Differences in the Early In Vitro Development of Preimplantation Human IVF Embryos Which Go on to Develop Congenital Heart Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Ethical Approval
2.3. Fertility Treatment and Embryo Culture
2.4. Insemination and Time-Lapse Imaging and Morphokinetic Data Collection
2.5. Morphokinetic Data Analysis
2.6. Identification of the Large Control Cohort
2.7. Identification of the CHD and Matched Control Groups
2.8. Data Stratification
2.9. Statistical Analysis
3. Results
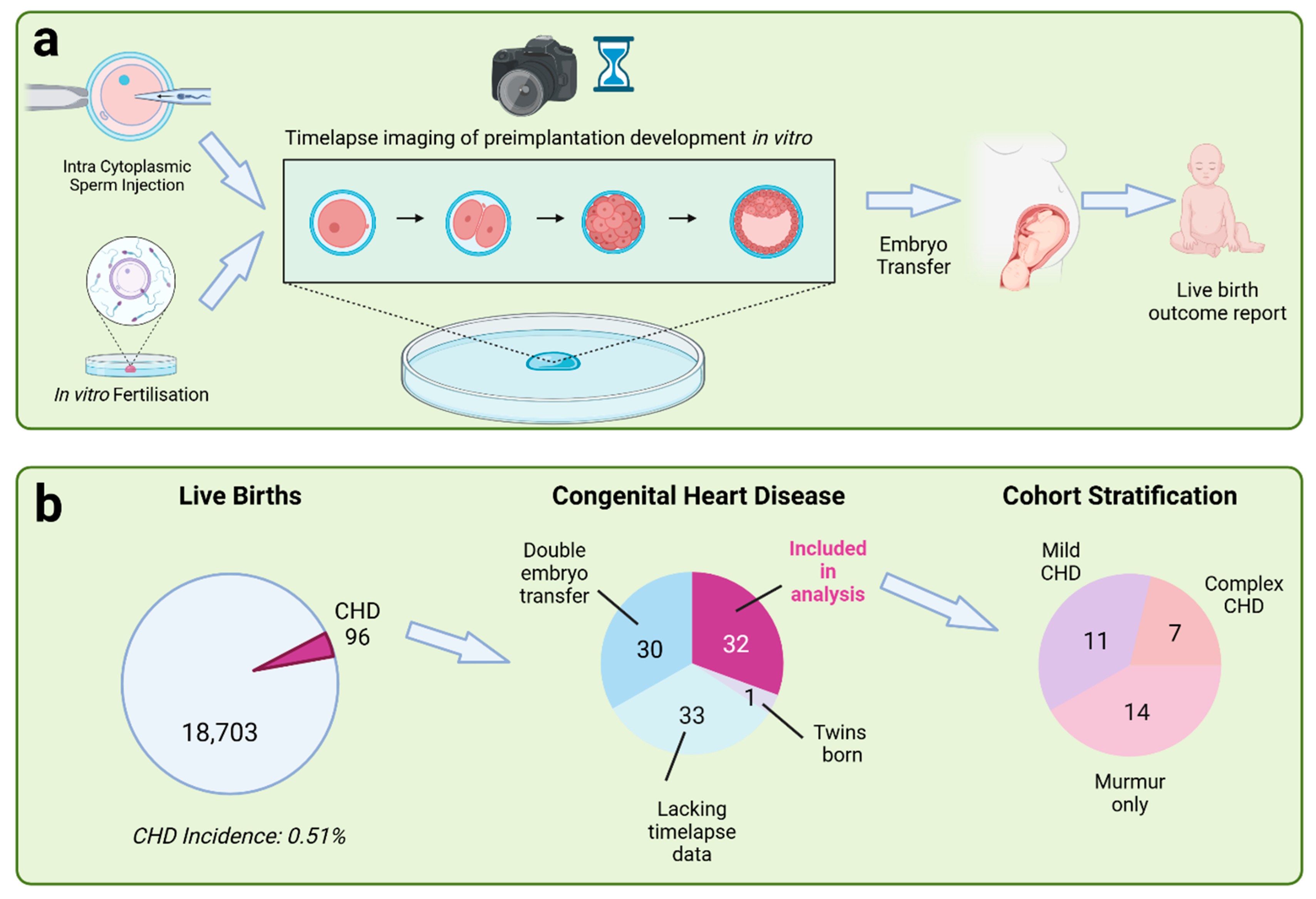
3.1. Study Design
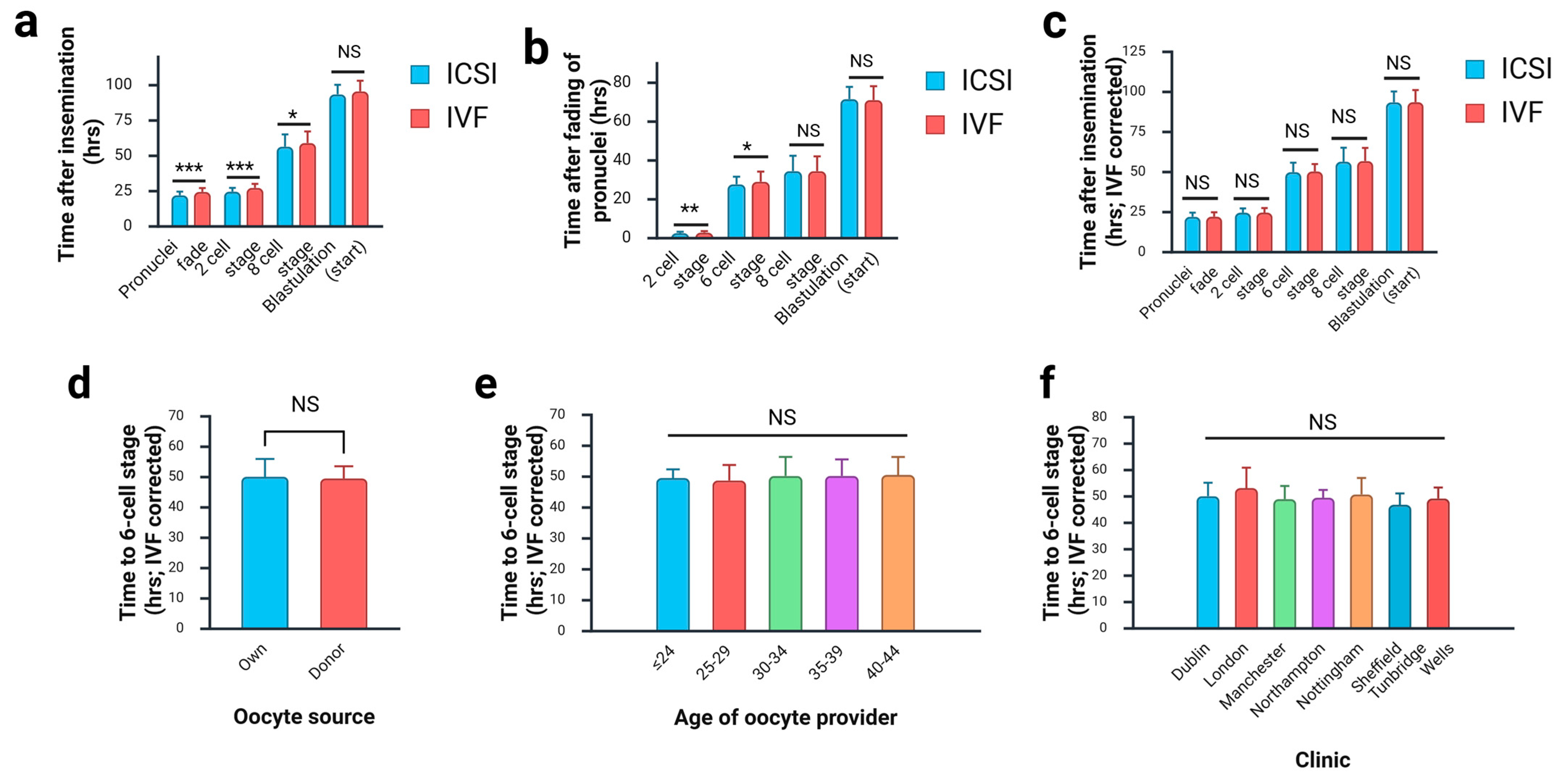
3.2. Effect of Fertilisation Method on Morphokinetics of Preimplantation Development
3.3. Identification of Congenital Heart Disease Cases
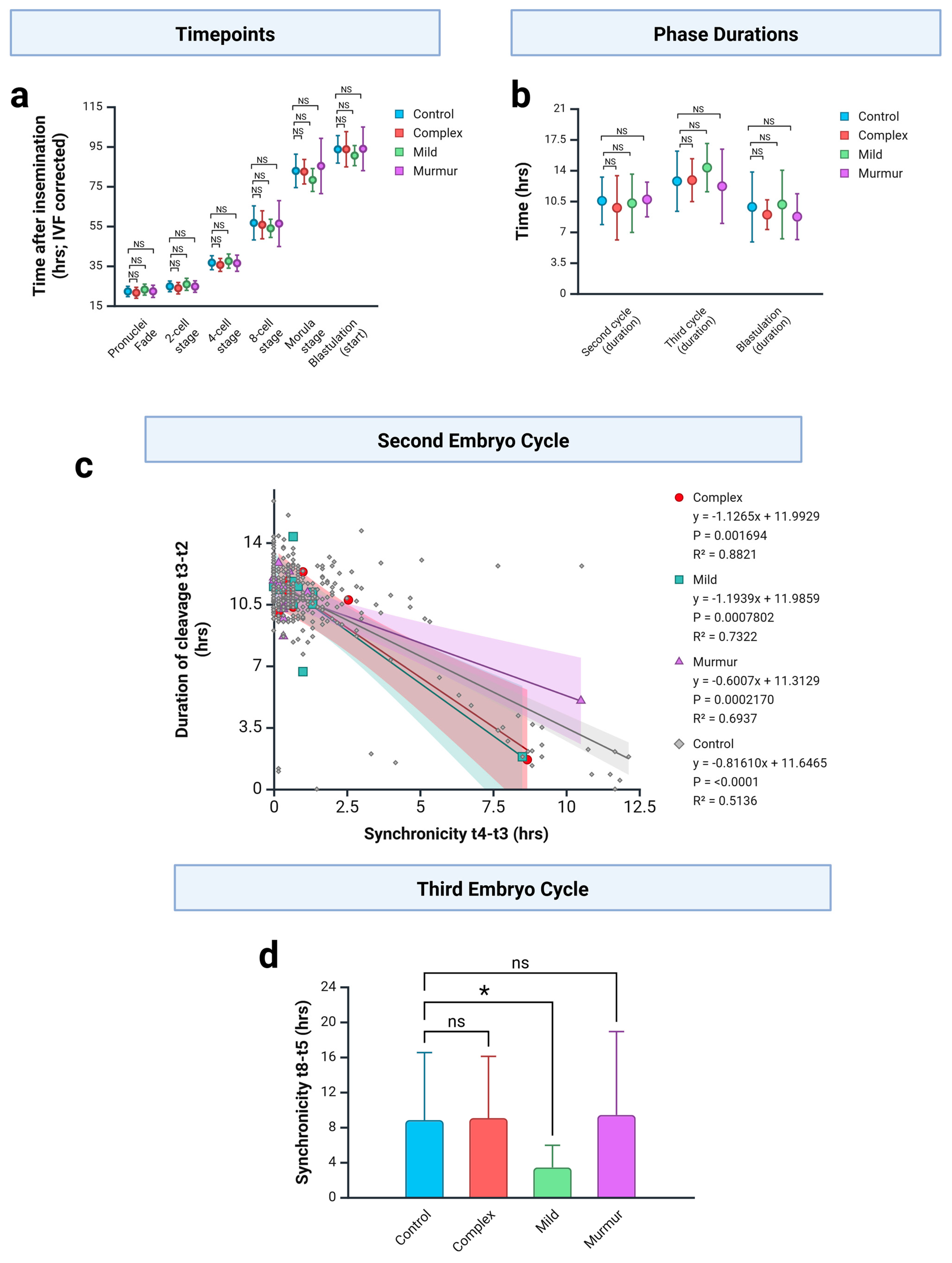
3.4. CHD Embryo Development Against Unaffected Controls
3.5. CHD Embryo Development Against Matched Unaffected Controls
4. Discussion
4.1. Changes in Cleavage Stage Embryos May Be Associated with Mild CHD
4.2. Changes in Blastulation Associated with Cyanotic CHD
4.3. CHD Incidence and Cohort Stratification
4.4. Delayed Development of IVF-Derived Preimplantation Embryos
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CHD | Congenital Heart Disease |
| ICSI | Intracytoplasmic Sperm Injection |
| IVF | In vitro fertilisation |
References
- Van Der Bom, T.; Zomer, A.C.; Zwinderman, A.H.; Meijboom, F.J.; Bouma, B.J.; Mulder, B.J. The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 2011, 8, 50–60. [Google Scholar] [CrossRef]
- Zimmerman, M.S.; Smith, A.G.C.; Sable, C.A.; Echko, M.M.; Wilner, L.B.; Olsen, H.E.; Atalay, H.T.; Awasthi, A.; Bhutta, Z.A.; Boucher, J.L.; et al. Global, regional, and national burden of congenital heart disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Child Adolesc. Health 2020, 4, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Thomas, R.; Mumme, M.; Golding, J.; Boyd, A.; Northstone, K.; Caputo, M.; Lawlor, D.A. Ascertaining and classifying cases of congenital anomalies in the ALSPAC birth cohort. Wellcome Open Res. 2021, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Giorgione, V.; Parazzini, F.; Fesslova, V.; Cipriani, S.; Candiani, M.; Inversetti, A.; Sigismondi, C.; Tiberio, F.; Cavoretto, P. Congenital heart defects in IVF/ICSI pregnancy: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2018, 51, 33–42. [Google Scholar] [CrossRef]
- Bouma, B.J.; Mulder, B.J.M. Changing Landscape of Congenital Heart Disease. Circ. Res. 2017, 120, 908–922. [Google Scholar] [CrossRef]
