.K.; et al. 58-kDa microspherule protein (MSP58) is novel Brahma-related gene 1 (BRG1)-associated protein that modulates p53/p21 senescence pathway. J. Biol. Chem. 2012, 287, 22533–22548. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Whyte, W.A.; Zepeda-Mendoza, C.J.; Milazzo, J.P.; Shen, C.; Roe, J.S.; Minder, J.L.; Mercan, F.; Wang, E.; Eckersley-Maslin, M.A.; et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013, 27, 2648–2662. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Benavides, T.; Nasipak, B.T.; Imbalzano, A.N. Brg1 Controls the Expression of Pax7 to Promote Viability and Proliferation of Mouse Primary Myoblasts. J. Cell. Physiol. 2015, 230, 2990–2997. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wang, Q.; Gu, J.; Xuan, Z.; Wu, J.I. SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development. Oncogene 2016, 35, 5746–5758. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Lajoie, B.R.; Fritz, A.J.; McCord, R.P.; Nickerson, J.A.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Dekker, J.; Stein, G.S.; et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016, 26, 1188–1201. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Koshiba-Takeuchi, K.; Tsuchiya, M.; Kojima, M.; Miyazawa, A.; Ito, K.; Ogawa, H.; Takeuchi, J.K. Expression analysis of Baf60c during heart regeneration in axolotls and neonatal mice. Dev. Growth Differ. 2016, 58, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Ooi, J.Y.; Lin, R.C.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Bezprozvannaya, S.; Williams, A.H.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008, 22, 3242–3254. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Wythe, J.D.; Xiao, T.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D.; Woo, S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011, 138, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Chiavacci, E.; Dolfi, L.; Verduci, L.; Meghini, F.; Gestri, G.; Evangelista, A.M.; Wilson, S.W.; Cremisi, F.; Pitto, L. MicroRNA 218 mediates the effects of Tbx5a over-expression on zebrafish heart development. PLoS ONE 2012, 7, e50536. [Google Scholar] [CrossRef] [PubMed]
- Chiavacci, E.; D’Aurizio, R.; Guzzolino, E.; Russo, F.; Baumgart, M.; Groth, M.; Mariani, L.; D’Onofrio, M.; Arisi, I.; Pellegrini, M.; et al. MicroRNA 19a replacement partially rescues fin and cardiac defects in zebrafish model of Holt Oram syndrome. Sci. Rep. 2015, 5, 18240. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.U.; Scherz, P.J.; Cordes, K.R.; Ivey, K.N.; Stainier, D.Y.; Srivastava, D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA 2008, 105, 17830–17835. [Google Scholar] [CrossRef] [PubMed]
- Banjo, T.; Grajcarek, J.; Yoshino, D.; Osada, H.; Miyasaka, K.Y.; Kida, Y.S.; Ueki, Y.; Nagayama, K.; Kawakami, K.; Matsumoto, T.; et al. Haemodynamically dependent valvulogenesis of zebrafish heart is mediated by flow-dependent expression of miR-21. Nat. Commun. 2013, 4, 1978. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wei, C.; Olena, A.F.; Patton, J.G. Regulation of endoderm formation and left-right asymmetry by miR-92 during early zebrafish development. Development 2011, 138, 1817–1826. [Google Scholar] [CrossRef] [PubMed]
- Yin, V.P.; Lepilina, A.; Smith, A.; Poss, K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012, 365, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, M.; Smith, A.; Yin, V.P. Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development 2015, 142, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.; Montserrat, N.; Zacchigna, S.; Nivet, E.; Hishida, T.; Krause, M.N.; Kurian, L.; Ocampo, A.; Vazquez-Ferrer, E.; Rodriguez-Esteban, C.; et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 2014, 15, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Crippa, S.; Nemir, M.; Ounzain, S.; Ibberson, M.; Berthonneche, C.; Sarre, A.; Boisset, G.; Maison, D.; Harshman, K.; Xenarios, I.; et al. Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways. Cardiovasc. Res. 2016, 110, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Mano, M.; Dal Ferro, M.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012, 492, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Lesizza, P.; Prosdocimo, G.; Martinelli, V.; Sinagra, G.; Zacchigna, S.; Giacca, M. Single-Dose Intracardiac Injection of Pro-Regenerative MicroRNAs Improves Cardiac Function After Myocardial Infarction. Circ. Res. 2017, 120, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, K.; Harding, R.L.; Bailey, T.; Patton, J.G.; Hyde, D.R. Dynamic miRNA expression patterns during retinal regeneration in zebrafish: Reduced dicer or miRNA expression suppresses proliferation of Muller glia-derived neuronal progenitor cells. Dev. Dyn. 2014, 243, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Fausett, B.V.; Goldman, D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 2010, 12, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Gupta, S.; Chaudhary, M.; Khursheed, M.A.; Mitra, S.; Kurup, A.J.; Ramachandran, R. let-7 MicroRNA-Mediated Regulation of Shh Signaling and the Gene Regulatory Network Is Essential for Retina Regeneration. Cell Rep. 2018, 23, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, K.; Harding, R.L.; Hyde, D.R.; Patton, J.G. miR-203 regulates progenitor cell proliferation during adult zebrafish retina regeneration. Dev. Biol. 2014, 392, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.M.; Gibbs, K.M.; Davila, J.; Campbell, N.; Sung, S.; Todorova, T.I.; Otsuka, S.; Sabaawy, H.E.; Hart, R.P.; Schachner, M. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 2011, 33, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Theis, T.; Yoo, M.; Park, C.S.; Chen, J.; Kugler, S.; Gibbs, K.M.; Schachner, M. Lentiviral Delivery of miR-133b Improves Functional Recovery After Spinal Cord Injury in Mice. Mol. Neurobiol. 2017, 54, 4659–4671. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Chen, M.; Yang, L.; Wagle, M.; Guo, S.; Hu, B. MicroRNA-133b Negatively Regulates Zebrafish Single Mauthner-Cell Axon Regeneration through Targeting tppp3 in Vivo. Front. Mol. Neurosci. 2017, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hu, J.; Karra, R.; Dickson, A.L.; Tornini, V.A.; Nachtrab, G.; Gemberling, M.; Goldman, J.A.; Black, B.L.; Poss, K.D. Modulation of tissue repair by regeneration enhancer elements. Nature 2016, 532, 201–206. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Xiao, C.; Xiong, J.-W. Epigenetic Regulation of Organ Regeneration in Zebrafish. J. Cardiovasc. Dev. Dis. 2018, 5, 57. https://doi.org/10.3390/jcdd5040057
Zhu X, Xiao C, Xiong J-W. Epigenetic Regulation of Organ Regeneration in Zebrafish. Journal of Cardiovascular Development and Disease. 2018; 5(4):57. https://doi.org/10.3390/jcdd5040057
Chicago/Turabian StyleZhu, Xiaojun, Chenglu Xiao, and Jing-Wei Xiong. 2018. "Epigenetic Regulation of Organ Regeneration in Zebrafish" Journal of Cardiovascular Development and Disease 5, no. 4: 57. https://doi.org/10.3390/jcdd5040057
APA StyleZhu, X., Xiao, C., & Xiong, J.-W. (2018). Epigenetic Regulation of Organ Regeneration in Zebrafish. Journal of Cardiovascular Development and Disease, 5(4), 57. https://doi.org/10.3390/jcdd5040057



