Nested PCR Detection of Pythium sp. from Formalin-Fixed, Paraffin-Embedded Canine Tissue Sections
Abstract
:Simple Summary
Abstract
1. Introduction
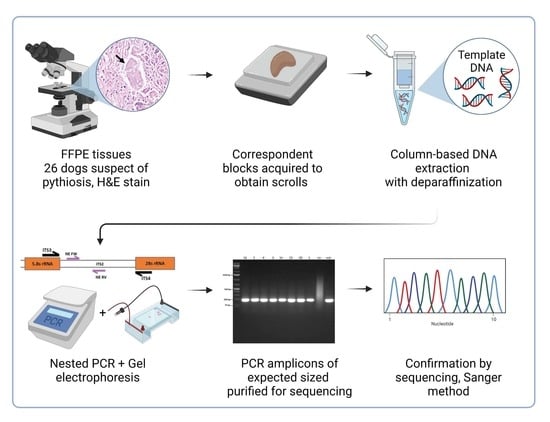
2. Materials and Methods
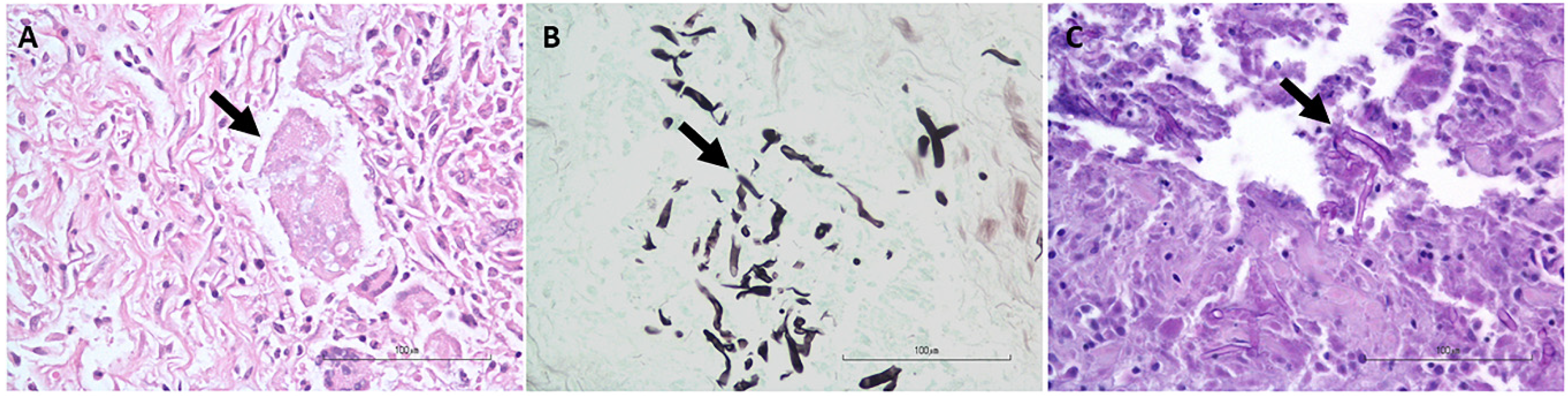
2.1. Samples
2.2. DNA Extraction
2.3. Pan-Fungal Conventional PCR Assay
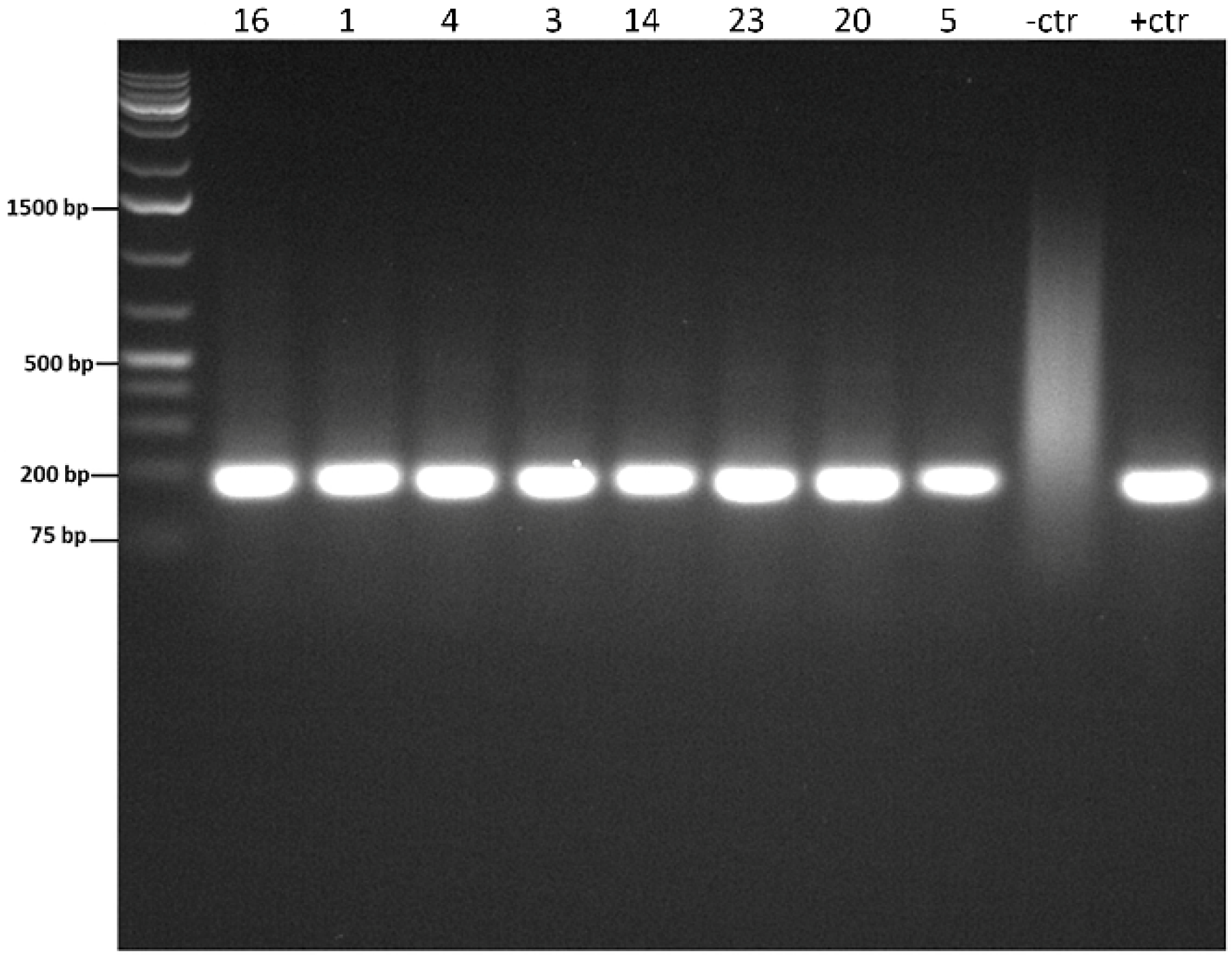
2.4. Nested PCR
2.5. Conventional PCR for the GAPDH Gene
2.6. Sequencing
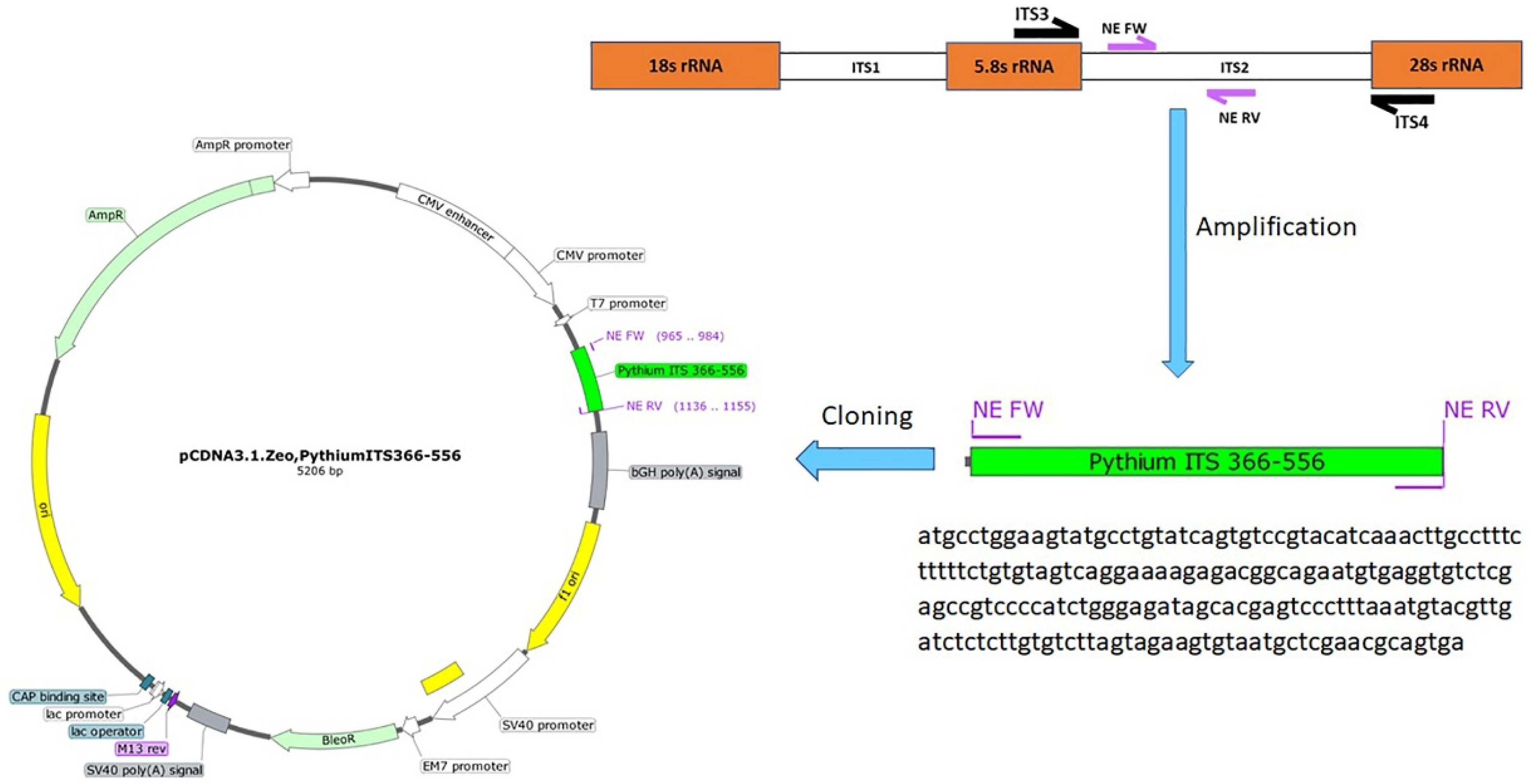
2.7. Cloning of the PCR Amplicons
2.8. Determination of PCR Sensitivity
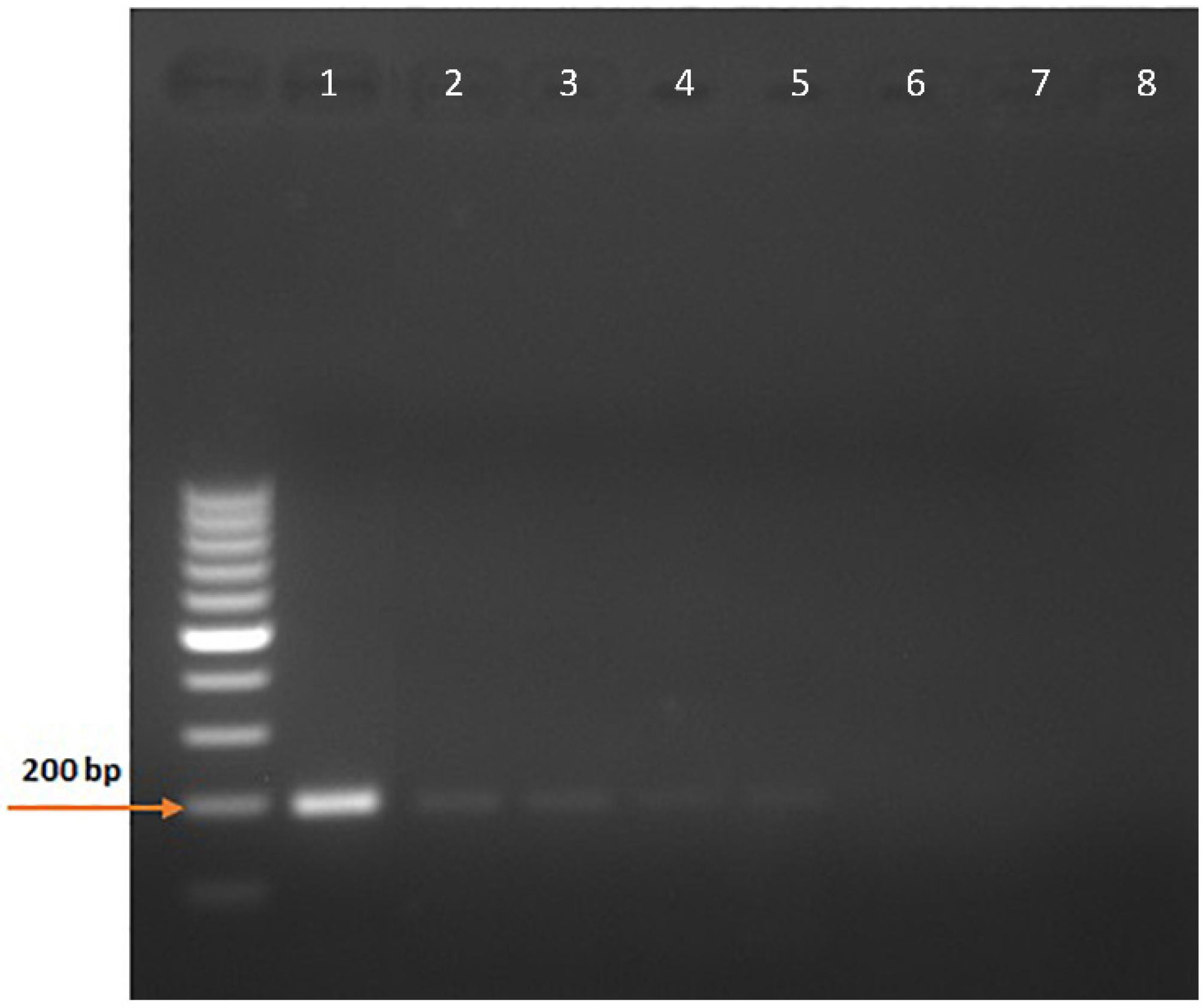
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paz, G.S.D.; Camargo, G.G.; Cury, J.E.; Apolonio, E.V.P.; Garces, H.G.; Prado, A.C.D.; Chechi, J.L.; Oliveira, A.L.; Watanabe, M.J.; Bagagli, E.; et al. Outbreak of equine pythiosis in a southeastern region of Brazil: Environmental isolation and phylogeny. Transbound. Emerg. Dis. 2022, 69, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.A. (Ed.) Pythiosis. In Clinical Veterinary Advisor; W.B. Saunders: Saint Louis, MO, USA, 2012; pp. 485–487. [Google Scholar]
- Mendoza, L.; Arias, M.; Colmenarez, V.; Perazzo, Y. Intestinal canine pythiosis in Venezuela confirmed by serological and sequencing analysis. Mycopathologia 2005, 159, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Grooters, A.M. Pythiosis, lagenidiosis, and zygomycosis in small animals. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 695–720. [Google Scholar] [CrossRef]
- Gaastra, W.; Lipman, L.J.; De Cock, A.W.; Exel, T.K.; Pegge, R.B.; Scheurwater, J.; Vilela, R.; Mendoza, L. Pythium insidiosum: An overview. Vet. Microbiol. 2010, 146, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gurnani, B.; Kaur, K.; Venugopal, A.; Srinivasan, B.; Bagga, B.; Iyer, G.; Christy, J.; Prajna, L.; Vanathi, M.; Garg, P.; et al. Pythium insidiosum keratitis—A review. Indian J. Ophthalmol. 2022, 70, 1107–1120. [Google Scholar] [CrossRef]
