A New Sampling Approach for the Detection of Swine Influenza a Virus on European Sow Farms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
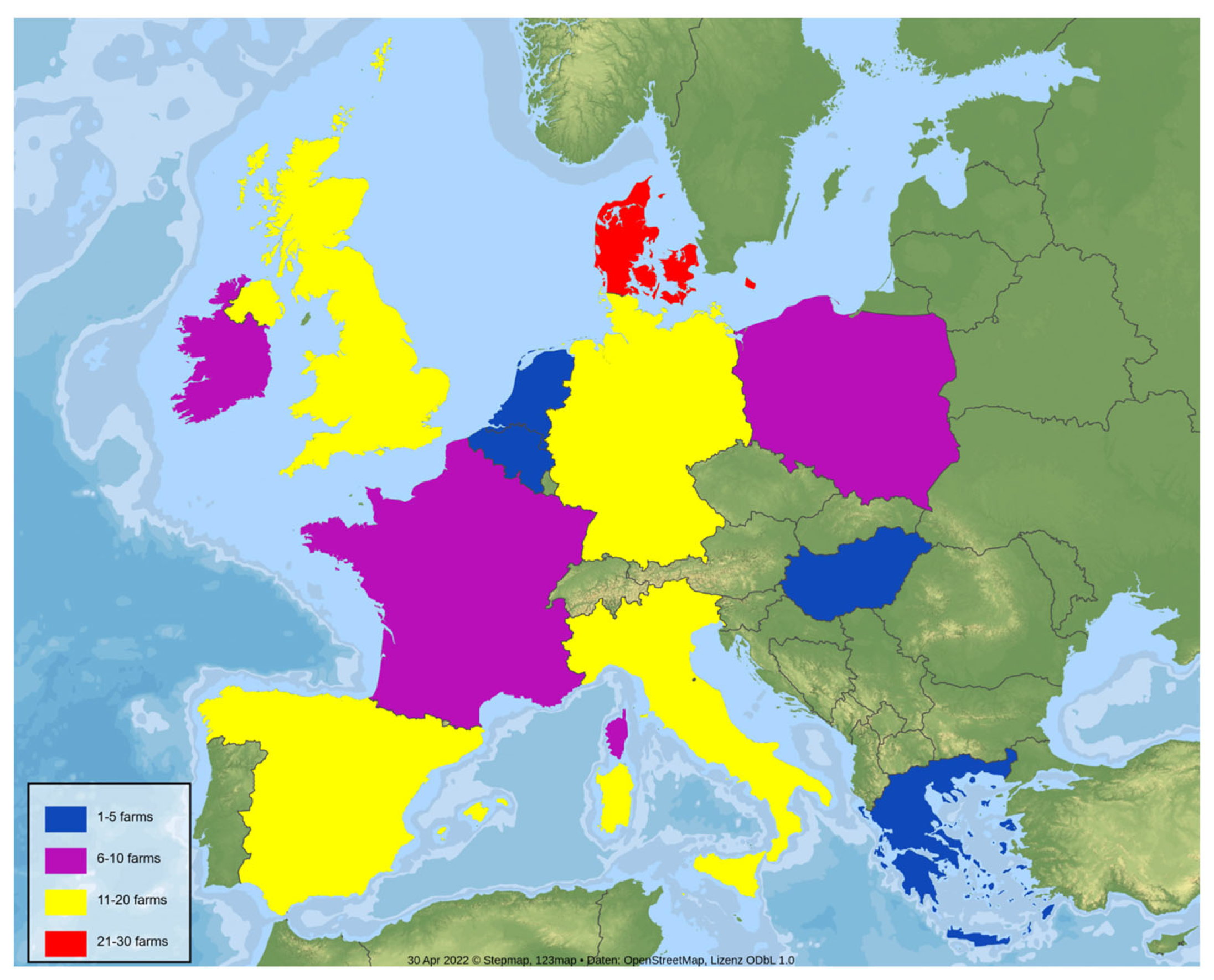
2.1. Farm Selection and Sampling Protocol
2.2. Statistical Analysis
3. Results
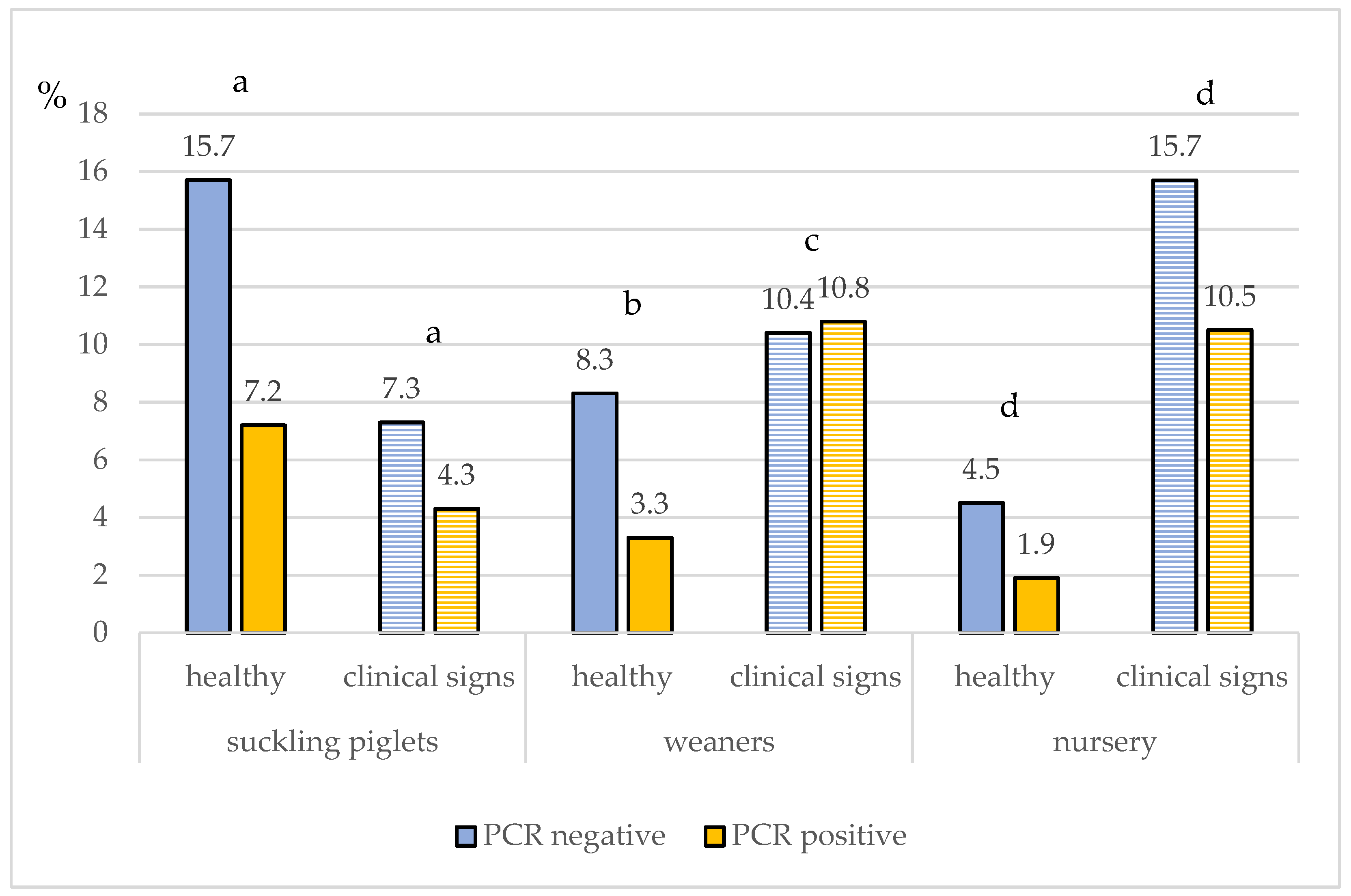
3.1. Clinical Signs
3.2. IAV rRT-PCR Results at Farm Level
3.3. IAV rRT-PCR Results on Sample Level
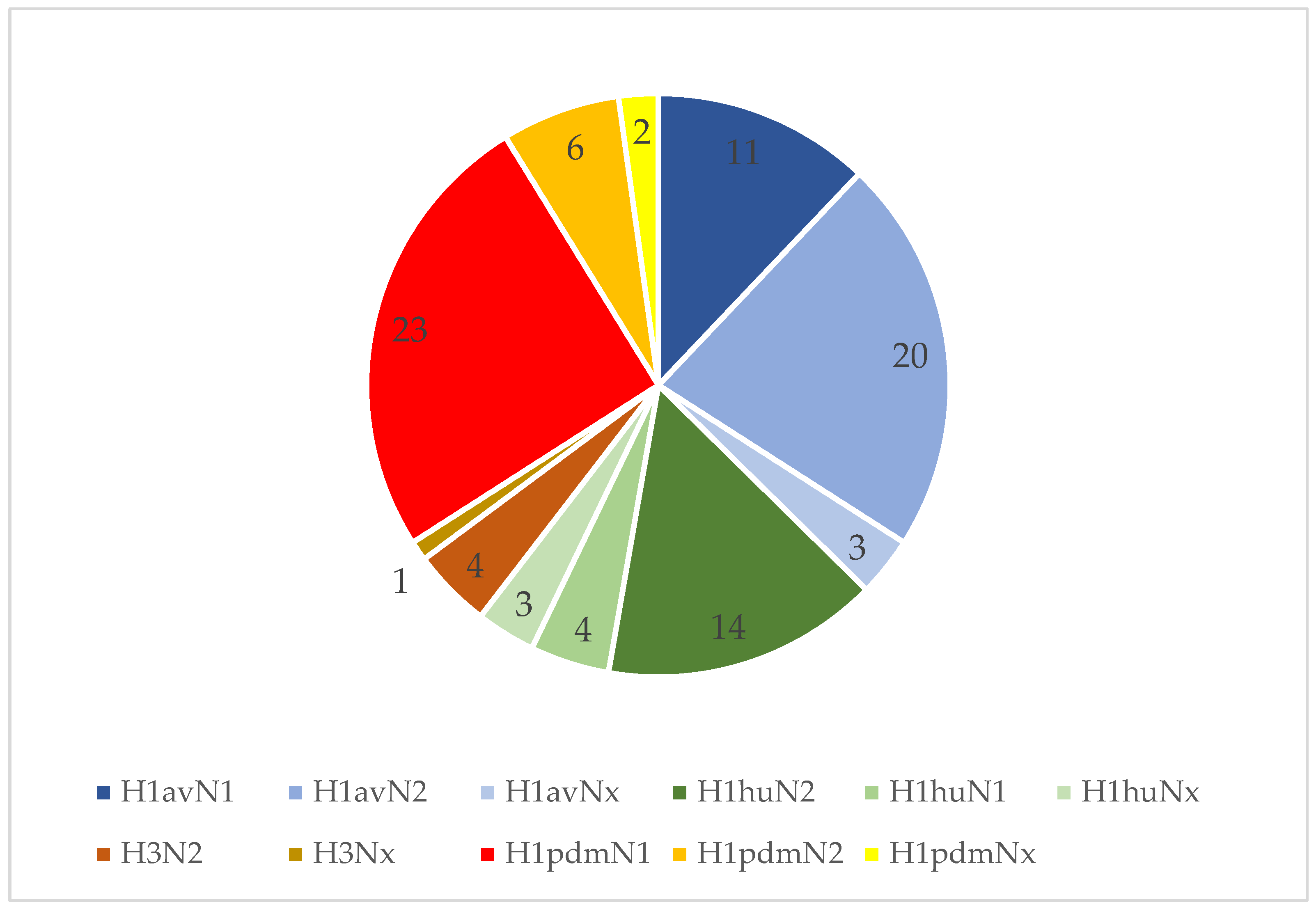
3.4. IAV Subtyping Results at Farm Level
3.5. IAV Subtyping Results on a Sample Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Er, C.; Skjerve, E.; Brun, E.; Hofmo, P.O.; Framstad, T.; Lium, B. Production Impact of Influenza A(H1N1)Pdm09 Virus Infection on Fattening Pigs in Norway1. J. Anim. Sci. 2016, 94, 751–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumbert, S.; Froehlich, S.; Rieger, A.; Stadler, J.; Ritzmann, M.; Zoels, S. Reproductive Performance of Pandemic Influenza A Virus Infected Sow Herds before and after Implementation of a Vaccine against the Influenza A (H1N1)Pdm09 Virus. Porc. Health Manag. 2020, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henritzi, D.; Petric, P.P.; Lewis, N.S.; Graaf, A.; Pessia, A.; Starick, E.; Breithaupt, A.; Strebelow, G.; Luttermann, C.; Parker, L.M.K.; et al. Surveillance of European Domestic Pig Populations Identifies an Emerging Reservoir of Potentially Zoonotic Swine Influenza A Viruses. Cell Host Microbe 2020, 28, 614–627.e6. [Google Scholar] [CrossRef] [PubMed]
- Scholtissek, C. Pigs as ‘Mixing Vessels’ for the Creation of New Pandemic Influenza A Viruses. Med. Princ. Pract. 1990, 2, 65–71. [Google Scholar] [CrossRef]