- Rao, P.S. Management of congenital heart disease: State of the art—Part II—Cyanotic heart defects. Children 2019, 6, 54. [Google Scholar] [CrossRef]
- Rao, P.S. Management of Congenital Heart Disease: State of the Art—Part I—ACYANOTIC Heart Defects. Children 2019, 6, 42. [Google Scholar] [CrossRef]
- Kurinczuk, J.J.; Bower, C. Birth defects in infants conceived by intracytoplasmic sperm injection: An alternative interpretation. Br. Med. J. 1997, 315, 1260–1265. [Google Scholar] [CrossRef]
- Gullo, G.; Scaglione, M.; Lagana, A.S.; Perino, A.; Andrisani, A.; Chiantera, V.; Cucinella, G.; Gitas, G.; Barra, F.; Riemma, G. Assisted reproductive techniques and risk of congenital heart diseases in children: A systematic review and meta-analysis. Reprod. Sci. 2023, 30, 2896–2906. [Google Scholar] [CrossRef] [PubMed]
- Reefhuis, J.; Honein, M.A.; Schieve, L.A.; Correa, A.; Hobbs, C.A.; Rasmussen, S.A. Assisted reproductive technology and major structural birth defects in the United States. Hum. Reprod. 2009, 24, 360–366. [Google Scholar] [CrossRef]
- Patil, A.S.; Nguyen, C.; Groff, K.; Wu, J.; Elliott, J.; Gunatilake, R.P. Severity of congenital heart defects associated with assisted reproductive technologies: Case series and review of the literature. Birth Defects Res. 2018, 110, 654–661. [Google Scholar] [CrossRef]
- Tararbit, K.; Houyel, L.; Bonnet, D.; De Vigan, C.; Lelong, N.; Goffinet, F.; Khoshnood, B. Risk of congenital heart defects associated with assisted reproductive technologies: A population-based evaluation. Eur. Heart J. 2011, 32, 500–508. [Google Scholar] [CrossRef]
- Valenzuela-Alcaraz, B.; Crispi, F.; Bijnens, B.; Cruz-Lemini, M.; Creus, M.; Sitges, M.; Bartrons, J.; Civico, S.; Balasch, J.; Gratacós, E. Assisted reproductive technologies are associated with cardiovascular remodeling in utero that persists postnatally. Circulation 2013, 128, 1442–1450. [Google Scholar] [CrossRef]
- Van Nisselrooij, A.; Teunissen, A.; Clur, S.; Rozendaal, L.; Pajkrt, E.; Linskens, I.; Rammeloo, L.; Van Lith, J.; Blom, N.; Haak, M. Why are congenital heart defects being missed? Ultrasound Obstet. Gynecol. 2020, 55, 747–757. [Google Scholar] [CrossRef]
- Sharland, G. Routine fetal cardiac screening: What are we doing and what should we do? Prenat. Diagn. Publ. Affil. Int. Soc. Prenat. Diagn. 2004, 24, 1123–1129. [Google Scholar] [CrossRef]