- Reis, J.L., Jr.; de Carvalho, E.C.; Nogueira, R.H.; Lemos, L.S.; Mendoza, L. Disseminated pythiosis in three horses. Vet. Microbiol. 2003, 96, 289–295. [Google Scholar] [CrossRef]
- Lohnoo, T.; Jongruja, N.; Rujirawat, T.; Yingyon, W.; Lerksuthirat, T.; Nampoon, U.; Kumsang, Y.; Onpaew, P.; Chongtrakool, P.; Keeratijarut, A.; et al. Efficiency comparison of three methods for extracting genomic DNA of the pathogenic oomycete Pythium insidiosum. J. Med. Assoc. Thai. 2014, 97, 342–348. [Google Scholar]
- Cridge, H.; Hughes, S.M.; Langston, V.C.; Mackin, A.J. Mefenoxam, Itraconazole, and Terbinafine Combination Therapy for Management of Pythiosis in Dogs (Six Cases). J. Am. Anim. Hosp. Assoc. 2020, 56, 307. [Google Scholar] [CrossRef]
- Salt, C.; Morris, P.J.; Wilson, D.; Lund, E.M.; German, A.J. Association between life span and body condition in neutered client-owned dogs. J. Vet. Intern. Med. 2019, 33, 89–99. [Google Scholar] [CrossRef]
- Hummel, J.; Grooters, A.; Davidson, G.; Jennings, S.; Nicklas, J.; Birkenheuer, A. Successful management of gastrointestinal pythiosis in a dog using itraconazole, terbinafine, and mefenoxam. Med. Mycol. 2011, 49, 539–542. [Google Scholar] [CrossRef]
- Denardi, L.B.; Weiblen, C.; Ianiski, L.B.; Stibbe, P.C.; Pinto, S.C.; Santurio, J.M. Anti-Pythium insidiosum activity of MSI-78, LL-37, and magainin-2 antimicrobial peptides. Braz. J. Microbiol. 