- Chastagner, A.; Hervé, S.; Quéguiner, S.; Hirchaud, E.; Lucas, P.; Gorin, S.; Béven, V.; Barbier, N.; Deblanc, C.; Blanchard, Y.; et al. Genetic and Antigenic Evolution of European Swine Influenza A Viruses of HA-1C (Avian-Like) and HA-1B (Human-Like) Lineages in France from 2000 to 2018. Viruses 2020, 12, 1304. [Google Scholar] [CrossRef]
- Everett, H.E.; Aramouni, M.; Coward, V.; Ramsay, A.; Kelly, M.; Morgan, S.; Tchilian, E.; Canini, L.; Woolhouse, M.E.J.; Gilbert, S.; et al. Vaccine-Mediated Protection of Pigs against Infection with Pandemic H1N1 2009 Swine Influenza A Virus Requires a Close Antigenic Match between the Vaccine Antigen and Challenge Virus. Vaccine 2019, 37, 2288–2293. [Google Scholar] [CrossRef]
- Deblanc, C.; Quéguiner, S.; Gorin, S.; Chastagner, A.; Hervé, S.; Paboeuf, F.; Simon, G. Evaluation of the Pathogenicity and the Escape from Vaccine Protection of a New Antigenic Variant Derived from the European Human-Like Reassortant Swine H1N2 Influenza Virus. Viruses 2020, 12, 1155. [Google Scholar] [CrossRef]
- Detmer, S.; Gramer, M.; Goyal, S.; Torremorell, M.; Torrison, J. Diagnostics and Surveillance for Swine Influenza. In Swine Influenza; Richt, J.A., Webby, R.J., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 370, pp. 85–112. ISBN 978-3-642-36870-7. [Google Scholar]