- Yasuhara, J.; Garg, V. Genetics of congenital heart disease: A narrative review of recent advances and clinical implications. Transl. Pediatr. 2021, 10, 2366–2386. [Google Scholar] [CrossRef]
- Yang, H.; DeWan, A.T.; Desai, M.M.; Vermund, S.H. Preimplantation genetic testing for aneuploidy: Challenges in clinical practice. Hum. Genom. 2022, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Bao, X.; Shi, H.; Niu, W.; Bu, Z.; Yang, J.; Zhang, Y.; Jin, H.; Song, W.; Sun, Y. Haplotyping-based preimplantation genetic testing for inherited cardiovascular disease: A multidisciplinary approach. Mol. Genet. Genom. 2025, 300, 11. [Google Scholar] [CrossRef]
- Rubio, I.; Galán, A.; Larreategui, Z.; Ayerdi, F.; Bellver, J.; Herrero, J.; Meseguer, M. Clinical validation of embryo culture and selection by morphokinetic analysis: A randomized, controlled trial of the EmbryoScope. Fertil. Steril. 2014, 102, 1287–1294.e5. [Google Scholar] [CrossRef] [PubMed]
- Fishel, S.; Campbell, A.; Montgomery, S.; Smith, R.; Nice, L.; Duffy, S.; Jenner, L.; Berrisford, K.; Kellam, L.; Smith, R.; et al. Time-lapse imaging algorithms rank human preimplantation embryos according to the probability of live birth. Reprod. Biomed. Online 2018, 37, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A.; Fishel, S.; Bowman, N.; Duffy, S.; Sedler, M.; Hickman, C.F. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod. Biomed. Online 2013, 26, 477–485. [Google Scholar] [CrossRef]
- Bamford, T.; Barrie, A.; Montgomery, S.; Dhillon-Smith, R.; Campbell, A.; Easter, C.; Coomarasamy, A. Morphological and morphokinetic associations with aneuploidy: A systematic review and meta-analysis. Hum. Reprod. Update 2022, 28, 656–686. [Google Scholar] [CrossRef] [PubMed]
- Figliuzzi, M.; Bori, L.; Ottolini, C.S.; Picchetta, L.; Caroselli, S.; Reverenna, M.; Poli, M.; Campbell, A.; Smith, R.; Coticchio, G.; et al. Human embryos with segmental aneuploidies display delayed early development: A multicenter morphokinetic analysis. Fertil. Steril. 2024, 123, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Junyent, S.; Meglicki, M.; Vetter, R.; Mandelbaum, R.; King, C.; Patel, E.M.; Iwamoto-Stohl, L.; Reynell, C.; Chen, D.-Y.; Rubino, P.; et al. The first two blastomeres contribute unequally to the human embryo. Cell 2024, 187, 2838–2854.e17. [Google Scholar] [CrossRef]