2022, 53, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Medhasi, S.; Chindamporn, A.; Worasilchai, N. A Review: Antimicrobial Therapy for Human Pythiosis. Antibiotics 2022, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Ianiski, L.B.; Stibbe, P.C.; Denardi, L.B.; Weiblen, C.; Soares, M.P.; Valente, J.S.S.; Sangioni, L.A.; Pereira, D.I.B.; Santurio, J.M.; Botton, S.A. In vitro anti-Pythium insidiosum activity of amorolfine hydrochloride and azithromycin, alone and in combination. Med. Mycol. 2021, 59, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Grooters, A.M.; Gee, M.K. Development of a nested polymerase chain reaction assay for the detection and identification of Pythium insidiosum. J. Vet. Intern. Med. 2002, 16, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Vanittanakom, N.; Supabandhu, J.; Khamwan, C.; Praparattanapan, J.; Thirach, S.; Prasertwitayakij, N.; Louthrenoo, W.; Chiewchanvit, S.; Tananuvat, N. Identification of emerging human-pathogenic Pythium insidiosum by serological and molecular assay-based methods. J. Clin. Microbiol. 2004, 42, 3970–3974. [Google Scholar] [CrossRef] [PubMed]
- Bosco Sde, M.; Bagagli, E.; Araújo, J.P., Jr.; Candeias, J.M.; de Franco, M.F.; Alencar Marques, M.E.; Mendoza, L.; de Camargo, R.P.; Alencar Marques, S. Human pythiosis, Brazil. Emerg. Infect. Dis. 2005, 11, 715–718. [Google Scholar] [CrossRef]
- Badenoch, P.R.; Coster, D.J.; Wetherall, B.L.; Brettig, H.T.; Rozenbilds, M.A.; Drenth, A.; Wagels, G. Pythium insidiosum keratitis confirmed by DNA sequence analysis. Br. J. Ophthalmol. 2001, 85, 502–503. [Google Scholar] [CrossRef]
- Znajda, N.R.; Grooters, A.M.; Marsella, R. PCR-based detection of Pythium and Lagendium DNA in frozen and ethanol-fixed animal tissues. Vet. Dermatol. 2002, 13, 187–194. [Google Scholar] [CrossRef]
