Simple Summary
Salmonellosis is a human and animal disease caused by Salmonella, a bacterial genus classified into different species, subspecies, and serological variants (serovars) according to adaptation to one or more different hosts (animals and humans), pathogenicity profiles, and antigenic properties. Some specific Salmonella serovars can spread more easily in the enteric microbiota of avian species, often causing disease in birds and/or being transmitted to humans through food (such as chicken and eggs). Antimicrobial resistance (AMR) has also been reported in poultry-associated Salmonella isolates due to the widespread use of antimicrobials on farms. The availability of comprehensive data on the emergence and spread of Salmonella serovars, as well as their AMR profiles in farms and food products in Brazil (a major producer of poultry in the World), is necessary to understand their relevance in all avian production chains and also occurrence in poultry-derived foods. This article aims to provide an overview of the genus Salmonella and the main serovars that emerged in Brazilian poultry over time (Gallinarum, Typhimurium, Enteritidis, Heidelberg, and Minnesota), reviewing the scientific literature and suggesting more effective prevention and control for the future.
Abstract
Salmonella infects poultry, and it is also a human foodborne pathogen. This bacterial genus is classified into several serovars/lineages, some of them showing high antimicrobial resistance (AMR). The ease of Salmonella transmission in farms, slaughterhouses, and eggs industries has made controlling it a real challenge in the poultry-production chains. This review describes the emergence, dissemination, and AMR of the main Salmonella serovars and lineages detected in Brazilian poultry. It is reported that few serovars emerged and have been more widely disseminated in breeders, broilers, and layers in the last 70 years. Salmonella Gallinarum was the first to spread on the farms, remaining as a concerning poultry pathogen. Salmonella Typhimurium and Enteritidis were also largely detected in poultry and foods (eggs, chicken, turkey), being associated with several human foodborne outbreaks. Salmonella Heidelberg and Minnesota have been more widely spread in recent years, resulting in frequent chicken/turkey meat contamination. A few more serovars (Infantis, Newport, Hadar, Senftenberg, Schwarzengrund, and Mbandaka, among others) were also detected, but less frequently and usually in specific poultry-production regions. AMR has been identified in most isolates, highlighting multi-drug resistance in specific poultry lineages from the serovars Typhimurium, Heidelberg, and Minnesota. Epidemiological studies are necessary to trace and control this pathogen in Brazilian commercial poultry production chains.
Keywords:
Salmonella; Brazil; Gallinarum; Enteritidis; Minnesota; Typhimurium; Heidelberg; antimicrobial resistance; poultry 1. Introduction
Meat consumption has been shifting towards poultry. It has been mainly driven by chicken (Gallus gallus) and turkey (Meleagris gallopavo), due to the relatively low cost of production [1]. Moreover, eggs are considered foods of high nutritional value for humans and they are widely consumed too [2].
Brazil is an important producer and exporter of poultry meat in the World, with volumes of 13.8 million and 4.2 million tons, respectively, in 2020. In addition, Brazil produced 172.3 thousand tons of turkey meat and 53.5 trillion eggs in this same year. Almost all stages of the poultry production chain (meat and eggs) are carried out inside the country, including farming the breeding birds (grandparent stock, hatchery, breeders), broilers, and layers. Furthermore, poultry foods are also largely processed in slaughterhouses and eggs industries in different Brazilian regions [3].
Foodborne bacteria have been detected in all poultry-producing regions in the World in the last seventy years [4]. Different bacteria species are the main pathogens of meat and eggs [5]. Poultry meat contamination occurs frequently by Salmonella spp., Campylobacter jejuni, Campylobacter coli and Clostridium perfringens [4]. Other frequent poultry foodborne bacteria include Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli [4,5]. The bacterial genera Pseudomonas, Hafnia, Serratia, Rahnella, Yersinia, and Buttiauxella have also been detected in poultry products [6,7]. Among all these pathogenic bacteria, Salmonella has been the most concerning to public health. Poultry foods most commonly associated with Salmonella outbreaks are eggs, chicken, and other meals mixed with poultry products [4]. In addition, some Salmonella serovars are also associated with specific poultry diseases with huge economic losses [8].
Prevention and control measures are adopted in producing farms and food processing industries to prevent the occurrence of the main Salmonella serovars worldwide. In Brazil, Salmonella is also routinely controlled on farms, with vaccination and constant laboratory diagnosis to monitor the infection of the flocks and prevent transmission to poultry-derived food [9]. Despite these measures, several poultry diseases and foodborne Salmonella outbreaks were reported in Brazil in recent decades. This review describes the main features of the Salmonella genus, including serovar classification and antimicrobial resistance (AMR), as well as the emergence and spread of serovars most frequently associated with poultry in Brazil.
2. Classification into Serovars
Salmonella belongs to the Enterobacteriaceae family. It is a Gram-negative bacillus, non-spore-forming, facultative anaerobic, and generally mobile due to the peritrichous flagella. Moreover, the genus is classified into the bacterial species S. enterica and S. bongori, with the first being divided into six subspecies: enterica, salamae, arizonae, diarizonae, houtenae, and indica. Salmonella isolates from all these species and subspecies are also classified according to antigenic characteristics and more than 2.650 serovars were already reported [10]. Differentiation into serovars is performed by the laboratory analysis of the O (membrane lipopolysaccharides), H (flagellar proteins), and Vi (capsular polysaccharide) bacterial antigens within the White–Kauffman–Le Minor scheme. All these antigens are expressed in a specific formula for each serovar, for example “1,4,[5],12:i:1,2” (Typhimurium) and “1,9,12:g,m“ (Enteritidis) [11].
Salmonella has also been classified according to two specific human clinical manifestations into typhoid and non-typhoid types. The first group is composed of the etiologic agents of enteric fever, and currently includes serovars Typhi and Paratyphi, while the second group is composed of all other serovars [12]. Similarly, salmonellosis in poultry is also divided into two main groups according to the pathogenesis and avian clinical manifestations: (i) typhoid, including generalized infection by Salmonella, resulting in fowl typhoid (FT) and pullorum disease (PD), both caused by the serovar Gallinarum biovars, Gallinarum and Pullorum, respectively, which are highly adapted and restrict transmission among chickens (Gallus gallus) and a few other bird species; (ii) paratyphoid, including all Salmonella associated with enteric infection in birds presenting or not presenting clinical disease under special circumstances (laying period, very young or old birds, viral co-infections). This last type of salmonellosis is caused by any serovar other than Gallinarum (such as Typhimurium, Enteritidis, Heidelberg, Minnesota, etc.), which also can be transmitted to humans by direct contact (on farms, slaughterhouses, etc.) or consumption of contaminated poultry foods [13] (Figure 1).
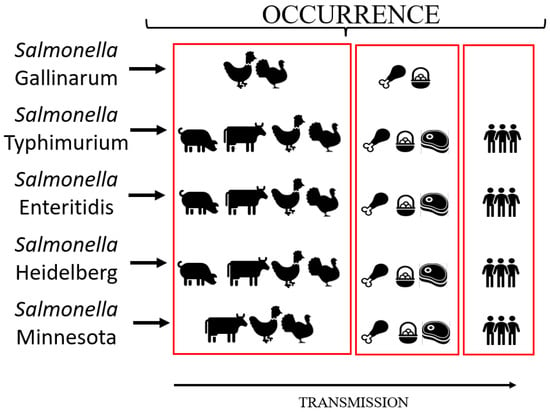
Figure 1.
Main sources of contamination and transmission of the Salmonella serovars frequently detected in the Brazilian poultry production chain.
Salmonella intra-serovar lineages laboratory identification is necessary for outbreak epidemiological investigations. The first method used to identify Salmonella isolates in outbreaks was phage typing in the 1980s [14]. Each Salmonella isolate from a given serovar was sorted into a unique phage type according to its reactivity against a set of specific viruses [15,16]. This procedure was more frequently used in the epidemiological surveillance of the concerning serovars Typhimurium and Enteritidis [15,16,17]. S. Typhimurium more disseminated definitive (phage) types (DTs) included DT49, DT104, DT135, and DT193 [18,19], while S. Enteritidis epidemiologically important phage types (PTs) were PT4, PT8, PT13a, and PT13 [20,21].
More recently, different molecular DNA-based methods have also been included in the arsenal of laboratory methods for epidemiological investigations, such as Pulsed-Field Gel Electrophoresis (PFGE), Multi-locus Sequence Typing (MLST), core genome Multi-locus Sequence Typing (cgMLST), Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), Multiple-Locus Variable-Number Tandem-Repeats Analysis (MLVA) and sequencing analysis of the Intergenic Spacer Regions (ISRs) of ribosomal RNA operons [22,23,24,25,26]. There are even some molecular techniques to identify the serovar from whole-genome sequences (WGS) data, such as Salmonella In Silico Typing Resource (SISTR) [24] and SeqSero [27]. Salmonella WGS data have also been increasingly used to evaluate single nucleotide polymorphisms (SNPs) in the complete genomes and to track specific lineages of many serovars in different stages of the poultry production chain [28,29].
PFGE is a genotyping method based on previous total DNA digestion of a specific bacterial isolate with few restriction enzymes followed by pulsed-field electrophoresis. It was largely used from the 1980s to the 2000s to track specific Salmonella lineages [30]. MLST allows the characterization of Salmonella isolates by sequencing seven housekeeping genes: aroC, dnaN, hemD, hisD, purE, sucA, and thrA [31,32]. It was refined and many studies are reporting the use of cgMLST, allowing the determination of the Salmonella sequence type based on all core genome genes [25]. According to these last two methods, Salmonella isolates have been currently classified into hundreds to thousands of different sequence types (STs) as an additional identification to serovar assignment [25,32]. As the number of Salmonella genomic sequences has increased, several STs were already reported and are now available in current specific databases. In addition, there are proposals to change the conventional nomenclature of Salmonella serovars, now using the genomic characteristics of the bacterial isolates and the specific ST groups [32,33].
3. Antimicrobial Resistance
Salmonella is also a matter of concern due to the occurrence of AMR in some specific serovars [34,35]. The excessive use of antimicrobials in animal/agricultural production and treatment of human/animal diseases has allowed the selection of many Salmonella strains resistant to one or more antimicrobials [36].
Antimicrobials have been administered in animal production with three main objectives: (1) to enhance animal performance, using low doses continuously throughout the feed; (2) to prevent the occurrence of pathogenic bacteria, using intermediate doses before or during critical transitions in the production process; and (3) to treat infectious animal diseases in the producing flocks/herds, usually with higher doses [37,38]. Aminoglycosides and other antimicrobial classes have been included in poultry production over time [39,40]. Tetracyclines and sulfonamides were banned as additives in animal feed in 1988, but their use for therapeutic purposes is still allowed, and are currently used to treat sick animals [41]. Data collected from 103 countries globally indicated that the Americas, Asia, and Eastern Oceania used 86% of 93,092 tons of antimicrobial agents for animals in 2017, with tetracycline and penicillin ranking at the top of the most used ones [42].
The prolonged use of antimicrobials has possibly increased Salmonella resistance to the most used classes in the poultry production chains [39,43,44]. Overall, 6 out of the 10 most frequently poultry-associated Salmonella serovars in the United States (Enteritidis, Montevideo, Schwarzengrund, Infantis, Thompson, and Mbandaka) have been demonstrated to be generally pan-susceptible or with resistance to few antimicrobials, whereas four (Heidelberg, Typhimurium, Kentucky, and Senftenberg) are more commonly reported as resistant to many of them [45]. Some Salmonella isolates from these last four serovars have also been reported as multidrug-resistant (MDR), which means, resistant to three or more antimicrobials classes [34,44,45,46,47].
Due to the intensive farming and the long history of antibiotic use, Brazil has reported the occurrence of AMR in different Salmonella serovars [48]. A study evaluating 930 WGS of different Salmonella serovars retrieved from the public database of the National Center for Biotechnology Information (NCBI) and published in the last four decades, demonstrated the prediction of the MDR phenotype in 58% (540/930) of the isolates, highlighting ciprofloxacin and nalidixic acid with the highest frequency rates [39]. Other recent reports have also demonstrated that MDR is frequent in Salmonella serovars Heidelberg and Minnesota isolated from broilers, layers, and poultry-derived food in Brazil [44,49,50,51,52].
4. Emergence and Dissemination
The intestines of birds are colonized by several microorganisms that make up the host’s microbiota in a state of equilibrium. Lactobacillus, Bifidobacterium, Streptococcus, Bacteroides, Fusobacterium, and Eubacterium are frequent bacterial genera of beneficial microbiota. Imbalance of the intestinal microbiota can occur and favor the colonization by pathogenic micro-organisms, including the genera Salmonella, Clostridium, Escherichia, Campylobacter, Staphylococcus, and Listeria [53,54].
Several factors affect colonization by Salmonella in poultry flocks on commercial farms, including host age, genetic susceptibility, stress due to overcrowding or secondary disease, level of exposure to pathogens, intestinal microbiota competitors, and bacterial genetic factors [55,56]. Furthermore, Salmonella needs first to multiply in the enteric tract of some poultry in the flock, after contaminating the environment with the excretion of high bacterial loads in the litter [57]. In the host infection, Salmonella has to compete with other micro-organisms for a niche that provides nutrients for replication and fights against the host’s immune defenses [58]. To successfully reach the host’s gut and start the infection, the bacteria need first survive in hostile environments. Salmonella has developed “biological tools” to be a good competitor, comprising a set of virulence factors, plasmids, prophages, and even mobile genetic elements acquired in its evolutionary history [59,60].
One specific and very important adaptation step in the evolutionary process was the acquisition of the genetic cluster known as Salmonella pathogenic island 1 (SPI-1). It produces a type III secretion system necessary for the enterocyte invasion. In addition, several other gains and losses of genes occurred over time and a total of 24 different SPIs were already reported scattered into different serovars [61]. In addition to SPI-1, four other SPIs have been more well-studied: SPIs-2 to 4, which are necessary for the bacteria to multiply and survive within the host, and SPI-5, which regulates inflammation and the secretion of metabolites for the enteric phase of the disease [62]. The remaining pathogenicity islands are also necessary according to other specific environments and hosts [61,62].