- Doron-Lalehzari, A.; Wainstock, T.; Szaingurten-Solodkin, I.; Richter, D.; Zeadna, A.; Harlev, A.; Lunenfeld, E.; Levitas, E.; Har-Vardi, I. Are morphokinetic parameters of embryo development associated with adverse perinatal outcomes following fresh blastocyst transfer? Reprod. Biomed. Online 2021, 42, 207–216. [Google Scholar] [CrossRef] [PubMed]
- van Duijn, L.; Rousian, M.; Kramer, C.S.; van Marion, E.S.; Willemsen, S.P.; Speksnijder, J.P.; Laven, J.S.E.; Steegers-Theunissen, R.P.M.; Baart, E.B. The Impact of Culture Medium on Morphokinetics of Cleavage Stage Embryos: An Observational Study. Reprod. Sci. 2022, 29, 2179–2189. [Google Scholar] [CrossRef]
- Nguyen, Q.; Sommer, S.; Greene, B.; Wrenzycki, C.; Wagner, U.; Ziller, V. Effects of opening the incubator on morphokinetics in mouse embryos. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 229, 64–69. [Google Scholar] [CrossRef]
- Barrie, A.; McDowell, G.; Troup, S. An investigation into the effect of potential confounding patient and treatment parameters on human embryo morphokinetics. Fertil. Steril. 2021, 115, 1014–1022. [Google Scholar] [CrossRef]
- Liu, Y.; Chapple, V.; Feenan, K.; Roberts, P.; Matson, P. Time-lapse videography of human embryos: Using pronuclear fading rather than insemination in IVF and ICSI cycles removes inconsistencies in time to reach early cleavage milestones. Reprod. Biol. 2015, 15, 122–125. [Google Scholar] [CrossRef]
- Ciray, H.N.; Campbell, A.; Agerholm, I.E.; Aguilar, J.; Chamayou, S.; Esbert, M.; Sayed, S.; Time-Lapse User Group. Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum. Reprod. 2014, 29, 2650–2660. [Google Scholar] [CrossRef]
- Schindler-Johnson, M.; Petridou, N.I. Collective effects of cell cleavage dynamics. Front. Cell Dev. Biol. 2024, 12, 1358971. [Google Scholar] [CrossRef]
- Casser, E.; Wdowik, S.; Israel, S.; Witten, A.; Schlatt, S.; Nordhoff, V.; Boiani, M. Differences in blastomere totipotency in 2-cell mouse embryos are a maternal trait mediated by asymmetric mRNA distribution. Mol. Hum. Reprod. 2019, 25, 729–744. [Google Scholar] [CrossRef]
- Piotrowska-Nitsche, K.; Perea-Gomez, A.; Haraguchi, S.; Zernicka-Goetz, M. Four-cell stage mouse blastomeres have different developmental properties. Development 2005, 132, 479–490. [Google Scholar] [CrossRef]
- Azizova, A.; Onder, O.; Arslan, S.; Ardali, S.; Hazirolan, T. Persistent left superior vena cava: Clinical importance and differential diagnoses. Insights Into Imaging 2020, 11, 110. [Google Scholar] [CrossRef]