- Garrido-Mantilla, J.; Alvarez, J.; Culhane, M.; Nirmala, J.; Cano, J.P.; Torremorell, M. Comparison of Individual, Group and Environmental Sampling Strategies to Conduct Influenza Surveillance in Pigs. BMC Vet. Res. 2019, 15, 61. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Wayne, S.; Culhane, M.R.; Perez, A.; Allerson, M.; Torremorell, M. Effect of Strain-Specific Maternally-Derived Antibodies on Influenza A Virus Infection Dynamics in Nursery Pigs. PLoS ONE 2019, 14, e0210700. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Moreno, G.; Davies, P.; Yang, M.; Culhane, M.R.; Corzo, C.A.; Li, C.; Rendahl, A.; Torremorell, M. Evidence of Influenza A Infection and Risk of Transmission between Pigs and Farmworkers. Zoonoses Public Health 2022, 00, 1–12. [Google Scholar] [CrossRef]
- Loeffen, W.L.A.; Heinen, P.P.; Bianchi, A.T.J.; Hunneman, W.A.; Verheijden, J.H.M. Effect of Maternally Derived Antibodies on the Clinical Signs and Immune Response in Pigs after Primary and Secondary Infection with an Influenza H1N1 Virus. Vet. Immunol. Immunopathol. 2003, 92, 23–35. [Google Scholar] [CrossRef]
- Deblanc, C.; Hervé, S.; Gorin, S.; Cador, C.; Andraud, M.; Quéguiner, S.; Barbier, N.; Paboeuf, F.; Rose, N.; Simon, G. Maternally-Derived Antibodies Do Not Inhibit Swine Influenza Virus Replication in Piglets but Decrease Excreted Virus Infectivity and Impair Post-Infectious Immune Responses. Vet. Microbiol. 2018, 216, 142–152. [Google Scholar] [CrossRef]
- Ferreira, J.B.; Grgić, H.; Friendship, R.; Wideman, G.; Nagy, É.; Poljak, Z. Longitudinal Study of Influenza A Virus Circulation in a Nursery Swine Barn. Vet. Res. 2017, 48, 63. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Huang, C.; Shi, J.; Wang, R.; Sun, X.; Liu, X.; Zhao, L.; Jin, M. Investigation of Pathogenesis of H1N1 Influenza Virus and Swine Streptococcus Suis Serotype 2 Co-Infection in Pigs by Microarray Analysis. PLoS ONE 2015, 10, e0124086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomorska-Mól, M.; Dors, A.; Kwit, K.; Czyżewska-Dors, E.; Pejsak, Z. Coinfection Modulates Inflammatory Responses, Clinical Outcome and Pathogen Load of H1N1 Swine Influenza Virus and Haemophilus Parasuis Infections in Pigs. BMC Vet. Res. 2017, 13, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougon, J.; Deblanc, C.; Renson, P.; Quéguiner, S.; Gorin, S.; Mahé, S.; Le Dimna, M.; Barbier, N.; Paboeuf, F.; Simon, G.; et al. Successive Inoculations of Pigs with Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) and Swine H1N2 Influenza Virus Suggest a Mutual Interference between the Two Viral Infections. Viruses 2021, 13, 2169. [Google Scholar] [CrossRef] [PubMed]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; et al. Coinfections and Their Molecular Consequences in the Porcine Respiratory Tract. Vet. Res. 2020, 51, 80. [Google Scholar] [CrossRef] [PubMed]
- Duerrwald, R.; Schlegel, M.; Bauer, K.; Vissiennon, T.; Wutzler, P.; Schmidtke, M. Efficacy of Influenza Vaccination and Tamiflu® Treatment—Comparative Studies with Eurasian Swine Influenza Viruses in Pigs. PLoS ONE 2013, 8, e61597. [Google Scholar] [CrossRef] [Green Version]
- Simon-Grifé, M.; Martín-Valls, G.E.; Vilar, M.J.; Busquets, N.; Mora-Salvatierra, M.; Bestebroer, T.M.; Fouchier, R.A.; Martín, M.; Mateu, E.; Casal, J. Swine Influenza Virus Infection Dynamics in Two Pig Farms; Results of a Longitudinal Assessment. Vet. Res. 2012, 43, 24. [Google Scholar] [CrossRef] [Green Version]