The emergence of a new Salmonella serovar in a specific ecological niche in poultry chains is frequently associated with the effective reduction in other bacteria populations (including other Salmonella serovars) and/or the lack of adequate immunization of the host. In the recent poultry intensive production history (over the last 100 years), “conquests and downfalls” seem to have occurred with the different serovars in the main places of poultry production in the world. In the mid-20th century, poultry diseases caused by Salmonella Gallinarum (including biovars Gallinarum and Pullorum) were the most concerning infections on commercial farms worldwide [63]. Some decades after, Salmonella Typhimurium and Enteritidis were massively detected in poultry flocks and foods (chicken, turkey, eggs), and were considered the most concerning serovars for public health [56,64,65]. Until the mid-1980s, S. Typhimurium was also one of the main serovars detected in animal production farms, and consequently in foods [64]. In the 1990s, S. Enteritidis predominated among the serovars frequently detected in avian farms and outbreaks due to the consumption of poultry foods in several countries [65,66]. More recently, other Salmonella serovars (such as Heidelberg, Kentucky, Montevideo, and Minnesota) were increasingly detected in specific poultry production chains and foods worldwide [67,68,69]. In Brazil, the most concerning serovars in poultry production along the time in the 20th century were the same of other poultry-producing western countries: Gallinarum, Typhimurium, and Enteritidis. Although two other serovars have been a matter of special concern in Brazilian broilers farms in this century: Heidelberg and Minnesota (Figure 2).
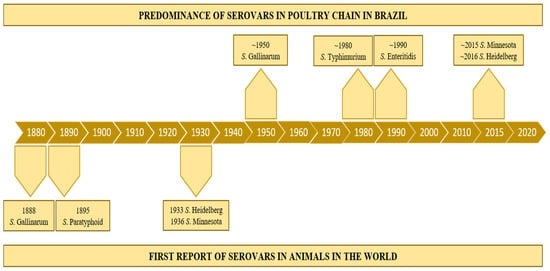
Figure 2.
Timeline of the emergence and dissemination of the main Salmonella serovars in Brazilian poultry-production chain.
4.1. Salmonella Gallinarum
Salmonellaenterica serovar Gallinarum (S. Gallinarum) is highly adapted to avian species, causing systemic diseases in birds from poultry farms worldwide [63]. It is considered a non-motile serovar due to the absence of peritrichous flagella in all bacterial isolates. Therefore, S. Gallinarum presents only O-surface antigens (antigenic formula 1,9,12:-:-). This serovar is further divided into Pullorum and Gallinarum biovars, which have specific genetic, metabolic, and physiological characteristics, in addition to being responsible for two well-characterized clinical diseases in chickens (Gallus gallus). Recent studies have also demonstrated that S. Galllinarum strains probably evolved from the same ancestor of serovar Enteritidis (antigenic formula 1,9,12:g:m). The main genomic alterations include deletions and mutations, including ones related to the absence of flagellin gene expression [70,71].
S. Gallinarum bv. Pullorum causes PD, a systemic infection in young birds, frequently related to transovarial/vertical transmission. Birds with PD present bacillary white diarrhea that can progress to septicemia and death. The macroscopic lesions can include hepatitis and splenitis with white necrotic foci and purulent airsacculitis. This disease presents a more clinically concerning outcome in two- to three-week-old chickens due to the high mortality rate. The surviving birds can become asymptomatic carriers and transmit the bacteria to other chickens [72]. S. Gallinarum bv. Gallinarum causes FT, an acute septicemic or chronic disease that occurs most often in adult birds through horizontal transmission. Acute septicemic and/or chronic FT is responsible for 40% of death in Brazilian poultry flocks [73,74]. FT also induces significant economic losses by reducing fertility and egg production [8].
The first S. Gallinarum isolate was probably from the biovar Pullorum and its emergence is estimated to have occurred around 914 DC, after dispersing worldwide [75]. The initial PD outbreaks were reported in the 19th century, while the bacterium was firstly characterized in the 1900s (Figure 2). It is noteworthy that the specific evolution of this biovar resulted in the emergence of novel virulent strains with a unique ability to induce arthritis in chickens, expanding its pathogenic profile [76]. Aiming to establish sanitary measures and to achieve the control/elimination of S. Gallinarum bv. Pullorum, official programs in the Western countries, highlighting the United States (with the NPIP, National Poultry Improvement Plan), were implemented at the beginning of the 20th century [77]. However, this biovar became endemic in the most important poultry-producing countries worldwide and it was associated with significant economic losses in poultry farms in the 20th century [78]. PD outbreaks were reported in North America, as well as in other continents [72,79,80]. In Brazil, PD was first diagnosed in 1928 with several official notifications over time, highlighting 15 outbreaks from 2009 to 2014 [81].
S. Gallinarum bv. Gallinarum appears to have a more recent origin, but definitive evolutionary analyses have not yet been performed. FT was rarely diagnosed in the 20th century in the United States due to the NPIP implemented to control S. Gallinarum bv. Pullorum. The last reported FT outbreak was in 1981, so this biovar is considered eradicated in the US [82]. It has also been rare to detect FT in Europe, with only two reports in Denmark and Germany due to breeder importation [83]. However, outbreaks have been frequently reported in Asia and South America, mainly in backyard birds [84]. In Brazil, reports have described the occurrence of FT in commercial poultry farms for a long time [22,84]. The first introduction is estimated to have occurred in the mid-19th century, followed by another in the mid-20th century [85]. This biovar also seems to be highly dispersed in Brazilian poultry farms and a total of 94 outbreaks were officially reported from 2005 to 2012 (51 in 2005, 5 in 2006, 24 in 2007, 2 in 2008, 1 in 2009, and 14 in 2012) [81]. Data about outbreaks occurring after 2012 are scarcer, but FT has been frequently reported by poultry farmers [86]. In addition to the horizontal transmission of field strains that are already hosted in poultry farms, industrial incubation of contaminated eggs has also been hypothesized as a key factor for the spreading of this biovar in the whole country [87].
Biosecurity programs, including the elimination of S. Gallinarum positive poultry flocks and massive vaccination, have been the main effective tools used to eradicate FT and PD. The official Brazilian poultry health control plan (PNSA, Programa Nacional de Sanidade Avícola) includes rules to safeguard flocks for this serovar. Poultry flocks with suspicion of FT or PD must be screened for S. Gallinarum by traditional methods (bacteriology culture, biochemical tests, serology), as well as molecular biology technologies, such as polymerase chain reaction (PCR). Positive flocks have to be immediately slaughtered, and the environment must be disinfected [9].
S. Gallinarum strains also present several different genomic profiles, including lineages with several prophages, plasmids, and gene clusters coding for different AMR mechanisms [75,88]. It has also been reported that S. Gallinarum has accumulated many pseudogenes and virulence genes associated with specificity to chicken hosts [89]. MLST analysis has demonstrated the occurrence of at least four STs of S. Gallinarum bv. Gallinarum (ST78, ST331, ST470, ST762) and two of S. Gallinarum bv. Pullorum (ST92 and ST747) [32] (Table 1).
AMR has not been frequently reported in S. Gallinarum. However, the resistance to nalidixic acid, gentamicin, ciprofloxacin, kanamycin, streptomycin, enrofloxacin, and ampicillin has already been reported in different countries [90,91,92]. In addition, genes coding for AMR, as well as specific mutations in the gyrA gene (associated with fluoroquinolone resistance) have been observed in the bacterial genomes of S. Gallinarum isolates [90,91]. In Brazil, S. Gallinarum appears to present AMR to nalidixic acid, ciprofloxacin, enrofloxacin, and tetracycline, but not for beta-lactams [93] (Table 1). S. Gallinarum bv. Gallinarum isolates have additionally demonstrated resistance to azithromycin and quinolone/fluoroquinolone [93,94]. Higher resistance in the more recent Salmonella isolates than in samples obtained in the past was also observed and associated with the increased use of antimicrobials in the poultry farms [93]. Noteworthy, the association of some resistant S. Gallinarum isolates with the historical indiscriminate use of antibiotics has already been demonstrated [75,92]. Antimicrobial therapy is also described to treat PD and FT, especially in small-scale commercial layer flocks [95].
4.2. Salmonella Typhimurium
Salmonellaenterica serovar Typhimurium (S. Typhimurium) infects a wide range of hosts and it is the most frequent serovar isolated from intensive-producing animals and foods worldwide. In addition to having several lineages, this serovar (antigenic formulae 1,4,[5],12:i:1,2) has also monophasic and aphasic flagellar variants with slightly different antigenic properties (1,4,[5],12:i:-/1,4,[5],12:-:1,2/1,4,[5],12:-:-), but very similar genetic and metabolically activities. These few antigenic differences have resulted in several problems in epidemiological investigations, making tracking these variants a real nightmare [96]. Importantly, S. Typhimurium is frequently motile due to its peritrichous flagella, which include the flagellin proteins FliC (phase 1 antigen) and FljB (phase 2 antigen) [97]. Most isolates are biphasic, expressing both flagellins in different physiological conditions. However, monophasic variants of phase 1 or phase 2 are frequently detected, since lacking the capacity of flagellar antigen production is a bacterial strategy to evade the immune system of the host animals [96]. Furthermore, this serovar also has a very long evolutionary history associated with human and animal infections, resulting in a high diversity of pathogenicity, virulence, AMR profiles, and host adaptation [98].
S. Typhimurium was the first non-typhoid concerning serovar isolated in humans infected by contaminated foods. In the mid-1950s, it was isolated from duck eggs consumed by patients presenting severe enteric infection in Europe [99]. It was also the most frequent serovar associated with animal and human outbreaks in the United Kingdom from 1941 to 1970, with a steady increase to a maximum of 85% of salmonellosis cases in 1954 [100]. Additionally, it was detected in cattle, poultry, swine, and sheep samples (Figure 1). In livestock, this serovar was the most frequent in turkeys and also presented in cattle [101,102]. It spread to other livestock before its global dissemination in the 1990s [102]. S. Typhimurium and its variants have continued to disseminate in animals and in humans being the top concerning serovar in most regions of the World [98,103,104,105].
Previous epidemiological investigations traced this serovar with phage typing and S. Typhimurium DT104 was one of the most disseminated worldwide. This type was associated with cattle hosts, but also detected in swine, birds, and wild animals [106]. In the 1990s, there was a global epidemic of DT104 in humans, being the most common DT in Europe, Asia, and America [102,106]. Human outbreaks by this phage type were caused by direct animal transmission, imported food, travel abroad, and environmental reservoirs [107]. Recent WGS analyses have provided more evidence that it probably emerged from an antimicrobial susceptible ancestor in ∼1948 and became MDR in ∼1972 through horizontal transfer of the 13 kb Salmonella genomic island 1 (SGI-1) [108]. Despite the epidemiological importance of S. Typhimurium DT104 for public health, other DTs (for example, DT193 and U288) have also been largely disseminated in livestock, foods, and, consequently, they infected humans too [109,110]. In addition, monophasic S. Typhimurium 1,4,[5],12:i:- has also been a matter of concern worldwide, including in Brazil [111,112]. In addition to being responsible for human salmonellosis outbreaks in America and Europe, it was also isolated from different animals and foods [113,114,115,116].
In poultry production, S. Typhimurium and its variants have already been detected in layers and broilers [117]. The contamination by this serovar can occur at multiple steps along the food chain, including production, processing, distribution, retail marketing, handling, and final preparation [118]. In Brazil, it is largely reported that S. Typhimurium was a very common isolate in non-human sources (including broilers, layers, and poultry-derived foods) before the 1990s [119]. With the wide spread of S. Enteritidis at the end of the 1980s and the beginning of the 1990s, S. Typhimurium isolations declined in non-human sources in Brazil for some years [64,120] (Figure 2). However, it was still classified as the most common serovar isolated from humans [121]. S. Typhimurium monophasic variants (mainly 1,4,5,12:i:-) were also frequently detected in human, animal, and food samples in this same period [111,112,116]. More recent studies have demonstrated the detection of this serovar in livestock and animal-derived foods, including poultry meat and eggs [122,123].
The implementation of biosecurity procedures in farms and industries to prevent food contamination has contributed to the reduction in the incidence of salmonellosis [56,124,125]. Additionally, the use of vaccines for S. Typhimurium in poultry flocks and swine herds has helped to prevent the dissemination of this pathogen [126,127,128].
S. Typhimurium genetic, antigenic, and metabolic diversity has been evidenced by the large amount of WGS data generated in recent years. Several lineages were characterized, some of them highly adapted to specific hosts. Now it is well-known that S. Typhimurium is a complex group of slightly different sequence types, such as ST19, ST313, ST213, ST128, and many others [32,49,129]. Some of the main lineages seem to be more adapted to specific hosts [98] (Table 1).
AMR has been reported in several S. Typhimurium isolates. Since the 1990s, MDR S. Typhimurium isolates have been frequently observed [102]. AMR seems to have benefitted S. Typhimurium DT104 to spread more quickly [108]. MDR isolates have also been associated with a higher risk of invasive infection, longer illness, increased frequency and duration of hospitalization, and a higher risk of death than antimicrobial susceptible strains [130,131]. In Brazil, MDR S. Typhimurium frequency has also increased over time [19]. Furthermore, MDR S. Typhimurium-specific lineages of swine-origin have been reported in farms and foods [132,133]. Most S. Typhimurium isolates obtained from poultry and chickens show resistance to several antimicrobials, including colistin [49,134,135,136]. The resistance genes detected in these isolates are presented in Table 1.
4.3. Salmonella Enteritidis
Salmonellaenterica serovar Enteritidis (S. Enteritidis) is highly adapted to avian species without causing severe clinical signs in most poultry-producing flocks. However, this non-typhoid serovar can occur in high concentrations in chicken meat and eggs, and can cause human foodborne outbreaks [137]. S. Enteritidis presents a specific surface structure with the expression of the two flagellar phases (antigenic formulae 1,9,12:g:m), so isolates are generally motile. Monophasic and even aphasic flagellar variants can be also rarely detected (1,9,12:g:-/1,9,12:-:m/1,9,12:-:-). Salmonella surveillance data showed that the occurrence of S. Enteritidis in foods increased worldwide from the 1970s to the 1980s [138]. Furthermore, epidemiological investigations have demonstrated that this global increase was related to the consumption of eggs and poultry meat [139].
S. Enteritidis emerged as the primary cause of foodborne outbreaks in the World in the mid-1980s [65]. The increased frequency of salmonellosis by this serovar was associated with the consumption of avian-source foods, such as eggs and undercooked chicken meat [56,138,139,140]. It was so hypothesized that S. Enteritidis filled the ecological niche vacated by the eradication of S. Gallinarum (biovars Pullorum and Gallinarum) from domestic fowl in many poultry-producing countries. Importantly, these two serovars share a common immuno-dominant somatic antigen. Enteric colonization, as well as flock immunity generated by the infection with the two biovars of S. Gallinarum, probably prevented an earlier emergence of S. Enteritidis in poultry flocks worldwide [65,141].