- Dykes, I.M. Left Right Patterning, Evolution and Cardiac Development. J. Cardiovasc. Dev. Dis. 2014, 1, 52–72. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Szumska, D.; Kuncheria, L.; Puliyadi, R.; Chen, C.M.; Papanayotou, C.; Lockstone, H.; Dubourg, C.; David, V.; Schneider, J.E.; et al. A Requirement for Zic2 in the Regulation of Nodal Expression Underlies the Establishment of Left-Sided Identity. Sci. Rep. 2018, 8, 10439. [Google Scholar] [CrossRef]
- Momma, K. Cardiovascular anomalies associated with chromosome 22q11.2 deletion syndrome. Am. J. Cardiol. 2010, 105, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Goldmuntz, E. 22q11.2 deletion syndrome and congenital heart disease. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2020, 184, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; van Bueren, K.L.; Ashmore, R.J.; Floss, T.; Wurst, W.; Szumska, D.; Bhattacharya, S.; Scambler, P.J. HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ. Res. 2014, 115, 23–31. [Google Scholar] [CrossRef]
- Galdini, A.; Fesslova, V.M.; Gaeta, G.; Candiani, M.; Pozzoni, M.; Chiarello, C.; Cavoretto, P.I. Prevalence of congenital heart defects in pregnancies conceived by assisted reproductive technology: A cohort study. J. Clin. Med. 2021, 10, 5363. [Google Scholar] [CrossRef]
- Shiratori, H.; Hamada, H. TGFbeta signaling in establishing left-right asymmetry. Semin. Cell Dev. Biol. 2014, 32, 80–84. [Google Scholar] [CrossRef]
- Ramsdell, A.F. Left-right asymmetry and congenital cardiac defects: Getting to the heart of the matter in vertebrate left-right axis determination. Dev. Biol. 2005, 288, 1–20. [Google Scholar] [CrossRef]
- Stefanovic, S.; Etchevers, H.C.; Zaffran, S. Outflow tract formation—Embryonic origins of conotruncal congenital heart disease. J. Cardiovasc. Dev. Dis. 2021, 8, 42. [Google Scholar] [CrossRef]
- Stainier, D.Y.; Lee, R.K.; Fishman, M.C. Cardiovascular development in the zebrafish I. Myocardial fate map and heart tube formation. Development 1993, 119, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Keegan, B.R.; Meyer, D.; Yelon, D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development 2004, 131, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Oteíza, P.; Köppen, M.; Concha, M.L.; Heisenberg, C.-P. Origin and shaping of the laterality organ in zebrafish. Development 2008, 135, 2807–2813. [Google Scholar] [CrossRef] [PubMed]
- Onjiko, R.M.; Nemes, P.; Moody, S.A. Altering metabolite distribution at Xenopus cleavage stages affects left–right gene expression asymmetries. Genesis 2021, 59, e23418. [Google Scholar] [CrossRef]