- Azevedo, M.I.; Botton, S.A.; Pereira, D.I.; Robe, L.J.; Jesus, F.P.; Mahl, C.D.; Costa, M.M.; Alves, S.H.; Santurio, J.M. Phylogenetic relationships of Brazilian isolates of Pythium insidiosum based on ITS rDNA and cytochrome oxidase II gene sequences. Vet. Microbiol. 2012, 159, 141–148. [Google Scholar] [CrossRef]
- Mar Htun, Z.; Laikul, A.; Pathomsakulwong, W.; Yurayart, C.; Lohnoo, T.; Yingyong, W.; Kumsang, Y.; Payattikul, P.; Sae-Chew, P.; Rujirawat, T.; et al. Identification and Biotyping of Pythium insidiosum Isolated from Urban and Rural Areas of Thailand by Multiplex PCR, DNA Barcode, and Proteomic Analyses. J. Fungi 2021, 7, 242. [Google Scholar] [CrossRef]
- Rivierre, C.; Laprie, C.; Guiard-Marigny, O.; Bergeaud, P.; Berthelemy, M.; Guillot, J. Pythiosis in Africa. Emerg. Infect. Dis. 2005, 11, 479–481. [Google Scholar] [CrossRef] [PubMed]
- Keeratijarut, A.; Lohnoo, T.; Yingyong, W.; Nampoon, U.; Lerksuthirat, T.; Onpaew, P.; Chongtrakool, P.; Krajaejun, T. PCR amplification of a putative gene for exo-1,3-β-glucanase to identify the pathogenic oomycete Pythium insidiosum. Asian Biomed. 2017, 8, 637–644. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. 38—Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Keeratijarut, A.; Lohnoo, T.; Yingyong, W.; Rujirawat, T.; Srichunrusami, C.; Onpeaw, P.; Chongtrakool, P.; Brandhorst, T.T.; Krajaejun, T. Detection of the oomycete Pythium insidiosum by real-time PCR targeting the gene coding for exo-1,3-β-glucanase. J. Med. Microbiol. 2015, 64, 971–977. [Google Scholar] [CrossRef]
- Meason-Smith, C.; Edwards, E.E.; Older, C.E.; Branco, M.; Bryan, L.K.; Lawhon, S.D.; Suchodolski, J.S.; Gomez, G.; Mansell, J.; Hoffmann, A.R. Panfungal Polymerase Chain Reaction for Identification of Fungal Pathogens in Formalin-Fixed Animal Tissues. Vet. Pathol. 