S. Enteritidis was initially traced by phage-typing and the different outbreaks were caused by PT4, PT8, and PT13a. In the United States, PT8 and PT13a were the most common in the northeast, south, and mid-west, while PT4 was predominant in the western states [142]. In Europe, PT4 was the most frequent one in S. Enteritidis isolated from chicken carcasses [140,143]. Therefore, most studies concluded that PT4 was more frequent in Europe, while PT8 and PT13 were prevalent in the United States [144]. In South America, the predominance of PT4 was demonstrated, followed by the less known PT7 and PT9 [145,146]. Overall, 150 S. Enteritidis foodborne disease outbreaks were reported only in Argentina between 1986 and 1993 [147]. In Brazil, human salmonellosis cases by this serovar were detected in all geographic regions in the late 1980s [64]. A recent study with WGS data reinforced the high increase in S. Enteritidis bacterial population in the second half of the 1980s and beginning of the 1990s, associated with the high frequency of this pathogen in the poultry production chain in this country [148].
Diagnostic tools, as well as biosecurity and control measures, were implemented to avoid the high spreading of S. Enteritidis. Monitoring and sanitization plans were introduced in all stages of poultry production, including breeding flocks, hatcheries, broiler flocks, and slaughter establishments [149]. In addition, layers and breeders’ flocks have been vaccinated, contributing to a significant reduction in S. Enteritidis contamination in the table egg industry and broiler processing plants [149,150,151,152]. In Brazil, preventive measures established by the PNSA were adopted in poultry farms, slaughterhouses, and eggs industries to reduce the prevalence of Salmonella spp. and to establish an adequate level of consumer protection [9]. Additionally, egg quality assurance programs were also implemented to avoid food contamination. All these procedures reduced the occurrence of S. Enteritidis in commercial farms and poultry-derived foods, improving public health [150,152].
S. Enteritidis strains present several different genomic profiles, including many lineages with some specific genetic and metabolic characteristics. MLST analysis has demonstrated the occurrence of more than 15 STs, including ST11 (the most frequent and largely disseminated), ST183, ST136, ST310, ST814, and others [32,148] (Table 1).
This serovar has also been characterized by low resistance to antimicrobials in Brazil. However, some S. Enteritidis isolates from foodborne outbreaks and hospitalized patients showed an MDR profile [122,131]. S. Enteritidis isolates from poultry-derived products and foods showed AMR mainly to sulfonamide, trimethoprim-sulfamethoxazole, nalidixic acid, streptomycin, gentamicin, and tetracycline [153,154,155,156]. AMR genes are further detailed in Table 1.
4.4. Salmonella Heidelberg
Salmonellaenterica serovar Heidelberg (S. Heidelberg) was firstly isolated from humans in the city of Heidelberg, Germany, in 1933 [157] (Figure 2). Later it was demonstrated to infect livestock, mainly poultry. It is frequently characterized as a biphasic motile serovar (antigenic formulae 1,4,[5],12:r:1,2), but monophasic and aphasic flagellar variants (1,4,[5],12:r:-/1,4,[5],12:-:1,2/1,4,[5],12:-:-) have also been reported [11].
Epidemiologically, S. Heidelberg is a serovar often detected in human foodborne salmonellosis [158,159,160]. Human infections are more frequently reported in hospitalized patients from North America than in other regions of the World [159,160]. It is estimated that more serious disease occurs in approximately 13% of human infections by this serovar [161]. However, the septicemic disease is rare, only occurring in some immunocompromised patients, the elderly, and young children [162,163].
Foodborne outbreaks are usually linked to the consumption of poultry-derived food [164]. S. Heidelberg has been frequently detected in chicken, eggs, and ground turkey [165,166,167,168,169]. However, it was also identified in other livestock and foods [170,171,172] (Figure 1). The outbreaks by this serovar have been reported worldwide [173,174,175,176,177,178,179,180].
In Brazil, this serovar was firstly detected in foods in 1962 [181]. It was also observed in different sources and regions of Brazil in the following four decades [181]. More recently, it has been increasingly detected in Brazilian broiler farms and, consequently, in chicken carcasses, generating economic losses for the poultry producers [182]. S. Heidelberg has also been very frequently isolated in poultry slaughterhouses. RASFF (Rapid Alert System for Food and Feed) reported that 50% of the Salmonella positive cases in chicken meat were from the serovar Heidelberg between 2013 and 2017 [183].
S. Heidelberg has a high ability to adapt to poultry farm environments [182,184]. Bacterial cells remain viable in the poultry litter for long periods, resisting a wide range of temperatures and pHs, as well as producing biofilms [184,185,186]. It has been a real challenge to remove this serovar from poultry farms and slaughterhouses in Brazil [182].
Recent epidemiological studies have used MLST to identify S. Heidelberg strains and four main STs have been reported: ST15, ST2071, ST7556, and ST3377 [29,34,44,49,177,187,188,189,190,191]. ST15 is the most frequent and it could be further divided into different lineages according to phylogenomic analyses [44]. This is also the most frequent ST detected in the Brazilian poultry production chains [29,34,44,49] (Table 1).
This serovar has also shown AMR to several antibiotics, raising concern among veterinary and public health authorities [69,191,192]. Most S. Heidelberg strains are resistant to tetracycline, nalidixic acid, and ampicillin, as well as some other antimicrobials [49,69,135,193,194,195,196,197,198] (Table 1). Furthermore, an MDR profile has been common in most isolates [49,135,194,195,197]. In a temporal comparative analysis, it was demonstrated that 46.1% of S. Heidelberg isolates were resistant to only one class of antimicrobials in 2005, while 100% of them were MDR in 2009 [199]. Brazilian S. Heidelberg isolates have demonstrated a high frequency of resistance (>50%) to streptomycin, nalidixic acid, tetracycline, cefotaxime, ampicillin, amoxicillin, cefoxitin, amoxicillin-clavulanate, and ceftiofur [197]. This high AMR has further been detected in raw chicken meat exported to Portugal and the Netherlands [29,34]. The most frequent AMR genes detected in S. Heidelberg are presented in Table 1.
4.5. Salmonella Minnesota
Salmonellaenterica serovar Minnesota (S. Minnesota) is a motile serovar with the expression of the two-phase flagellar antigens (antigenic formulae 21:b:e,n,x) frequently detected in the natural environment. So, it has been detected in different sources, such as natural water samples, plants, livestock farms, and foods [200,201] (Figure 1). It can also infect humans and animals, but reports of outbreaks of this serovar are not so common [202,203].
Historical data demonstrate an association of this serovar with a few small outbreaks [202,203]. More recently, it was associated with other foodborne salmonellosis cases [204,205]. In addition to the usual gastroenteritis, typhoid-like illnesses can rarely occur [206].
S. Minnesota was first isolated in a turkey from a poultry farm located in the state of Minnesota, United States, in 1936 [207] (Figure 2). In Europe, it was first isolated from spray-dried eggs in England in 1947 [208]. It was also the most prevalent serovar in broilers from Belgium poultry farms [209]. S. Minnesota has raised concerns after the increase in the number of isolations in the poultry production chain in Brazil [50,69]. This serovar was the tenth most identified in avian salmonellosis from 2004 to 2008, but it became the second one in 2010 in Brazil. More recently, it was the most serovar isolated from drag swabs in broiler farms [69]. There are also additional reports of S. Minnesota in samples of Brazilian chicken meat [210].
The ability to produce biofilms has also been evaluated in isolates of S. Minnesota from poultry sources. Most isolates showed invA, lpfA, and agfA genes. In addition, genes linked to apoptosis induction (avrA), oxidative stress (sodC), and quorum sensing (luxS) were also identified in samples of broiler slaughtering plants in Brazil, demonstrating adapting to adverse conditions. Furthermore, it has been demonstrated that S. Minnesota is a moderate-intensity biofilm producer [211].
Five main STs have been assigned to S. Minnesota: ST548, ST7557, ST7558, ST285, and ST3088 [34,47,49,50]. ST548 is the most frequent one, but ST3088 has also been detected in the poultry production chains [40,49,50] (Table 1). It has also been demonstrated that S. Minnesota strains currently circulating in Brazilian poultry can be divided into two lineages: SM-PLI and SM-PLII [50].
S. Minnesota strains more recently isolated have shown AMR to several antibiotics, including amoxicillin, ampicillin, cefazoline, cefoxitin, ceftazidime, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, clavulanic acid, gentamicin, nalidixic acid, neomycin, penicillin, streptomycin, sulfamethoxazole, sulfonamide, tetracycline, and trimethoprim [40,49,212,213,214,215,216] (Table 1). Furthermore, most isolates have shown an MDR profile [40,49,213,214,215]. Bacterial resistance genes have also been reported (Table 1).

Table 1.
Phenotypic resistance, genotypic resistance, and ST already reported of the main Salmonella serovars from the Brazilian poultry-production chain.
Table 1.
Phenotypic resistance, genotypic resistance, and ST already reported of the main Salmonella serovars from the Brazilian poultry-production chain.
| Serovar and Variants | Phenotypic Resistance | Genotypic Resistance | STs | References |
|---|---|---|---|---|
| Gallinarum (1,9,12:-:-) | Ampicillin, azithromycin, ciprofloxacin, enrofloxacin, fluoroquinolone, gentamicin, kanamycin, nalidixic acid, streptomycin, and tetracycline. | gyrA, aadA and aadB | 78, 92, 331, 470, 762, 747 | [32,90,91,92,93,94,95] |
| Typhimurium (1,4,[5],12:i:1,2/ 1,4,[5],12:i:-/ 1,4,[5],12:-:1,2/ 1,4,[5],12:-:-) | Aminoglycoside, ampicillin, aztreonam, cefepime, ceftriaxone, chloramphenicol, ciprofloxacin, colistin, doxycycline, fluoroquinolone, gentamicin, nalidixic acid, streptomycin, sulfamethoxazole, sulfonamide, tetracycline, and trimethoprim. | aac(3)-lla, aac(3)-lld, aadA1, aadA2, aph(6)-ld, blaCTX-M-2, blaTEM-1B, dfrA1, floR, mrc-1, strA, strB, sul1, sul2, tet(A), and tet(B) | 19, 128, 213, 313 | [32,49,130,136,137] |
| Enteritidis (1,9,12:g:-/ 1,9,12:-:m/ 1,9,12:-:-) | Gentamicin, nalidixic acid, streptomycin, sulfonamide, tetracycline, and trimethoprim-sulfamethoxazole. | aac(3)-Iva, aac(6′)-Iaa, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, mdf(A), tet(34), tet(A) | 11, 183, 136, 310, 814 | [32,149,154,155,156,157] |
| Heidelberg (1,4,[5],12:r:1,2/ 1,4,[5],12:r:-/ 1,4,[5],12:-:1,2/ 1,4,[5],12:-:-) | Amoxicillin, ampicillin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, clavulanic acid, colistin, doxycycline, florfenicol, gentamicin, meropenem, nalidixic acid, pefloxacin, penicillin, quinolone, streptomycin, sulfamethoxazole, sulfonamide, tetracycline, tobramycin, and trimethoprim. | aac(3)-Via, aadA1, aadA8, aph(3′)-Ia, blaCMY-2, blaCTX-M, blaCTX-M-2, blaCTX-M-8, blaTEM-1B, cmlA1, dfrA15, fosA7, mdf(A), mphB, qnrB1, strA, strB, sul1, sul2, sul3, tet(34), tet(A) | 15, 2071, 3377, 7556 | [29,34,44,49,69,136,178,185,190,191,193,195,196,197,198,199] |
| Minnesota (21:b:e,n,x) | Amoxicillin, ampicillin, cefazoline, cefoxitin, ceftazidime, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, clavulanic acid, gentamicin, nalidixic acid, neomycin, penicillin, streptomycin, sulfamethoxazole, sulfonamide, tetracycline, and trimethoprim. | aadA1, ant(3″)-Ia, aph(3′)-Ia, aphA1, blaCMY-2, blaCTX-M, blaCTX-M-8, blaTEM, mdf(A), qnrB19, qnrB5, sul2, tet(A) | 285, 548, 3088, 7557, 7558 | [34,47,49,50,213,214,215,217] |
4.6. Other Salmonella Serovars
Several more serovars were already detected in the poultry production chains in Brazil in recent decades. Serovar Infantis appears to be the most frequent one, followed by Schwarzengrund, Senftenberg, Mbandaka, Hadar, and Newport [49,69,121,124,213,216].
Salmonellaenterica serovar Infantis (antigenic formulae 6,7,14:r:1,5) has been reported in humans, poultry farm environments and foods [69,124,218,219]. It is one of the 15 most isolated serovars from human sources [217,218,219]. In the Brazilian poultry chain, S. Infantis has been isolated at a low frequency [27,49,69,216,220]. The bacterial isolates showed AMR to different antibiotics, such as sulfonamide, tetracycline, and amoxicillin [219].
Salmonellaenterica serovar Schwarzengrund (antigenic formulae 1,4,12,27:d:1,7) was already isolated from chicken carcasses, broiler farms, frozen chicken cuts, chicken, poultry slaughterhouses, and feed factories in different Brazilian states [69,213,216,221]. The isolates presented AMR to sulfonamide, tetracycline, and amoxicillin [213], as well as a frequent MDR profile [49,216].
Salmonellaenterica serovar Senftenberg (antigenic formulae 1,3,19:g,[s],t:-) has also been detected in chicken carcasses, broiler farms, poultry environments, slaughterhouses, and food in Brazil [69,121,213,216,221,222]. Isolates were resistant to cefoxitin, ciprofloxacin, enrofloxacin, nalidixic acid, and trimethoprim-sulfamethoxazole [216,222].
Salmonellaenterica serovar Mbandaka (antigenic formulae 6,7,14:z10:e,n,z15) was detected in Brazilian broiler farms, poultry slaughterhouses, feed factories, and chicken carcasses [69,213,216,221]. The isolates showed resistance to sulfonamide, norfloxacin, and amoxicillin [213].
Salmonellaenterica serovar Hadar (antigenic formulae 6,8:z10:e,n,x) has been isolated from foodstuff, broiler chicken, poultry slaughterhouses, and chicken carcasses [213,216,223]. AMR was observed in amoxicillin, chloramphenicol, nalidixic acid, nitrofurantoin, tetracycline, streptomycin, sulfazotrim, and sulfonamide [213,223].
Salmonellaenterica serovar Newport (antigenic formulae 6,8,20:e,h:1,2) was already isolated from turkeys, broiler chicken, and poultry slaughterhouses [124,213,216]. The isolates showed AMR to different antimicrobials and MDR profile [213,216].