- Sendra, M.; Domínguez, J.N.; Torres, M.; Ocaña, O.H. Dissecting the complexity of early heart progenitor cells. J. Cardiovasc. Dev. Dis. 2021, 9, 5. [Google Scholar] [CrossRef]
- Satoh, M.; Teramoto, R.; Victorio, R.; Goto, S.; Zhu, W.; Homilius, M.; Kaveh, A.; Chiang, D.; Saha, K.; Zu, Y. Cell physiology and multi-omics analysis revealed disordered embryogenesis as early as mesoderm specification in models of Holt-Oram Syndrome. Circulation 2024, 150, A4120919. [Google Scholar] [CrossRef]
- Frank, J.E.; Jacobe, K.M. Evaluation and management of heart murmurs in children. Am. Fam. Physician 2011, 84, 793–800. [Google Scholar]
- Seignior, H.L. What is the significance of hearing a heart murmur during the newborn physical examination? Br. J. Midwifery 2021, 29, 158–170. [Google Scholar] [CrossRef]
- Campbell, A.; Smith, R.; Petersen, B.; Moore, L.; Khan, A.; Barrie, A. O-125 Application of artificial intelligence using big data to devise and train a machine learning model on over 63,000 human embryos to automate time-lapse embryo annotation. Hum. Reprod. 2022, 37, deac105-025. [Google Scholar] [CrossRef]

| Cohort | n | Maternal Age at Oocyte Collection (yrs; Mean ± SD) | Maternal Age at Embryo Transfer (yrs; Mean ± SD) | IVF % (n) | ICSI % (n) | Donor Oocyte % (n) | Donor Sperm % (n) |
|---|---|---|---|---|---|---|---|
| Large unaffected control | 352 | 33.8 ± 4.5 | 35.3 ± 4.5 | 21.9 (77) | 78.1 (275) | 14.2 (50) | 8.5 (30) |
| Complex CHD | 7 | 32.9 ± 6.9 | 34.4 ± 9.6 | 57.1 (4) | 42.9 (3) | 28.6 (2) | 28.6 (2) |
| Complex CHD: Matched control | 7 | 31.7 ± 2.3 | 31.7 ± 2.3 | 57.1 (4) | 42.9 (3) | 0 (0) | 14.3 (1) |
| Mild CHD | 11 | 35.9 ± 4.9 | 36.3 ± 5.0 | 45.5 (5) | 54.5 (6) | 9.1 (1) | 0 (0) |
| Mild CHD: Matched control | 11 | 34.5 ± 3.9 | 34.5 ± 3.9 | 45.5 (5) | 54.5 (6) | 0 (0) | 18.2 (2) |
| Murmur | 14 | 33.1 ± 5.6 | 35.8 ± 5.5 | 21.4 (3) | 78.6 (11) | 14.3 (2) | 7.1 (1) |
| Murmur: Matched control | 14 | 33.8 ± 4.2 | 33.8 ± 4.2 | 21.4 (3) | 78.6 (11) | 0 (0) | 14.3 (2) |
| tPNf | t2 | t3 | t4 | t5 | t6 | t8 | tM | tSB | tB | |
|---|---|---|---|---|---|---|---|---|---|---|
| ICSI | 21.80 | 24.40 | 35.43 | 36.55 | 48.13 | 50.03 | 54.89 | 81.66 | 92.75 | 102.89 |
| IVF | 24.10 | 27.04 | 37.85 | 39.23 | 51.65 | 52.70 | 57.10 | 84.52 | 94.76 | 103.24 |
| Delay | 2.30 | 2.63 | 2.42 | 2.68 | 3.52 | 2.67 | 2.21 | 2.86 | 2.01 | 0.35 |
| Cohort | Case | Cardiovascular Phenotype | Other Birth Defects | Genetic/Chromosomal Testing Results |