2017, 54, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Cadavid, C.; Rudd, S.; Zaki, S.R.; Patel, M.; Moser, S.A.; Brandt, M.E.; Gómez, B.L. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: Comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 2010, 48, 2147–2153. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Friehs, K. Plasmid copy number and plasmid stability. Adv. Biochem. Eng. Biotechnol. 2004, 86, 47–82. [Google Scholar] [CrossRef]
- Sayedahmed, E.E.; Mittal, S.K. A potential approach for assessing the quality of human and nonhuman adenoviral vector preparations. Can. J. Vet. Res. 2020, 84, 314–318. [Google Scholar]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, D34–D38. [Google Scholar] [CrossRef]
- Yolanda, H.; Krajaejun, T. Global Distribution and Clinical Features of Pythiosis in Humans and Animals. J. Fungi 2022, 8, 182. [Google Scholar] [CrossRef]
- Supabandhu, J.; Vanittanakom, P.; Laohapensang, K.; Vanittanakom, N. Application of immunoblot assay for rapid diagnosis of human pythiosis. J. Med. Assoc. Thai. 2009, 92, 1063–1071. [Google Scholar] [PubMed]
- Mendoza, L.; Prasla, S.H.; Ajello, L. Orbital pythiosis: A non-fungal disease mimicking orbital mycotic infections, with a retrospective review of the literature. Mycoses 2004, 47, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Bentinck-Smith, J.; Padhye, A.A.; Maslin, W.R.; Hamilton, C.; McDonald, R.K.; Woody, B.J. Canine pythiosis—Isolation and identification of Pythium insidiosum. J. Vet. Diagn. Investig. 1989, 1, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Jena, S.K.; Desai, A.; Agarwal, S. Pythium insidiosum Keratitis: Histopathology and Rapid Novel Diagnostic Staining Technique. Cornea 2017, 36, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D. A microchemical reaction resulting in the staining of polysaccharide structures in fixed tissue preparations. Arch. Biochem. 1948, 16, 131–141. [Google Scholar]