Salmonella from several more serovars (Abatetuba, Abony, Carrau, Grumpensis, Idikan, Isangi, Orion, Ouakam Rochdale, Saphra, etc.) have also been rarely isolated from food (chicken), food-producing animals (broiler), and environmental samples (slaughterhouse) collected in a Brazil [224]. In general, these serovars also presented AMR genes encoding resistance to quinolones, third-generation cephalosporin, tetracycline, aminoglycoside, sulfonamide, and fosfomycin, respectively [224].
5. Prevention and Control
In Brazil, official standards to prevent and control Salmonella are ruled by the National Poultry Health Program of the Ministry of Agriculture, Livestock, and Supply (MAPA, Ministério da Agricultura, Pecuária e Abastecimento) [9]. Feces and drag or boot swabs from the flocks are routinely collected in poultry farms and submitted to laboratory analysis with different methods (bacteriology culture, biochemical tests, serology, molecular biology assays, etc.) to detect and identify Salmonella serovars. In slaughterhouses and egg industries, self-control programs must be carried out to monitor the contamination by Salmonella spp. from the acquisition of the feedstock to the final food products [9].
Some antimicrobials have also been banned as additives and antibiotic-free (ABF) strategies have been implemented in the poultry production chains [225,226,227,228]. There are several alternatives to be used as growth promoters, such as medicinal plants, probiotics, prebiotics, and organic acids [225]. Formic acid, an extensively studied organic acid, has been reported to limit infection with Salmonella and other foodborne pathogens when used in the poultry diet [229]. In addition, it has also been recommended that rigorous management on the farms to avoid Salmonella infection, including quality control of the water consumed, biosecurity efforts, and the overall organization of the flocks [225].
ABF strategies can include feeding-based and non-feeding-based strategies to control Salmonella infection in poultry flocks. The first includes prebiotics, probiotics, synbiotics, postbiotics, and phytobiotics; while the second focuses on the use of bacteriophages, in ovo applications, and vaccines [228]. Prebiotics are usually administered to induce a modulating effect on the gut microbiota, increasing the growth of resident beneficial bacteria [230,231]. Probiotic bacteria (alone or in combination) have been used to control Salmonella infections during poultry production [232,233], improving production performance [234]. Strategies based on symbiotics may trigger some mechanisms involved in the inhibition or reduction in clinical signs caused by Salmonella [228]. Postbiotics involve the use of non-viable bacteria to provide benefits to poultry health [235]. Phytobiotics have also been shown to contribute to improving poultry performance, increasing nutrient uptake and carcass quality [236]. Bacteriophages have already been used against S. Enteritidis, S. Hadar, and S. Typhimurium, presenting interesting results [237]. Vaccination is part of the biosecurity protocol on farms to prevent the spread of diseases, such as Salmonella Gallinarum, Enteritidis, and Typhimurium serovars [238,239,240]. The administration of probiotics, prebiotics, and vaccines in ovo were shown to be effective to control Salmonella [228]. New technologies, including all the methods to study the omics (genomics, metagenomics, transcriptomics, proteomics, metabolomics) are useful tools for a better understanding of Salmonella metabolic arsenal [228].
Other studies have also compared ABF with conventional production, including animal welfare analysis [226,241,242,243]. As expected, the rate of Salmonella with MDR isolated from flocks of laying hens fed excluding antibiotics was significantly lower than that of chickens fed with them in conventional diets [242]. In addition, the prevalence of Salmonella with MDR in retail chicken meat was lower in packages categorized as “low or no antibiotic use” [243].
6. Conclusions
The dissemination of many Salmonella serovars has caused a huge impact on livestock production chains worldwide for decades. The five main poultry-associated serovars detected in Brazil over time (Gallinarum, Typhimurium, Enteritidis, Heidelberg, and Minnesota) demonstrated to be adapted to chicken hosts with different pathogenicity and antigenic properties. They have also presented different AMR profiles. Salmonella serovars presented specific dissemination waves in some restricted geographic regions or worldwide in the past, requiring plans to control the epidemics as well as to avoid economic losses on the farms and/or the occurrence of concerning foodborne outbreaks. In Brazilian poultry production, programs were also paramount for controlling the different Salmonella serovars epidemics.
Furthermore, current intensive poultry production in Brazil presents a perfect scenario for the emergence of novel concerning Salmonella serovars. Continuous epidemiological surveillance is necessary to track all serovars and lineages, mainly those presenting AMR. It is also necessary to eliminate the use of antimicrobials in poultry production to reduce the emergence and dissemination of Salmonella lineages with AMR.
Author Contributions
D.K., A.K.M., S.D.C., A.M.C., and V.R.L. reviewed the literature and wrote the original draft preparation. D.K., A.K.M., S.D.C., A.M.C., A.F.S., A.S.K.F., N.I., and V.R.L. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)-Finance Code 001. This study was also financed by Simbios Biotecnologia. V.R.L. was financially supported by the National Council for Scientific and Technological Development from Brazil (Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq]; process number 308445/2020-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank the University of Caxias do Sul (UCS) and the Lutheran University of Brazil (ULBRA) for support in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- OECD (Organisation for Economic Co-Operation and Development). Meat Consumption. 2022. Available online: https://data.oecd.org/agroutput/meat-consumption.htm (accessed on 13 January 2022).
- Réhault-Godbert, S.; Guyot, N.; Nys, Y. The golden egg: Nutritional value, bioactivities, and emerging benefits for human health. Nutrients 2019, 11, 684. [Google Scholar] [CrossRef] [PubMed]
- ABPA (Associação Brasileira de Proteína Animal). Relatório Anual. 2021. Available online: http://abpa-br.org/wp-content/uploads/2021/04/ABPA_Relatorio_Anual_2021_web.pdf (accessed on 15 January 2022).
- Gomes, B.C.; Franco, B.D.; De Martinis, E.C. Microbiological food safety issues in Brazil: Bacterial pathogens. Foodborne Pathog. Dis. 2013, 10, 197–205. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on major foodborne zoonotic bacterial pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef] [PubMed]
- Arnaut-Rollier, I.; Vauterin, L.; De Vos, P.; Massart, D.L.; Devriese, L.A.; De Zutter, L.; Van Hoof, J. A numerical taxonomic study of the Pseudomonas flora isolated from poultry meat. J. Appl. Microbiol. 1999, 87, 15–28. [Google Scholar] [CrossRef]
- Säde, E.; Murros, A.; Björkroth, J. Predominant enterobacteria on modified-atmosphere packaged meat and poultry. Food Microbiol. 2013, 34, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, H.L. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 2000, 19, 405–424. [Google Scholar] [CrossRef] [PubMed]
- MAPA (Ministério da Agricultura, Pecuária e Abastecimento). Instrução Normativa No 78, de 3 de Novembro de 2003. 2003. Available online: https://www.gov.br/agricultura/pt-br/assuntos/sanidade-animal-e-vegetal/saude-animal/programas-de-saude-animal/pnsa/2003_78.INconsolidada.pdf (accessed on 20 January 2021).
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.X. Supplement 2008–2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F. Antigenic Formulae of the Salmonella Serovars, 9th ed.; Institut Pasteur, WHO Collaborating Center for Reference and Research on Salmonella: Paris, France, 2007. [Google Scholar]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef]
- Uzzau, S.; Brown, D.J.; Wallis, T.; Rubino, S.; Leori, G.; Bernard, S.; Casadesús, J.; Platt, D.J.; Olsen, J.E. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 2000, 125, 229–255. [Google Scholar] [CrossRef] [PubMed]
- Crook, P.D.; Aguilera, J.F.; Threlfall, E.J.; O’Brien, S.J.; Sigmundsdóttir, G.; Wilson, D.; Fisher, I.S.; Ammon, A.; Briem, H.; Cowden, J.M.; et al. A European outbreak of Salmonella enterica serotype Typhimurium definitive phage type 204b in 2000. Clin. Microbiol. Infect. 2003, 9, 839–845. [Google Scholar] [CrossRef]
- Barco, L.; Barrucci, F.; Olsen, J.E.; Ricci, A. Salmonella source attribution based on microbial subtyping. Int. J. Food Microbiol. 2013, 163, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Reeves, P.R.; Octavia, S. Population structure, origins and evolution of major Salmonella enterica clones. Infect. Genet. Evol. 2009, 9, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Mughini-Gras, L.; Franz, E.; van Pelt, W. New paradigms for Salmonella source attribution based on microbial subtyping. Food Microbiol. 2018, 71, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Glynn, M.K.; Bopp, C.; Dewitt, W.; Dabney, P.; Mokhtar, M.; Angulo, F.J. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 1998, 338, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Ghilardi, A.C.; Tavechio, A.T.; Fernandes, S.A. Antimicrobial susceptibility, phage types, and pulse types of Salmonella Typhimurium, in São Paulo, Brazil. Memórias Inst. Oswaldo Cruz 2006, 101, 281–286. [Google Scholar] [CrossRef]
- Dos Santos, L.R.; do Nascimento, V.P.; de Oliveira, S.D.; Rodrigues, D.P.; dos Reis, E.M.; Seki, L.M.; Ribeiro, A.R.; Fernandes, S.A. Phage types of Salmonella enteritidis isolated from clinical and food samples, and from broiler carcasses in southern Brazil. Rev. Inst. Med. Trop. São Paulo 2003, 45, 1–4. [Google Scholar] [CrossRef]
- Nunes, I.A.; Helmuth, R.; Schroeter, A.; Mead, G.C.; Santos, M.A.; Solari, C.A.; Silva, O.R.; Ferreira, A.J. Phage typing of Salmonella enteritidis from different sources in Brazil. J. Food Prot. 2003, 66, 324–327. [Google Scholar] [CrossRef]
- Gast, R.K. Bacterial diseases: Salmonella infection. In Diseases of Poultry, 12th ed.; Saif, Y.M., Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Swayne, D.E., Eds.; Blackwell Publishing: Oxford, UK, 2008; pp. 619–636. [Google Scholar]
- Bergamini, F.; Iori, A.; Massi, P.; Pongolini, S. Multilocus variable-number of tandem-repeats analysis of Salmonella enterica serotype Gallinarum and comparison with pulsed-field gel electrophoresis genotyping. Vet. Microbiol. 2011, 149, 430–436. [Google Scholar] [CrossRef]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Zhou, Z.; Sergeant, M.J.; Achtman, M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018, 14, e1007261. [Google Scholar] [CrossRef]
- Kipper, D.; Hellfeldt, R.M.; De Carli, S.; Lehmann, F.K.M.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Salmonella serotype assignment by sequencing analysis of intergenic regions of ribosomal RNA operons. Poult. Sci. 2019, 98, 5989–5998. [Google Scholar] [CrossRef]
- Zhang, S.; den Bakker, H.C.; Li, S.; Chen, J.; Dinsmore, B.A.; Lane, C.; Lauer, A.C.; Fields, P.I.; Deng, X. SeqSero2: Rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl. Environ. Microbiol. 2019, 85, e01746-19. [Google Scholar] [CrossRef]
- Pijnacker, R.; Dallman, T.J.; Tijsma, A.S.L.; Hawkins, G.; Larkin, L.; Kotila, S.M.; Amore, G.; Amato, E.; Suzuki, P.M.; Denayer, S.; et al. An international outbreak of Salmonella enterica serotype Enteritidis linked to eggs from Poland: A microbiological and epidemiological study. Lancet Infect. Dis. 2019, 19, 778–786. [Google Scholar] [CrossRef]
- Van den Berg, R.R.; Dissel, S.; Rapallini, M.L.B.A.; van der Weijden, C.C.; Wit, B.; Heymans, R. Characterization and whole genome sequencing of closely related multidrug-resistant Salmonella enterica serovar Heidelberg isolates from imported poultry meat in the Netherlands. PLoS ONE 2019, 14, e0219795. [Google Scholar] [CrossRef]
- Singh, A.; Goering, R.V.; Simjee, S.; Foley, S.L.; Zervos, M.J. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 2006, 19, 512–530. [Google Scholar] [CrossRef]
- Li, W.; Raoult, D.; Fournier, P.E. Bacterial strain typing in the genomic era. FEMS Microbiol. Rev. 2009, 33, 892–916. [Google Scholar] [CrossRef]
- Achtman, M.; Wain, J.; Weill, F.X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Enterica MLST Study Group. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef]
- Chattaway, M.A.; Langridge, G.C.; Wain, J. Salmonella nomenclature in the genomic era: A time for change. Sci. Rep. 2021, 11, 7494. [Google Scholar] [CrossRef]
- Silveira, L.; Nunes, A.; Pista, Â.; Isidro, J.; Belo Correia, C.; Saraiva, M.; Batista, R.; Castanheira, I.; Machado, J.; Gomes, J.P. Characterization of multidrug-resistant isolates of Salmonella enterica serovars Heidelberg and Minnesota from fresh poultry meat imported to Portugal. Microb. Drug Resist. 2021, 27, 87–98. [Google Scholar] [CrossRef]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, Clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef]
- Swartz, M.N. Human diseases caused by foodborne pathogens of animal origin. Clin. Infect. Dis. 2002, 34, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Bokma-Bakker, M.H.; Bondt, N.; Neijenhuis, F.; Mevius, D.J.; Ruiter, S.J.M. Antibiotic Use in Brazilian Broiler and Pig Production: An Indication and Forecast of Trends; Report 714; Wageningen UR Livestock Research: Lelystad, The Netherlands, 2014; pp. 1–25. [Google Scholar]
- Chattopadhyay, M.K. Use of antibiotics as feed additives: A burning question. Front. Microbiol. 2014, 5, 334. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.L.; Panzenhagen, P.; Ferrari, R.G.; Dos Santos, A.; Paschoalin, V.M.F.; Conte-Junior, C.A. Frequency of antimicrobial resistance genes in Salmonella from Brazil by in silico whole-genome sequencing analysis: An overview of the last four decades. Front. Microbiol. 2020, 11, 1864. [Google Scholar] [CrossRef]
- Dias de Oliveira, S.; Siqueira Flores, F.; dos Santos, L.R.; Brandelli, A. Antimicrobial resistance in Salmonella enteritidis strains isolated from broiler carcasses, food, human and poultry-related samples. Int. J. Food Microbiol. 2005, 97, 297–305. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Potter, L.; Vaz, C.S.; Pereira, D.I.; Sangioni, L.A.; Vargas, Á.C.; de Avila Botton, S. Antimicrobial resistance in nontyphoidal Salmonella isolated from human and poultry-related samples in Brazil: 20-Year meta-analysis. Foodborne Pathog. Dis. 2017, 14, 116–124. [Google Scholar] [CrossRef]
- OIE (World Organization for Animal Health). OIE Annual Report on Antimicrobial Agents Intended for Use in Animals. 2021. Available online: https://www.oie.int/app/uploads/2021/05/a-fifth-annual-report-amr.pdf (accessed on 24 January 2022).