|---|---|---|---|---|
| Complex CHD | A1 | 1. Congenital heart disease (details not specified) 2. Operated on at 6 months and died from bleeding out 3. Congenital heart disease was not cause of death | Not specified | 1. Euploid 2. same birth mother as case C2 but derived from oocytes from different donors |
| A2 | 1. Pan systolic murmur 2. Large atrial septal defect with no significant septum 3. Moderate to large perimembraneous ventricular septal defect measuring 6–7 cm 4. Left arch stenosis 5. Bilateral superior vena cava with normal pulmonary venous drainage 6. No restricted right or left shunt 7. No coarctation adductus arteriosis 8. Required surgical intervention at 3–4 months of age | Not specified | Not specified | |
| A3 | 1. Tetralogy of Fallot 2. Died perinatally | Numerous other conditions including kidney issues. | Chorionic villus sample tested: 1. Euploid. 2. Negative for DiGeorge Syndrome test | |
| A4 | 1. Mitral valve atresia 2. Right-sided aortic arch 3. Bilateral superior vena cava 4. Atrial septal defect with oval fossa | Not specified | Not specified | |
| A5 | 1. Right sided aortic arch 2. Left subclavian artery arises aberrantly from the right aortic arch 3. Small patent foramen ovale 4. Tetralogy of Fallot and coarctation of aorta suspected at foetal anomaly scan but not seen at birth | Born prematurely at 22 + 3 weeks | Karyotyped by microarray analysis to exclude chromosome 22 microdeletion: detected a female profile with no copy number imbalances that could be considered clinically significant | |
| A6 | 1. Interrupted inferior vena cava | Not specified | Not specified | |
| A7 | 1. CHD diagnosed at foetal anomaly scan at 23 weeks 2. Born with serious congenital heart defect (details not specified) 3. Required open-heart surgery after birth | Born prematurely at 33 weeks | May be linked to genetic/chromosomal conditions (details not specified) | |
| Mild CHD | B1 | Hole in heart | Born prematurely at 34 weeks | Not specified |
| B2 | Heart defect (details not specified) | Not specified | Not specified | |
| B3 | Small hole in the heart (not a point of concern) | Not specified | Not specified | |
| B4 | Heart defect (details not specified) | Not specified | Not specified | |
| B5 | Heart murmur and small hole in the heart | Not specified | Not specified | |
| B6 | Heart defect (details not specified) | Not specified | Not specified | |