- Bagga, B.; Vishwakarma, P.; Sharma, S.; Jospeh, J.; Mitra, S.; Mohamed, A. Sensitivity and specificity of potassium hydroxide and calcofluor white stain to differentiate between fungal and Pythium filaments in corneal scrapings from patients of Pythium keratitis. Indian J. Ophthalmol. 2022, 70, 542–545. [Google Scholar] [CrossRef]
- Pesavento, P.A.; Barr, B.; Riggs, S.M.; Eigenheer, A.L.; Pamma, R.; Walker, R.L. Cutaneous pythiosis in a nestling white-faced ibis. Vet. Pathol. 2008, 45, 538–541. [Google Scholar] [CrossRef]
- Supabandhu, J.; Fisher, M.C.; Mendoza, L.; Vanittanakom, N. Isolation and identification of the human pathogen Pythium insidiosum from environmental samples collected in Thai agricultural areas. Med. Mycol. 2008, 46, 41–52. [Google Scholar] [CrossRef]
- Balows, A. Manual of clinical microbiology 8th edition: P. R. Murray, E. J. Baron, J. H. Jorgenson, M. A. Pfaller, and R. H. Yolken, eds., ASM Press, 2003, 2113 pages, 2 vol, 2003. Diagn. Microbiol. Infect. Dis. 2003, 47, 625–626. [Google Scholar] [CrossRef]
- Botton, S.A.; Pereira, D.I.; Costa, M.M.; Azevedo, M.I.; Argenta, J.S.; Jesus, F.P.; Alves, S.H.; Santurio, J.M. Identification of Pythium insidiosum by nested PCR in cutaneous lesions of Brazilian horses and rabbits. Curr. Microbiol. 2011, 62, 1225–1229. [Google Scholar] [CrossRef]
- Nguyen, D.; Vilela, R.; Miraglia, B.M.; Vilela, G.; Jasem-Alali, N.; Rohn, R.; Glass, R.; Hansen, R.D.; Mendoza, L. Geographic distribution of Pythium insidiosum infections in the United States. J. Am. Vet. Med. Assoc. 2021, 260, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Oldenhoff, W.; Grooters, A.; Pinkerton, M.E.; Knorr, J.; Trepanier, L. Cutaneous pythiosis in two dogs from Wisconsin, USA. Vet. Dermatol. 2014, 25, 52-e21. [Google Scholar] [CrossRef] [PubMed]