- Saraiva, M.M.S.; Lim, K.; do Monte, D.F.M.; Givisiez, P.E.N.; Alves, L.B.R.; de Freitas Neto, O.C.; Kariuki, S.; Júnior, A.B.; de Oliveira, C.J.B.; Gebreyes, W.A. Antimicrobial resistance in the globalized food chain: A One Health perspective applied to the poultry industry. Braz. J. Microbiol. 2022, 53, 465–486. [Google Scholar] [CrossRef]
- Kipper, D.; Orsi, R.H.; Carroll, L.M.; Mascitti, A.K.; Streck, A.F.; Fonseca, A.S.K.; Ikuta, N.; Tondo, E.C.; Wiedmann, M.; Lunge, V.R. Recent evolution and genomic profile of Salmonella enterica serovar Heidelberg isolates from poultry flocks in Brazil. Appl. Environ. Microbiol. 2021, 87, e0103621. [Google Scholar] [CrossRef]
- Shah, D.H.; Paul, N.C.; Sischo, W.C.; Crespo, R.; Guard, J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 2017, 96, 687–702. [Google Scholar] [CrossRef]
- Andrysiak, A.K.; Olson, A.B.; Tracz, D.M.; Dore, K.; Irwin, R.; Ng, L.K.; Gilmour, M.W. Canadian Integrated Program for Antimicrobial Resistance Surveillance Collaborative. Genetic characterization of clinical and agri-food isolates of multi drug resistant Salmonella enterica serovar Heidelberg from Canada. BMC Microbiol. 2008, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Moura, Q.; Fernandes, M.R.; Cerdeira, L.; Ienne, S.; Souza, T.A.; Negrão, F.J.; Lincopan, N. Draft genome sequence of a multidrug-resistant CMY-2-producing Salmonella enterica subsp. enterica serovar Minnesota ST3088 isolated from chicken meat. J. Glob. Antimicrob. Resist. 2017, 8, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Monte, D.F.; Lincopan, N.; Berman, H.; Cerdeira, L.; Keelara, S.; Thakur, S.; Fedorka-Cray, P.J.; Landgraf, M. Genomic features of high-priority Salmonella enterica serovars circulating in the food production chain, Brazil, 2000–2016. Sci. Rep. 2019, 9, 11058. [Google Scholar] [CrossRef]
- Kipper, D.; Carroll, L.M.; Mascitti, A.K.; Streck, A.F.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Genomic characterization of Salmonella Minnesota clonal lineages associated with poultry production in Brazil. Animals 2020, 10, 2043. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.B.B.E.; Silva, R.L.; Menezes, J.; Machado, S.C.A.; Rodrigues, D.P.; Pomba, C.; Abreu, D.L.C.; Nascimento, E.R.; Aquino, M.H.C.; Pereira, V.L.A. High prevalence of multidrug-resistant nontyphoidal Salmonella recovered from broiler chickens and chicken carcasses in Brazil. Braz. J. Poult. Sci. 2020, 22, 1–6. [Google Scholar] [CrossRef]
- Rau, R.B.; Ribeiro, A.R.; dos Santos, A.; Barth, A.L. Antimicrobial resistance of Salmonella from poultry meat in Brazil: Results of a nationwide survey. Epidemiol. Infect. 2021, 149, E228. [Google Scholar] [CrossRef]
- Maki, J.J.; Klima, C.L.; Sylte, M.J.; Looft, T. The microbial pecking order: Utilization of intestinal microbiota for poultry health. Microorganisms 2019, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.S. Integrated colonization control of Salmonella in poultry. Poult. Sci. 1988, 67, 928–932. [Google Scholar] [CrossRef]
- Foley, S.L.; Nayak, R.; Hanning, I.B.; Johnson, T.J.; Han, J.; Ricke, S.C. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl. Environ. Microbiol. 2011, 77, 4273–4279. [Google Scholar] [CrossRef]
- Dunkley, K.D.; Callaway, T.R.; Chalova, V.I.; McReynolds, J.L.; Hume, M.E.; Dunkley, C.S.; Kubena, L.F.; Nisbet, D.J.; Ricke, S.C. Foodborne Salmonella ecology in the avian gastrointestinal tract. Anaerobe 2009, 15, 26–35. [Google Scholar] [CrossRef]
- Tanner, J.R.; Kingsley, R.A. Evolution of Salmonella within hosts. Trends Microbiol. 2018, 26, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Mebrhatu, M.T.; Cenens, W.; Aertsen, A. An overview of the domestication and impact of the Salmonella mobilome. Crit. Rev. Microbiol. 2014, 40, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Staes, I.; Passaris, I.; Cambré, A.; Aertsen, A. Population heterogeneity tactics as driving force in Salmonella virulence and survival. Food Res. Int. 2019, 125, 108560. [Google Scholar] [CrossRef]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing diversity: Differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front. Microbiol. 2019, 10, 1368. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.L.; Brumell, J.H.; Pfeifer, C.G.; Finlay, B.B. Salmonella pathogenicity islands: Big virulence in small packages. Microbes Infect. 2000, 2, 145–156. [Google Scholar] [CrossRef]
- Shivaprasad, H.L. Pullorum disease and fowl typhoid. In Diseases of Poultry, 11th ed.; Saif, Y.M., Ed.; Iowa State Press: Ames, IA, USA, 2003. [Google Scholar]
- Tavechio, A.T.; Fernandes, S.A.; Neves, B.C.; Dias, A.M.; Irino, K. Changing patterns of Salmonella serovars: Increase of Salmonella enteritidis in São Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo 1996, 38, 315–322. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Hargis, B.M.; Tsolis, R.M. Tracing the origins of Salmonella outbreaks. Science 2000, 287, 50–52. [Google Scholar] [CrossRef]
- Silva, E.N.; Duarte, A. Salmonella enteritidis em aves: Retrospectiva no Brasil. Rev. Bras. Cienc. Avic. 2002, 4, 85–100. [Google Scholar] [CrossRef]
- Foley, S.L.; Lynne, A.M.; Nayak, R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 2008, 86, E149–E162. [Google Scholar] [CrossRef]
- Pulido-Landínez, M.; Sánchez-Ingunza, R.; Guard, J.; Pinheiro do Nascimento, V. Assignment of serotype to Salmonella enterica isolates obtained from poultry and their environment in southern Brazil. Lett. Appl. Microbiol. 2013, 57, 288–294. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Vaz, C.S.; Alves, L.; Coldebella, A.; Leão, J.A.; Rodrigues, D.P.; Back, A. A temporal study of Salmonella enterica serotypes from broiler farms in Brazil. Poult. Sci. 2015, 94, 433–441. [Google Scholar] [CrossRef]
- Langridge, G.C.; Fookes, M.; Connor, T.R.; Feltwell, T.; Feasey, N.; Parsons, B.N.; Seth-Smith, H.M.; Barquist, L.; Stedman, A.; Humphrey, T.; et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc. Natl. Acad. Sci. USA 2015, 112, 863–868. [Google Scholar] [CrossRef]
- Wigley, P. Salmonella enterica serovar Gallinarum: Addressing fundamental questions in bacteriology sixty years on from the 9R vaccine. Avian Pathol. 2017, 46, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.; Sommer, F.; Liebhart, D.; Bilic, I.; Hess, M.; Hess, C. An outbreak of Pullorum Disease in a young layer parent flock in Austria presented with central nervous system signs. Avian Dis. 2021, 65, 159–164. [Google Scholar] [CrossRef]
- Freitas Neto, O.C.; Arroyave, W.; Alessi, A.C.; Fagliari, J.J.; Berchieri, A. Infection of commercial laying hens with Salmonella Gallinarum: Clinical, anatomopathological and haematological studies. Braz. J. Poult. Sci. 2002, 9, 133–141. [Google Scholar] [CrossRef]
- Zanetti, N.S.; De Carli, S.; Souza, M.N.; Lehmann, F.K.M.; Kipper, D.; Dias, K.K.R.; Fonseca, A.S.K.; Lunge, V.R.; Ikuta, N. Molecular detection and characterization of Salmonella Gallinarum from poultry farms in Brazil. J. Appl. Poult. 2019, 28, 1335–1341. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Z.; Qiang, B.; Xu, Y.; Chen, X.; Li, Q.; Jiao, X. Loss and gain in the evolution of the Salmonella enterica serovar Gallinarum biovar Pullorum genome. mSphere 2019, 4, e00627-18. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Li, Z.; Zhou, X.; Huang, C.; Hu, Y.; Geng, S.; Chen, X.; Li, Q.; Pan, Z.; Jiao, X. Induction of arthritis in chickens by infection with novel virulent Salmonella Pullorum strains. Vet. Microbiol. 2019, 228, 165–172. [Google Scholar] [CrossRef]
- Schat, K.A.; Nagaraja, K.V.; Saif, Y.M. Pullorum Disease: Evolution of the eradication strategy. Avian Dis. 2021, 65, 227–236. [Google Scholar] [CrossRef]
- Bullis, K. The history of avian medicine in the U.S. II: Pullorum disease and fowl typhoid. Avian Dis. 1977, 21, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhuang, L.; Wang, C.; Zhang, P.; Zhang, T.; Shao, H.; Han, X.; Gong, J. Virulence gene distribution of Salmonella Pullorum isolates recovered from chickens in China (1953–2015). Avian Dis. 2018, 62, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Le Bouquin, S.; Bonifait, L.; Thépault, A.; Ledein, T.; Guillon, F.; Rouxel, S.; Souillard, R.; Chemaly, M. Epidemiological and bacteriological investigations using whole-genome sequencing in a recurrent outbreak of Pullorum Disease on a quail farm in France. Animals 2020, 11, 29. [Google Scholar] [CrossRef]
- OIE (World Organization for Animal Health). Animal Desease Events. 2016. Available online: https://wahis.oie.int/#/events?viewAll=true (accessed on 1 February 2022).
- Foley, S.L.; Johnson, T.J.; Ricke, S.C.; Nayak, R.; Danzeisen, J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 2013, 77, 582–607. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.P.; Skov, M.N.; Hinz, K.H.; Bisgaard, M. Salmonella enterica serovar Gallinarum biovar gallinarum in layers: Epidemiological investigations of a recent outbreak in Denmark. Avian Pathol. 1994, 23, 489–501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barrow, P.A.; Freitas Neto, O.C. Pullorum disease and fowl typhoid—New thoughts on old diseases: A review. Avian Pathol. 2011, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- De Carli, S.; Gräf, T.; Kipper, D.; Lehmann, F.K.M.; Zanetti, N.; Siqueira, F.M.; Cibulski, S.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Molecular and phylogenetic analyses of Salmonella Gallinarum trace the origin and diversification of recent outbreaks of fowl typhoid in poultry farms. Vet. Microbiol. 2017, 212, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Koerich, P.K.V.; Fonseca, B.B.; Balestrin, E.; Tagliari, V.; Hoepers, P.G.; Ueira-Vieira, C.; Oldoni, I.; Rauber, R.H.; Ruschel, L.; Nascimento, V.P. Salmonella Gallinarum field isolates and its relationship to vaccine strain SG9R. Br. Poult. Sci. 2018, 59, 154–159. [Google Scholar] [CrossRef]
- Celis-Estupiñan, A.L.D.P.; Batista, D.F.A.; Cardozo, M.V.; Secundo de Souza, A.I.; Rodrigues Alves, L.B.; Maria de Almeida, A.; Barrow, P.A.; Berchieri, A., Jr.; Caetano de Freitas Neto, O. Further investigations on the epidemiology of fowl typhoid in Brazil. Avian Pathol. 2017, 46, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Vaid, R.K.; Thakur, Z.; Anand, T.; Kumar, S.; Tripathi, B.N. Comparative genome analysis of Salmonella enterica serovar Gallinarum biovars Pullorum and Gallinarum decodes strain specific genes. PLoS ONE 2021, 16, e0255612. [Google Scholar] [CrossRef]
- Kim, N.H.; Ha, E.J.; Ko, D.S.; Lee, C.Y.; Kim, J.H.; Kwon, H.J. Molecular evolution of Salmonella enterica subsp. enterica serovar Gallinarum biovar Gallinarum in the field. Vet. Microbiol. 2019, 235, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Li, X.; Wang, Y.; Wang, F.; Ge, H.; Pan, Z.; Xu, Y.; Wang, Y.; Jiao, X.; Chen, X. Epidemic patterns of antimicrobial resistance of Salmonella enterica serovar Gallinarum biovar Pullorum isolates in China during the past half-century. Poult. Sci. 2021, 100, 100894. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, K.S.; Kim, J.H.; Tak, R.B. Salmonella gallinarum gyrA mutations associated with fluoroquinolone resistance. Avian Pathol. 2004, 33, 251–257. [Google Scholar] [CrossRef]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial resistance in bacterial poultry pathogens: A review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef]
- Seo, K.W.; Kim, J.J.; Mo, I.P.; Lee, Y.J. Molecular characteristic of antimicrobial resistance of Salmonella Gallinarum isolates from chickens in Korea, 2014 to 2018. Poult. Sci. 2019, 98, 5416–5423. [Google Scholar] [CrossRef] [PubMed]
- Penha Filho, R.A.C.; Ferreira, J.C.; Kanashiro, A.M.B.; Darini, A.L.C.; Berchieri, A., Jr. Antimicrobial susceptibility of Salmonella Gallinarum and Salmonella Pullorum isolated from ill poultry in Brazil. Ciência Rural. 2016, 46, 513–518. [Google Scholar] [CrossRef]
- Rizzo, N.N.; Pottker, E.S.; Webber, B.; Borges, K.A.; Duarte, S.C.; Levandowski, R.; Ruschel, L.R.; Rodrigues, L.B. Effect of two lytic bacteriophages against multidrug-resistant and biofilm-forming Salmonella Gallinarum from poultry. Br. Poult. Sci. 2020, 61, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.K.M.Z.; Akter, M.R.; Islam, S.K.S.; Alam, J.; Neogi, S.B.; Yamasaki, S.; Kabir, S.M.L. Salmonella Gallinarum in small-scale commercial layer flocks: Occurrence, molecular diversity and antibiogram. Vet. Sci. 2021, 8, 71. [Google Scholar] [CrossRef]
- Bawn, M.; Alikhan, N.F.; Thilliez, G.; Kirkwood, M.; Wheeler, N.E.; Petrovska, L.; Dallman, T.J.; Adriaenssens, E.M.; Hall, N.; Kingsley, R.A. Evolution of Salmonella enterica serotype Typhimurium driven by anthropogenic selection and niche adaptation. PLoS Genet. 2020, 16, e1008850. [Google Scholar] [CrossRef] [PubMed]
- Chevance, F.F.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008, 6, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, S.; Gu, W.; den Bakker, H.; Boxrud, D.; Taylor, A.; Roe, C.; Driebe, E.; Engelthaler, D.M.; Allard, M.; et al. Zoonotic source attribution of Salmonella enterica serotype Typhimurium using genomic surveillance data, United States. Emerg. Infect. Dis. 2019, 25, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.A. Human Salmonella typhimurium infection due to duck eggs, with special reference to flocks of ducks. Br. Med. J. 1952, 2, 125–127. [Google Scholar] [CrossRef][Green Version]
- McCoy, J.H. Trends in Salmonella food poisoning in England and Wales 1941–72. J. Hyg. 1975, 74, 271–282. [Google Scholar] [CrossRef]
- Sadler, W.W.; Yamamoto, R.; Adler, H.E.; Stewart, G.F. Survey of market poultry for Salmonella infection. Appl. Microbiol. 1961, 9, 72–76. [Google Scholar] [CrossRef]
- Helms, M.; Ethelberg, S.; Mølbak, K. DT104 Study Group. International Salmonella Typhimurium DT104 infections, 1992–2001. Emerg. Infect. Dis. 2005, 11, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Branchu, P.; Bawn, M.; Kingsley, R.A. Genome variation and molecular epidemiology of Salmonella enterica serovar Typhimurium pathovariants. Infect. Immun. 2018, 86, e00079-18. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.O.D.; Souza, M.N.; Cecconi, M.C.P.; Timm, L.; Ikuta, N.; Simon, D.; Wolf, J.M.; Lunge, V.R. Increasing prevalence and dissemination of invasive nontyphoidal Salmonella serotype Typhimurium with multidrug resistance in hospitalized patients from southern Brazil. Braz. J. Infect. Dis. 2018, 22, 424–432. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Poppe, C.; Smart, N.; Khakhria, R.; Johnson, W.; Spika, J.; Prescott, J. Salmonella Typhimurium DT104: A virulent and drug-resistant pathogen. Can. Vet. J. 1998, 39, 559–565. [Google Scholar]