| B7 | Heart murmur and small patent foramen ovale Did not require surgery | Not specified | Not specified | |
| B8 | Hole in heart at birth, but this closed naturally | Born prematurely at 28 + 1 weeks | Not specified | |
| B9 | Hole in heart | Not specified | Not specified | |
| B10 | Heart defect (details not specified) | Not specified | Not specified | |
| B11 | Hole in heart | Not specified | Not specified | |
| Murmur | C1 | Heart murmur | Not specified | Not specified |
| C2 | Heart murmur | Not specified | Not specified | |
| C3 | Heart murmur | Not specified | Not specified | |
| C4 | Heart murmur | Not specified | Not specified | |
| C5 | Heart murmur: likely to be innocent | Not specified | Not specified | |
| C6 | Heart murmur | Not specified | Not specified | |
| C7 | Heart murmur | Not specified | Not specified | |
| C8 | Heart murmur and ectopic heartbeat | Not specified | Not specified | |
| C9 | Heart murmur | Not specified | Not specified | |
| C10 | Heart murmur | Not specified | Not specified | |
| C11 | Heart murmur | Not specified | Not specified | |
| C12 | Heart murmur | flappy larynx Born at 37 weeks | Not specified | |
| C13 | Heart murmur | Not specified | Not specified | |
| C14 | Heart murmur | Not specified | Not specified |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markham, S.; Campbell, A.; Montgomery, S.; Dykes, I.M. Differences in the Early In Vitro Development of Preimplantation Human IVF Embryos Which Go on to Develop Congenital Heart Disease. J. Cardiovasc. Dev. Dis. 2025, 12, 370. https://doi.org/10.3390/jcdd12090370
Markham S, Campbell A, Montgomery S, Dykes IM. Differences in the Early In Vitro Development of Preimplantation Human IVF Embryos Which Go on to Develop Congenital Heart Disease. Journal of Cardiovascular Development and Disease. 2025; 12(9):370. https://doi.org/10.3390/jcdd12090370
Chicago/Turabian StyleMarkham, Sophie, Alison Campbell, Sue Montgomery, and Iain M. Dykes. 2025. "Differences in the Early In Vitro Development of Preimplantation Human IVF Embryos Which Go on to Develop Congenital Heart Disease" Journal of Cardiovascular Development and Disease 12, no. 9: 370. https://doi.org/10.3390/jcdd12090370
APA StyleMarkham, S., Campbell, A., Montgomery, S., & Dykes, I. M. (2025). Differences in the Early In Vitro Development of Preimplantation Human IVF Embryos Which Go on to Develop Congenital Heart Disease. Journal of Cardiovascular Development and Disease, 12(9), 370. https://doi.org/10.3390/jcdd12090370