- Berryessa, N.A.; Marks, S.L.; Pesavento, P.A.; Krasnansky, T.; Yoshimoto, S.K.; Johnson, E.G.; Grooters, A.M. Gastrointestinal pythiosis in 10 dogs from California. J. Vet. Intern. Med. 2008, 22, 1065–1069. [Google Scholar] [CrossRef] [PubMed]
- Yolanda, H.; Krajaejun, T. Review of methods and antimicrobial agents for susceptibility testing against Pythium insidiosum. Heliyon 2020, 6, e03737. [Google Scholar] [CrossRef]
- Duclos, D. Canine pododermatitis. Vet. Clin. N. Am. Small Anim. Pract. 2013, 43, 57–87. [Google Scholar] [CrossRef]
| Case Number | Age (Years) | Sex | Organ (s) Affected | H & E | GMS | PAS | Nested PCR |
|---|---|---|---|---|---|---|---|
| 1 | 1.5 | F | Pancreas | + | - | + | + |
| 2 | 2 | F | Stomach | + | + | + | + |
| 3 | 1 | M | Duodenum | + | - | + | + |
| 4 | 1.5 | M | Stomach | - | - | + | + |
| 5 | 2 | F | Stomach, lymph nodes | + | + | + | + |
| 6 | 2.5 | FS | Liver, mesenteric nodule | - | - | - | - |
| 7 | 0.10 | M | Mesenteric mass | - | + | - | + |
| 8 | 1 | F | Small intestine | + | + | + | - |
| 9 | 1 | M | Small intestine | - | + | - | - |
| 10 | 3 | M | Mesentery | - | - | - | - |
| 11 | 2 | MN | Large Intestine | N/A | + | - | + |
| 12 | 2 | MN | Small intestine, colon, rectum | + | + | + | - |
| 13 | 3 | M | Mesenteric lymph node | + | + | + | + |
| 14 | 4 | MN | Proximal duodenum, liver | - | - | - | + |
| 15 | 8.5 | F | Small Intestine, pancreas | - | - | - | + |
| 16 | 2 | M | Pylorus, small intestine, mesenteric lymph node | + | + | + | + |
| 17 | 3.5 | FS | Jejunum | + | + | + | + |
| 18 | 10 | FS | Ileum | - | - | - | - * |