- Mather, A.E.; Reid, S.W.; Maskell, D.J.; Parkhill, J.; Fookes, M.C.; Harris, S.R.; Brown, D.J.; Coia, J.E.; Mulvey, M.R.; Gilmour, M.W.; et al. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 2013, 341, 1514–1517. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Hendriksen, R.S.; Le Hello, S.; Weill, F.X.; Baggesen, D.L.; Jun, S.R.; Ussery, D.W.; Lund, O.; Crook, D.W.; Wilson, D.J.; et al. Global genomic epidemiology of Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 2016, 82, 2516–2526. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Doblies, D.; Speed, K.; Davies, R.H. A retrospective analysis of Salmonella serovars isolated from pigs in Great Britain between 1994 and 2010. Prev. Vet. Med. 2013, 110, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Parsons, B.N.; Crayford, G.; Humphrey, T.J.; Wigley, P. Infection of chickens with antimicrobial-resistant Salmonella enterica Typhimurium DT193 and monophasic Salmonella Typhimurium-like variants: An emerging risk to the poultry industry? Avian Pathol. 2013, 42, 443–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Echeita, M.; Herrera, S.; Usera, M. Atypical, fljB-negative Salmonella enterica strain of serovar 4,5,12:i:– appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 2001, 39, 2981–2983. [Google Scholar] [CrossRef] [PubMed]
- Tavechio, A.T.; Ghilardi, A.C.; Fernandes, S.A. Multiplex PCR identification of the atypical and monophasic Salmonella enterica subsp. enterica serotype 1,4,[5],12:i:- in São Paulo State, Brazil: Frequency and antibiotic resistance patterns. Rev. Inst. Med. Trop. São Paulo 2004, 46, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Mossong, J.; Marques, P.; Ragimbeau, C.; Huberty-Krau, P.; Losch, S.; Meyer, G.; Moris, G.; Strottner, C.; Rabsch, W.; Schneider, F. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:- in Luxembourg, 2006. Eurosurveillance 2007, 12, E11–E12. [Google Scholar] [CrossRef]
- Agasan, A.; Kornblum, J.; Williams, G.; Pratt, C.C.; Fleckenstein, P.; Wong, M.; Ramon, A. Profile of Salmonella enterica subsp. enterica (subspecies I) serotype 4,5,12:i:- strains causing food-borne infections in New York City. J. Clin. Microbiol. 2002, 40, 1924–1929. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zamperini, K.; Soni, V.; Waltman, D.; Sanchez, S.; Theriault, E.C.; Bray, J.; Maurer, J.J. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant Typhimurium serovar. Avian Dis. 2007, 51, 958–964. [Google Scholar] [CrossRef]
- Tavechio, A.T.; Fernandes, S.A.; Ghilardi, A.C.; Soule, G.; Ahmed, R.; Melles, C.E. Tracing lineage by phenotypic and genotypic markers in Salmonella enterica subsp. enterica serovar 1,4,[5],12:i:- and Salmonella Typhimurium isolated in state of São Paulo, Brazil. Memórias Inst. Oswaldo Cruz 2009, 104, 1042–1046. [Google Scholar] [CrossRef] [PubMed][Green Version]
- EFSA (European Food Safety Authority). Report of the Task Force on Zoonoses Data Collection on the Analysis of the baseline study on the prevalence of Salmonella in holdings of laying hen flocks of Gallus gallus. EFSA J. 2007, 5, 97r. [Google Scholar] [CrossRef]
- Dookeran, M.M.; Baccus-Taylor, G.S.; Akingbala, J.O.; Tameru, B.; Lammerding, A.M. Transmission of Salmonella on broiler chickens and carcasses from production to retail in Trinidad and Tobago. J. Agric. Biodivers. Res. 2012, 1, 78–84. [Google Scholar]
- Taunay, A.E.; Fernandes, S.A.; Tavechio, A.T.; Neves, B.C.; Dias, A.M.; Irino, K. The role of public health laboratory in the problem of salmonellosis in São Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo 1996, 38, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Tavechio, A.T.; Ghilardi, A.C.; Peresi, J.T.; Fuzihara, T.O.; Yonamine, E.K.; Jakabi, M.; Fernandes, S.A. Salmonella serotypes isolated from nonhuman sources in São Paulo, Brazil, from 1996 through 2000. J. Food Prot. 2002, 65, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.A.; Tavechio, A.T.; Ghilardi, A.C.; Dias, A.M.; Almeida, I.A.; Melo, L.C. Salmonella serovars isolated from humans in São Paulo State, Brazil, 1996-2003. Rev. Inst. Med. Trop. São Paulo 2006, 48, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Capalonga, R.; Ramos, R.C.; Both, J.M.; Soeiro, M.L.; Longaray, S.M.; Haas, S.; Tondo, E.C. Salmonella serotypes, resistance patterns, and food vehicles of salmonellosis in southern Brazil between 2007 and 2012. J. Infect. Dev. Ctries. 2014, 8, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Neto, W.S.; Leotti, V.B.; Pires, S.M.; Hald, T.; Corbellini, L.G. Non-typhoidal human salmonellosis in Rio Grande do Sul, Brazil: A combined source attribution study of microbial subtyping and outbreak data. Int. J. Food Microbiol. 2021, 338, 108992. [Google Scholar] [CrossRef] [PubMed]
- Barrow, P.A.; Jones, M.A.; Smith, A.L.; Wigley, P. The long view: Salmonella the last forty years. Avian Pathol. 2012, 41, 413–420. [Google Scholar] [CrossRef]
- Hugas, M.; Beloeil, P. Controlling Salmonella along the food chain in the European Union—Progress over the last ten years. Eurosurveillance 2014, 19, 20804. [Google Scholar] [CrossRef]
- Hassan, J.O.; Curtiss, R., 3rd. Development and evaluation of an experimental vaccination program using a live avirulent Salmonella Typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect. Immun. 1994, 62, 5519–5527. [Google Scholar] [CrossRef] [PubMed]
- Groves, P.J.; Sharpe, S.M.; Muir, W.I.; Pavic, A.; Cox, J.M. Live and inactivated vaccine regimens against caecal Salmonella Typhimurium colonisation in laying hens. Aust. Vet. J. 2016, 94, 387–393. [Google Scholar] [CrossRef]
- Yang, Y.; Wolfenden, A.; Mandal, R.K.; Faulkner, O.; Hargis, B.; Kwon, Y.M.; Bielke, L. Evaluation of recombinant Salmonella vaccines to provide cross-serovar and cross-serogroup protection. Poult. Sci. 2017, 96, 4352–4360. [Google Scholar] [CrossRef] [PubMed]
- De Melo, A.N.F.; Monte, D.F.M.; de Souza Pedrosa, G.T.; Balkey, M.; Jin, Q.; Brown, E.; Allard, M.; de Oliveira, T.C.R.M.; Cao, G.; Magnani, M.; et al. Genomic investigation of antimicrobial resistance determinants and virulence factors in Salmonella enterica serovars isolated from contaminated food and human stool samples in Brazil. Int. J. Food Microbiol. 2021, 343, 109091. [Google Scholar] [CrossRef] [PubMed]
- Mouttotou, N.; Ahmad, S.; Kamran, Z.; Koutoulis, K.C. Prevalence, risks and antibiotic resistance of Salmonella in poultry production chain. In Current Topics in Salmonella and Salmonellosis; InTechOpen: London, UK, 2017; pp. 215–234. [Google Scholar] [CrossRef]
- Reis, R.O.; Cecconi, M.C.; Timm, L.; Souza, M.N.; Ikuta, N.; Wolf, J.M.; Lunge, V.R. Salmonella isolates from urine cultures: Serotypes and antimicrobial resistance in hospital settings. Braz. J. Microbiol. 2019, 50, 445–448. [Google Scholar] [CrossRef]
- Bessa, M.C.; Michael, G.B.; Canu, N.; Canal, C.W.; Cardoso, M.; Rabsch, W.; Rubino, S. Phenotypic and genetic characterization of Salmonella enterica subsp. enterica serovar Typhimurium isolated from pigs in Rio Grande do Sul, Brazil. Res. Vet. Sci. 2007, 83, 302–310. [Google Scholar] [CrossRef]
- Almeida, F.; Medeiros, M.I.; Kich, J.D.; Falcão, J.P. Virulence-associated genes, antimicrobial resistance and molecular typing of Salmonella Typhimurium strains isolated from swine from 2000 to 2012 in Brazil. J. Appl. Microbiol. 2016, 120, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.; Seribelli, A.A.; Medeiros, M.I.C.; Rodrigues, D.D.P.; de MelloVarani, A.; Luo, Y.; Allard, M.W.; Falcão, J.P. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLoS ONE. 2018, 13, e0201882. [Google Scholar] [CrossRef]
- Perin, A.P.; Martins, B.T.F.; Barreiros, M.A.B.; Yamatogi, R.S.; Nero, L.A.; Dos Santos Bersot, L. Occurrence, quantification, pulse types, and antimicrobial susceptibility of Salmonella sp. isolated from chicken meat in the state of Paraná, Brazil. Braz. J. Microbiol. 2020, 51, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.B.; de Lima-Morales, D.; Wink, P.L.; Ribeiro, A.R.; Barth, A.L. Salmonella enterica mcr-1 positive from food in Brazil: Detection and characterization. Foodborne Pathog. Dis. 2020, 17, 202–208. [Google Scholar] [CrossRef]
- Galanis, E.; Lo Fo Wong, D.M.; Patrick, M.E.; Binsztein, N.; Cieslik, A.; Chalermchikit, T.; Aidara-Kane, A.; Ellis, A.; Angulo, F.J.; Wegener, H.C.; et al. Webbased surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 2006, 12, 381–388. [Google Scholar] [CrossRef]
- Rodrigue, D.C.; Tauxe, R.V.; Rowe, B. International increase in Salmonella Enteritidis: A new pandemic? Epidemiol. Infect. 1990, 105, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Guard-Petter, J. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 2001, 3, 421–430. [Google Scholar] [CrossRef]
- Rampling, A.; Anderson, J.R.; Upson, R.; Peters, E.; Ward, L.R.; Rowe, B. Salmonella enteritidis phage type 4 infection of broiler chickens: A hazard to public health. Lancet 1989, 2, 436–438. [Google Scholar] [CrossRef]
- Rabsch, W.; Hargis, B.M.; Tsolis, R.M.; Kingsley, R.A.; Hinz, K.H.; Tschäpe, H.; Bäumler, A.J. Competitive exclusion of Salmonella enteritidis by Salmonella gallinarum in poultry. Emerg. Infect. Dis. 2000, 6, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.E.; Adcock, P.M.; Gomez, T.M.; Altekruse, S.F.; Holland, B.H.; Tauxe, R.V.; Swerdlow, D.L. Salmonella Enteritidis infections, United States, 1985–1999. Emerg. Infect. Dis. 2004, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, T.J.; Baskerville, A.; Mawer, S.; Rowe, B.; Hopper, S. Salmonella enteritidis phage type 4 from the contents of intact eggs: A study involving naturally infected hens. Epidemiol. Infect. 1989, 103, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Rampling, A. Salmonella enteritidis five years on. Lancet 1993, 342, 317–318. [Google Scholar] [CrossRef]
- Alexandre, M.; Pozo, C.; González, V.; Martínez, M.C.; Prat, S.; Fernández, A.; Fica, A.; Fernández, J.; Heitmann, I. Detection of Salmonella enteritidis in samples of poultry products for human consumption in the Chilean metropolitan area. Rev. Med. Chil. 2000, 128, 1075–1083. [Google Scholar] [PubMed]
- Kottwitz, L.B.M.; Scheffer, M.C.; Dalla-Costa, L.M.; Farah, S.M.S.S.; Moscalewski, W.S.B.; Magnani, M.; de Oliveira, T.C.R.M. Molecular characterization and resistance profile of Salmonella Enteritidis PT4 and PT9 strains isolated in Brazil. J. Med. Microbiol. 2011, 60, 1026–1031. [Google Scholar] [CrossRef]
- Caffer, M.I.; Eiguer, T. Salmonella enteritidis in Argentina. Int. J. Food Microbiol. 1994, 21, 15–19. [Google Scholar] [CrossRef]
- Mascitti, A.K.; Kipper, D.; Dos Reis, R.O.; da Silva, J.S.; Fonseca, A.S.K.; Ikuta, N.; Tondo, E.C.; Lunge, V.R. Retrospective whole-genome comparison of Salmonella enterica serovar Enteritidis from foodborne outbreaks in Southern Brazil. Braz. J. Microbiol. 2021, 52, 1523–1533. [Google Scholar] [CrossRef]
- de Freitas Neto, O.C.; Mesquita, A.L.; de Paiva, J.B.; Zotesso, F.; Berchieri Júnior, A. Control of Salmonella enterica serovar Enteritidis in laying hens by inactivated Salmonella Enteritidis vaccines. Braz. J. Microbiol. 2008, 39, 390–396. [Google Scholar] [CrossRef]
- Obe, T.; Nannapaneni, R.; Schilling, W.; Zhang, L.; McDaniel, C.; Kiess, A. Prevalence of Salmonella enterica on poultry processing equipment after completion of sanitization procedures. Poult. Sci. 2020, 99, 4539–4548. [Google Scholar] [CrossRef] [PubMed]
- Penha Filho, R.A.; de Paiva, J.B.; Arguello, Y.M.; da Silva, M.D.; Gardin, Y.; Resende, F.; Berchieri, A.B., Jr.; Sesti, L. Efficacy of several vaccination programmes in commercial layer and broiler breeder hens against experimental challenge with Salmonella enterica serovar Enteritidis. Avian Pathol. 2009, 38, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Huberman, Y.D.; Velilla, A.V.; Terzolo, H.R. Evaluation of different live Salmonella Enteritidis vaccine schedules administered during layer hen rearing to reduce excretion, organ colonization, and egg contamination. Poult. Sci. 2019, 98, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, F.A.; Brandelli, A.; Tondo, E.C. Antimicrobial resistance in Salmonella enteritidis from foods involved in human salmonellosis outbreaks in southern Brazil. New Microbiol. 2006, 29, 49–54. [Google Scholar]
- Vaz, C.S.; Streck, A.F.; Michael, G.B.; Marks, F.S.; Rodrigues, D.P.; Dos Reis, E.M.; Cardoso, M.R.; Canal, C.W. Antimicrobial resistance and subtyping of Salmonella enterica subspecies enterica serovar Enteritidis isolated from human outbreaks and poultry in southern Brazil. Poult. Sci. 2010, 89, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Moratto Bergamini, A.M.; Falcão, J.P. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012, 32, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Souza, R.A.; Martins, V.V.; Stehling, E.G.; Bergamini, A.M.M.; Falcão, J.P. Prevalence of gyrA mutations in nalidixic acid-resistant strains of Salmonella Enteritidis isolated from humans, food, chickens, and the farm environment in Brazil. Microb. Drug Resist. 2017, 23, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Habbs, V.H. About a new type of bacteria from the paratyphoid enteritic group. J. Bacterial. 1933, 130, 374–396. [Google Scholar]
- Deblais, L.; Lorentz, B.; Scaria, J.; Nagaraja, K.V.; Nisar, M.; Lauer, D.; Voss, S.; Rajashekara, G. Comparative genomic studies of Salmonella Heidelberg isolated from chicken- and turkey-associated farm environmental samples. Front. Microbiol. 2018, 9, 1841. [Google Scholar] [CrossRef]
- Eurosurveillance Editorial Team. European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food 2012 published. Eurosurveillance 2014, 19, 20748. [Google Scholar] [CrossRef][Green Version]
- CDC (Centers for Disease Control and Prevention). Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Contact with Dairy Calves (Final Update). 2018. Available online: http://www.cdc.gov/Salmonella/heidelberg-11-16/index.html (accessed on 15 January 2022).
- Jones, T.F.; Ingram, L.A.; Cieslak, P.R.; Vugia, D.J.; Tobin-D’Angelo, M.; Hurd, S.; Medus, C.; Cronquist, A.; Ângulo, F.J. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 2008, 198, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.A.; Craven, C.; Wells, J.G. Salmonella Heidelberg enteritis and bacteremia. An epidemic on two pediatric wards. Am. J. Med. 1976, 60, 509–516. [Google Scholar] [CrossRef]
- Asmar, B.I.; Abdel-Haq, N. Nontyphoidal Salmonella infection in children: Relation to bacteremia, age, and infecting serotype. Infect. Dis. 2016, 48, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Chittick, P.; Sulka, A.; Tauxe, R.V.; Fry, A.M. A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: Clues for disease prevention. J. Food Prot. 2006, 69, 1150–1153. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, T.W.; Cheng, L.H.; Kassenborg, H.; Ahuja, S.D.; Mohle-Boetani, J.; Marcus, R.; Shiferaw, B.; Angulo, F.J. Emerging Infections Program FoodNet Working Group. Egg consumption is the principal risk factor for sporadic Salmonella serotype Heidelberg infections: A case-control study in FoodNet sites. Clin. Infect. Dis. 2004, 38, S237–S243. [Google Scholar] [CrossRef]
- CDC (Centers for Disease Control and Prevention). Outbreak of Salmonella Heidelberg infections linked to a single poultry producer—13 states, 2012–2013. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 553–556. [Google Scholar]
- Routh, J.A.; Pringle, J.; Mohr, M.; Bidol, S.; Arends, K.; Adams-Cameron, M.; Hancock, W.T.; Kissler, B.; Rickert, R.; Folster, J.; et al. Nationwide outbreak of multidrug-resistant Salmonella Heidelberg infections associated with ground turkey: United States, 2011. Epidemiol. Infect. 2015, 143, 3227–3234. [Google Scholar] [CrossRef] [PubMed]
- Gieraltowski, L.; Higa, J.; Peralta, V.; Green, A.; Schwensohn, C.; Rosen, H.; Libby, T.; Kissler, B.; Marsden-Haug, N.; Booth, H.; et al. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS ONE 2016, 11, e0162369. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Bearson, S.M.D.; Looft, T.; Cai, G.; Shippy, D.C. Characterization of a multidrug-resistant Salmonella enterica serovar Heidelberg outbreak strain in commercial turkeys: Colonization, transmission, and host transcriptional response. Front. Vet. Sci. 2017, 4, 156. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.; Gollarza, L.; Sockett, D.; Aulik, N.; Patton, E.; Francois Watkins, L.K.; Gambino-Shirley, K.J.; Folster, J.P.; Chen, J.C.; Tagg, K.A.; et al. Outbreak of multidrug-resistant Salmonella Heidelberg infections linked to dairy calf exposure, United States, 2015–2018. Foodborne Pathog. Dis. 2022, 19, 199–208. [Google Scholar] [CrossRef]
- Fontaine, R.E.; Cohen, M.L.; Martin, W.T.; Vernon, T.M. Epidemic salmonellosis from cheddar cheese: Surveillance and prevention. Am. J. Epidemiol. 1980, 111, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Antony, L.; Behr, M.; Sockett, D.; Miskimins, D.; Aulik, N.; Christopher-Hennings, J.; Nelson, E.; Allard, M.W.; Scaria, J. Genome divergence and increased virulence of outbreak associated Salmonella enterica subspecies enterica serovar Heidelberg. Gut Pathog. 2018, 10, 53. [Google Scholar] [CrossRef]
- Green, A.; Defibaugh-Chavez, S.; Douris, A.; Vetter, D.; Atkinson, R.; Kissler, B.; Khroustalev, A.; Robertson, K.; Sharma, Y.; Becker, K.; et al. Intensified sampling in response to a Salmonella Heidelberg outbreak associated with multiple establishments within a single poultry corporation. Foodborne Pathog. Dis. 2018, 15, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.T.; Rives, D.V.; Carey, J.B. Salmonella contamination in commercial eggs and an egg production facility. Poult. Sci. 1995, 74, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Currie, A.; MacDougall, L.; Aramini, J.; Gaulin, C.; Ahmed, R.; Isaacs, S. Frozen chicken nuggets and strips and eggs are leading risk factors for Salmonella Heidelberg infections in Canada. Epidemiol. Infect. 2005, 133, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, L.R.; van der Graaf-van Bloois, L.; Donado-Godoy, P.; León, M.; Clavijo, V.; Arévalo, A.; Bernal, J.F.; Mevius, D.J.; Wagenaar, J.A.; Zomer, A.; et al. Genomic characterization of extended-spectrum cephalosporin-resistant Salmonella enterica in the Colombian poultry chain. Front. Microbiol. 2018, 9, 2431. [Google Scholar] [CrossRef]
- Aravena, C.; Valencia, B.; Villegas, A.; Ortega, M.; Fernández, R.A.; Araya, R.P.; Saavedra, A.; Del Campo, R. Caracterización de cepas clínicas y ambientales de Salmonella enterica subsp. enterica serovar Heidelberg aisladas en Chile [Characterization of Salmonella Heidelberg strains isolated in Chile]. Rev. Med. Chil. 2019, 147, 24–33. [Google Scholar] [CrossRef]
- Gelaw, A.K.; Nthaba, P.; Matle, I. Detection of Salmonella from animal sources in South Africa between 2007 and 2014. J. S. Afr. Vet. Assoc. 2018, 89, e1–e10. [Google Scholar] [CrossRef] [PubMed]
- Mammina, C.; Talini, M.; Pontello, M.; Di Noto, A.M.; Nastasi, A. Clonal circulation of Salmonella enterica serotype Heidelberg in Italy? Eurosurveillance 2003, 8, 222–225. [Google Scholar] [CrossRef]
- Hofer, E.; da Silva Filho, S.J.; dos Reis, E.M.F. Prevalência de sorovares de Salmonella isolados de aves no Brasil. Pesqui. Vet. Bras. 1997, 17, 55–62. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Kramer, B.; Silva, V.S.; Rebelatto, R.; Abreu, P.G.; Coldebella, A.; Vaz, C.S.L. Longitudinal study reveals persistent environmental Salmonella Heidelberg in Brazilian broiler farms. Vet. Microbiol. 2019, 233, 118–123. [Google Scholar] [CrossRef] [PubMed]
- RASFF (The Rapid Alert System for Food and Feed). Food and Feed Safety Alerts. 2018. Available online: https://webgate.ec.europa.eu/rasff-window/portal/ (accessed on 20 January 2022).
- Melo, R.T.; Galvão, N.N.; Guidotti-Takeuchi, M.; Peres, P.A.B.M.; Fonseca, B.B.; Profeta, R.; Azevedo, V.A.C.; Monteiro, G.P.; Brenig, B.; Rossi, D.A. Molecular characterization and survive abilities of Salmonella Heidelberg strains of poultry origin in Brazil. Front. Microbiol. 2021, 12, 674147. [Google Scholar] [CrossRef] [PubMed]
- Borges, K.A.; Furian, T.Q.; Souza, S.N.; Menezes, R.; Tondo, E.C.; Salle, C.T.; Moraes, H.L.; Nascimento, V.P. Biofilm formation capacity of Salmonella serotypes at different temperature conditions. Pesqui. Vet. Bras. 2018, 38, 71–76. [Google Scholar] [CrossRef]
- Lucca, V.; Apellanis Borges, K.; Quedi Furian, T.; Borsoi, A.; Pippi Salle, C.T.; de Souza Moraes, H.L.; Pinheiro do Nascimento, V. Influence of the norepinephrine and medium acidification in the growth and adhesion of Salmonella Heidelberg isolated from poultry. Microb. Pathog. 2020, 138, 103799. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.; Usongo, V.; Berry, C.; Tremblay, D.M.; Moineau, S.; Yousfi, K.; Doualla-Bell, F.; Fournier, E.; Nadon, C.; Goodridge, L.; et al. Comparison of advanced whole genome sequence-based methods to distinguish strains of Salmonella enterica serovar Heidelberg involved in foodborne outbreaks in Québec. Food Microbiol. 2018, 73, 99–110. [Google Scholar] [CrossRef]
- Kerr, E.J.; Stafford, R.; Rathnayake, I.U.; Graham, R.M.A.; Fearnley, E.; Gregory, J.; Glasgow, K.; Wright, R.; Sintchenko, V.; Wang, Q.; et al. Multistate Outbreak of Salmonella enterica Serovar Heidelberg with Unidentified Source, Australia, 2018–2019. Emerg. Infect. Dis. 2022, 28, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Edirmanasinghe, R.; Finley, R.; Parmley, E.J.; Avery, B.P.; Carson, C.; Bekal, S.; Golding, G.; Mulvey, M.R. A Whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob. Agents Chemother. 2017, 61, e01919-16. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Li, J.; Zhang, X.; Gao, F.; Benton, C.S.; Andam, C.P. Diverse lineages of multidrug resistant clinical Salmonella enterica and a cryptic outbreak in New Hampshire, USA revealed from a year-long genomic surveillance. Infect. Genet. Evol. 2021, 87, 104645. [Google Scholar] [CrossRef] [PubMed]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–54. [Google Scholar] [CrossRef]
- Fitch, F.M.; Carmo-Rodrigues, M.S.; Oliveira, V.G.; Gaspari, M.V.; Dos Santos, A.; de Freitas, J.B.; Pignatari, A.C. β-Lactam resistance genes: Characterization, epidemiology, and first detection of blaCTX-M-1 and blaCTX-M-14 in Salmonella spp. isolated from poultry in Brazil-Brazil Ministry of Agriculture’s Pathogen Reduction Program. Microb. Drug Resist. 2016, 22, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Borsoi, A.; Santin, E.; Santos, L.R.; Salle, C.T.; Moraes, H.L.; Nascimento, V.P. Inoculation of newly hatched broiler chicks with two Brazilian isolates of Salmonella Heidelberg strains with different virulence gene profiles, antimicrobial resistance, and pulsed field gel electrophoresis patterns to intestinal changes evaluation. Poult. Sci. 2009, 88, 750–758. [Google Scholar] [CrossRef]
- Medeiros, M.A.; Oliveira, D.C.; Rodrigues Ddos, P.; Freitas, D.R. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev. Panam. Salud Pública 2011, 30, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Giuriatti, J.; Stefani, L.M.; Brisola, M.C.; Crecencio, R.B.; Bitner, D.S.; Faria, G.A. Salmonella Heidelberg: Genetic profile of its antimicrobial resistance related to extended spectrum β-lactamases (ESBLs). Microb. Pathog. 2017, 109, 195–199. [Google Scholar] [CrossRef]
- Santin, E.; Hayashi, R.M.; Wammes, J.C.; Gonzalez-Esquerra, R.; Carazzolle, M.F.; Freire, C.C.M.; Monzani, P.S.; da Cunha, A.F. Phenotypic and genotypic features of a Salmonella Heidelberg strain isolated in broilers in Brazil and their possible association to antibiotics and short-chain organic acids resistance and susceptibility. Front. Vet. Sci. 2017, 4, 184. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.I.S.; Saraiva, M.M.S.; Casas, M.R.T.; Oliveira, G.M.; Cardozo, M.V.; Benevides, V.P.; Barbosa, F.O.; Freitas Neto, O.C.; Almeida, A.M.; Berchieri, A.J. High occurrence of β-lactamase-producing Salmonella Heidelberg from poultry origin. PLoS ONE 2020, 15, e0230676. [Google Scholar] [CrossRef] [PubMed]
- Gomes, V.T.M.; Moreno, L.Z.; Silva, A.P.S.; Thakur, S.; La Ragione, R.; Mather, A.E.; Moreno, A.M. Characterization of Salmonella enterica contamination in pork and poultry meat from São Paulo/Brazil: Serotypes, genotypes and antimicrobial resistance profiles. Pathogens 2022, 11, 358. [Google Scholar] [CrossRef]
- Jiménez, M.; Martínez-Urtaza, J.; Chaidez, C. Geographical and temporal dissemination of Salmonellae isolated from domestic animal hosts in the Culiacan Valley, Mexico. Microb. Ecol. 2011, 61, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, M.; Martinez-Urtaza, J.; Rodriguez-Alvarez, M.X.; Leon-Felix, J.; Chaidez, C. Prevalence and genetic diversity of Salmonella spp. in a river in a tropical environment in Mexico. J. Water Health 2014, 12, 874–884. [Google Scholar] [CrossRef]
- McCracken, D.A. Salmonella minnesota infection in Northamptonshire. J. R. Sanit. Inst. 1954, 74, 1091–1100. [Google Scholar] [CrossRef]
- Al-Ghamdi, M.; Al-Sabty, S.; Kannan, A.; Rowe, B. An outbreak of food poisoning in a workers’ camp in Saudi Arabia caused by Salmonella minnesota. J. Diarrhoeal Dis. Res. JSTOR 1989, 7, 18–20. [Google Scholar]
- CDC (Centers for Disease Control and Prevention). Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne diseases active surveillance network, 10 U.S. sites, 1996–2012. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 283–287. [Google Scholar]
- EFSA (European Food Safety Authority). Scientific report of EFSA and ECDC: The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks. 2012EFSA J. 2014, 12, 3547. [Google Scholar]
- Steinebrunner, N.; Sandig, C.; Zimmermann, S.; Stremmel, W.; Eisenbach, C.; Mischnik, A. Salmonella enterica serovar Minnesota urosepsis in a patient with Crohn’s disease in the absence of recent or current gastrointestinal symptoms. J. Med. Microbiol. 2013, 62, 1360–1362. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.R.; Bruner, D.W. Two new Salmonella types isolated from fowls. J. Hyg. 1938, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Solowey, M.; McFarlane, V.H.; Spaulding, E.H.; Chemerda, C. Microbiology of spray-dried whole egg: II. Incidence and types of Salmonella. Am. J. Public Health Nation’s Health 1947, 37, 971–982. [Google Scholar] [CrossRef]
- Tabo, D.A.; Diguimbaye, C.D.; Granier, S.A.; Moury, F.; Brisabois, A.; Elgroud, R.; Millemann, Y. Prevalence and antimicrobial resistance of non-typhoidal Salmonella serotypes isolated from laying hens and broiler chicken farms in N’Djamena, Chad. Vet. Microbiol. 2013, 166, 293–298. [Google Scholar] [CrossRef]
- RASFF (The Rapid Alert System for Food and Feed). Annual Report 2017. 2017. Available online: https://op.europa.eu/en/publication-detail/-/publication/f4adf22f-4f7c-11e9-a8ed-01aa75ed71a1/language-en (accessed on 12 January 2022).