| 19 | 4 | MN | Stomach, duodenum, lung, lymph node | + | + | + | + |
| 20 | 5.5 | MN | Colon, mesenteric root, mesenteric lymph nodes | - | - | - | + |
| 21 | 2 | MN | Colon, ileocecal junction, mesenteric lymph node | + | + | + | + |
| 22 | 6 | M | Intestine, omentum, lymph node | - | - | - | + |
| 23 | 7 | FS | Left ventral thoracic lesion | + | + | + | + |
| 24 | 1 | FS | Ileum, mesenteric lymph node | - | - | - | + |
| 25 | 0.5 | U | Skin | + | + | + | + |
| 26 | 0.10 | FS | Proximal jejunum, liver abdominal mass, intestinal mass, mesenteric lymph node | - | + | - | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshafie, N.O.; Hanlon, J.; Malkawi, M.; Sayedahmed, E.E.; Guptill, L.F.; Jones-Hall, Y.L.; Santos, A.P. Nested PCR Detection of Pythium sp. from Formalin-Fixed, Paraffin-Embedded Canine Tissue Sections. Vet. Sci. 2022, 9, 444. https://doi.org/10.3390/vetsci9080444
Elshafie NO, Hanlon J, Malkawi M, Sayedahmed EE, Guptill LF, Jones-Hall YL, Santos AP. Nested PCR Detection of Pythium sp. from Formalin-Fixed, Paraffin-Embedded Canine Tissue Sections. Veterinary Sciences. 2022; 9(8):444. https://doi.org/10.3390/vetsci9080444
Chicago/Turabian StyleElshafie, Nelly O., Jessica Hanlon, Mays Malkawi, Ekramy E. Sayedahmed, Lynn F. Guptill, Yava L. Jones-Hall, and Andrea P. Santos. 2022. "Nested PCR Detection of Pythium sp. from Formalin-Fixed, Paraffin-Embedded Canine Tissue Sections" Veterinary Sciences 9, no. 8: 444. https://doi.org/10.3390/vetsci9080444
APA StyleElshafie, N. O., Hanlon, J., Malkawi, M., Sayedahmed, E. E., Guptill, L. F., Jones-Hall, Y. L., & Santos, A. P. (2022). Nested PCR Detection of Pythium sp. from Formalin-Fixed, Paraffin-Embedded Canine Tissue Sections. Veterinary Sciences, 9(8), 444. https://doi.org/10.3390/vetsci9080444