- Silva, P.L.A.P.A.; Goulart, L.R.; Reis, T.F.M.; Mendonça, E.P.; Melo, R.T.; Penha, V.A.S.; Peres, P.A.B.M.; Hoepers, P.G.; Beletti, M.E.; Fonseca, B.B. Biofilm formation in different Salmonella serotypes isolated from poultry. Curr. Microbiol. 2019, 76, 124–129. [Google Scholar] [CrossRef]
- Costa, R.G.; Festivo, M.L.; Araujo, M.S.; Reis, E.M.; Lázaro, N.S.; Rodrigues, D.P. Antimicrobial susceptibility and serovars of Salmonella circulating in commercial poultry carcasses and poultry products in Brazil. J. Food Prot. 2013, 76, 2011–2017. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.P.; de Melo, R.T.; Nalevaiko, P.C.; Monteiro, G.P.; Fonseca, B.B.; Galvão, N.N.; Giombelli, A.; Rossi, D.A. Spread of the serotypes and antimicrobial resistance in strains of Salmonella spp. isolated from broiler. Braz. J. Microbiol. 2019, 50, 515–522. [Google Scholar] [CrossRef]
- Moreira, J.P.F.F.; do Monte, D.F.M.; Lima, C.A.; de Oliveira, C.J.B.; da Silva Martins, N.R.; Berchieri, A., Jr.; de Freitas Neto, O.C. Molecular genotyping reveals inter-regional relatedness among antimicrobial resistant Salmonella Minnesota strains isolated from poultry farm and humans, Brazil. Braz. J. Microbiol. 2022, 53, 503–508. [Google Scholar] [CrossRef]
- Agostinho Davanzo, E.F.; Dos Santos, R.L.; Castro, V.H.L.; Palma, J.M.; Pribul, B.R.; Dallago, B.S.L.; Fuga, B.; Medeiros, M.; Titze de Almeida, S.S.; da Costa, H.M.B.; et al. Molecular characterization of Salmonella spp. and Listeria monocytogenes strains from biofilms in cattle and poultry slaughterhouses located in the federal District and State of Goiás, Brazil. PLoS ONE 2021, 16, e0259687. [Google Scholar] [CrossRef]
- Penha Filho, R.A.C.; Ferreira, J.C.; Kanashiro, A.M.I.; Berchieri, A., Jr.; Darini, A.L.D.C. Emergent multidrug-resistant nontyphoidal Salmonella serovars isolated from poultry in Brazil coharboring blaCTX-M-2 and qnrB or blaCMY-2 in large plasmids. Diagn. Microbiol. Infect. Dis. 2019, 95, 93–98. [Google Scholar] [CrossRef]
- Almeida, F.; Pitondo-Silva, A.; Oliveira, M.A.; Falcão, J.P. Molecular epidemiology and virulence markers of Salmonella Infantis isolated over 25 years in São Paulo State, Brazil. Infect. Genet. Evol. 2013, 19, 145–151. [Google Scholar] [CrossRef]
- Merino, L.A.; Ronconi, M.C.; Navia, M.M.; Ruiz, J.; Sierra, J.M.; Cech, N.B.; Lodeiro, N.S.; Vila, J. Analysis of the clonal relationship among clinical isolates of Salmonella enterica serovar Infantis by different typing methods. Rev. Inst. Med. Trop. São Paulo 2003, 45, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.P.; Melo, R.T.; Oliveira, M.R.; Monteiro, G.P.; Peres, P.A.; Fonseca, B.B.; Giombelli, A.; Rossi, D.A. Characteristics of virulence, resistance and genetic diversity of strains of Salmonella Infantis isolated from broiler chicken in Brazil. Pesqui. Veterinária Bras. 2020, 40, 29–38. [Google Scholar] [CrossRef]
- Cunha-Neto, A.D.; Carvalho, L.A.; Carvalho, R.C.T.; Dos Prazeres Rodrigues, D.; Mano, S.B.; Figueiredo, E.E.S.; Conte, C.A., Jr. Salmonella isolated from chicken carcasses from a slaughterhouse in the state of Mato Grosso, Brazil: Antibiotic resistance profile, serotyping, and characterization by repetitive sequence-based PCR system. Poult. Sci. 2018, 97, 1373–1381. [Google Scholar] [CrossRef]
- Baptista, D.Q.; Santos, A.F.; Aquino, M.H.C.; Abreu, D.L.; Rodrigues, D.P.; Nascimento, E.R.; Pereira, V.L. Prevalência e susceptibilidade antimicrobiana de sorotipos de Salmonella spp. isolados de frangos vivos e carcaças no estado do Rio de Janeiro. Pesqui. Vet. Bras. 2018, 38, 1278–1285. [Google Scholar] [CrossRef]
- Mattiello, S.P.; Drescher, G.; Barth, V.C., Jr.; Ferreira, C.; Oliveira, S.D. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie van Leeuwenhoek 2015, 108, 1227–1238. [Google Scholar] [CrossRef]
- Ribeiro, A.R.; Kellermann, A.; dos Santos, L.R.; Fittél, A.P.; do Nascimento, V.P. Resistência antimicrobiana em Salmonella enterica subsp. enterica sorovar Hadar isoladas de carcaças de frango. Arq. Inst. Biológico 2022, 73, 357–360. [Google Scholar] [CrossRef]
- Monte, D.F.M.; Nethery, M.A.; Barrangou, R.; Landgraf, M.; Fedorka-Cray, P.J. Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol. 2021, 93, 103601. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.H.; Sarker, S.; Islam, M.S.; Islam, M.A.; Karim, M.R.; Kayesh, M.E.H.; Shiddiky, M.J.A.; Anwer, M.S. Sustainable antibiotic-free broiler meat production: Current trends, challenges, and possibilities in a developing country perspective. Biology 2020, 9, 411. [Google Scholar] [CrossRef]
- Iannetti, L.; Romagnoli, S.; Cotturone, G.; Podaliri Vulpiani, M. Animal welfare assessment in antibiotic-free and conventional broiler chicken. Animals 2021, 11, 2822. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Ruvalcaba-Gómez, J.M.; Villagrán, Z.; Valdez-Alarcón, J.J.; Martínez-Núñez, M.; Gomez-Godínez, L.J.; Ruesga-Gutiérrez, E.; Anaya-Esparza, L.M.; Arteaga-Garibay, R.I.; Villarruel-López, A. Non-antibiotics strategies to control Salmonella infection in poultry. Animals 2022, 12, 102. [Google Scholar] [CrossRef]
- Ricke, S.C.; Dittoe, D.K.; Richardson, K.E. Formic acid as an antimicrobial for poultry production: A review. Front. Vet. Sci. 2020, 7, 563. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Moore, R.J.; Stanley, D.; Chousalkar, K.K. The gut microbiota of laying hens and its manipulation with prebiotics and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 2020, 86, e00600-20. [Google Scholar] [CrossRef]
- Ricke, S.C.; Lee, S.I.; Kim, S.A.; Park, S.H.; Shi, Z. Prebiotics and the poultry gastrointestinal tract microbiome. Poult. Sci. 2020, 99, 670–677. [Google Scholar] [CrossRef]
- Jha, R.; Das, R.; Oak, S.; Mishra, P. Probiotics (direct-fed microbials) in poultry nutrition and their effects on nutrient utilization, growth and laying performance, and gut health: A systematic review. Animals 2020, 10, 1863. [Google Scholar] [CrossRef]
- Khan, S.; Chousalkar, K.K. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J. Anim. Sci. Biotechnol. 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Arif, M.; Sajjad, N.; Al-Ghadi, M.Q.; Alagawany, M.; Abd El-Hack, M.E.; Alhimaidi, A.R.; Elnesr, S.S.; Almutairi, B.O.; Amran, R.A.; et al. Dietary effect of probiotics and prebiotics on broiler performance, carcass, and immunity. Poult. Sci. 2020, 99, 6946–6953. [Google Scholar] [CrossRef]
- Sit, C.S.; Vederas, J.C. Approaches to the discovery of new antibacterial agents based on bacteriocins. Biochem. Cell Biol. 2008, 86, 116–123. [Google Scholar] [CrossRef]
- Alagawany, M.; Elnesr, S.S.; Farag, M.R.; Abd El-Hack, M.E.; Barkat, R.A.; Gabr, A.A.; Foda, M.A.; Noreldin, A.E.; Khafaga, A.F.; El-Sabrout, K.; et al. Potential role of important nutraceuticals in poultry performance and health—A comprehensive review. Res. Vet. Sci. 2021, 137, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Atterbury, R.J.; Van Bergen, M.A.; Ortiz, F.; Lovell, M.A.; Harris, J.A.; De Boer, A.; Wagenaar, J.A.; Allen, V.M.; Barrow, P.A. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 2007, 73, 4543–4549. [Google Scholar] [CrossRef]
- Smith, H.W. The use of live vaccines in experimental Salmonella gallinarum infection in chickens with observations on their interference effect. J. Hyg. 1956, 54, 419–432. [Google Scholar] [CrossRef]
- Desin, T.S.; Köster, W.; Potter, A.A. Salmonella vaccines in poultry: Past, present and future. Expert Rev. Vaccines 2013, 12, 87–96. [Google Scholar] [CrossRef]
- Crouch, C.F.; Pugh, C.; Patel, A.; Brink, H.; Wharmby, C.; Watts, A.; van Hulten, M.C.W.; de Vries, S.P.W. Reduction in intestinal colonization and invasion of internal organs after challenge by homologous and heterologous serovars of Salmonella enterica following vaccination of chickens with a novel trivalent inactivated Salmonella vaccine. Avian Pathol. 2020, 49, 666–677. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, H.S.; Yim, J.H.; Kim, Y.J.; Kim, D.H.; Chon, J.W.; Kim, H.; Om, A.S.; Seo, K.H. Comparison of the isolation rates and characteristics of Salmonella isolated from antibiotic-free and conventional chicken meat samples. Poult. Sci. 2017, 96, 2831–2838. [Google Scholar] [CrossRef]
- Cui, L.; Liu, Q.; Jiang, Z.; Song, Y.; Yi, S.; Qiu, J.; Hao, G.; Sun, S. Characteristics of Salmonella from chinese native chicken breeds fed on conventional or antibiotic-free diets. Front. Vet. Sci. 2021, 8, 607491. [Google Scholar] [CrossRef]
- Yin, X.; M’ikanatha, N.M.; Nyirabahizi, E.; McDermott, P.F.; Tate, H. Antimicrobial resistance in non-Typhoidal Salmonella from retail poultry meat by antibiotic usage-related production claims—United States, 2008–2017. Int. J. Food Microbiol. 2021, 342, 109044. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).