- Henritzi, D.; Zhao, N.; Starick, E.; Simon, G.; Krog, J.S.; Larsen, L.E.; Reid, S.M.; Brown, I.H.; Chiapponi, C.; Foni, E.; et al. Rapid Detection and Subtyping of European Swine Influenza Viruses in Porcine Clinical Samples by Haemagglutinin- and Neuraminidase-specific Tetra- and Triplex Real-time RT—PCR s. Influenza Other Respir. Viruses 2016, 10, 504–517. [Google Scholar] [CrossRef] [Green Version]
- Takemae, N.; Shobugawa, Y.; Nguyen, P.T.; Nguyen, T.; Nguyen, T.N.; To, T.L.; Thai, P.D.; Nguyen, T.D.; Nguyen, D.T.; Nguyen, D.K.; et al. Effect of Herd Size on Subclinical Infection of Swine in Vietnam with Influenza A Viruses. BMC Vet. Res. 2016, 12, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, W. A Structured Approach for Analysing Survey Data and Making Useful Causal Inferences. Epidémiol. Et St. Anim. 1997, 8, 31–32. [Google Scholar]
- Ryt-Hansen, P.; Larsen, I.; Kristensen, C.S.; Krog, J.S.; Wacheck, S.; Larsen, L.E. Longitudinal Field Studies Reveal Early Infection and Persistence of Influenza A Virus in Piglets despite the Presence of Maternally Derived Antibodies. Vet. Res. 2019, 50, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mickey, R.M.; Greenland, S. The Impact of Confounder Selection Criteria on Effect Estimation. Am. J. Epidemiol. 1989, 129, 125–137. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression; Wiley Series in Probability and Mathematical Statistics; Wiley: New York, NY, USA, 1989; ISBN 978-0-471-61553-8. [Google Scholar]
- Reeth, K.V.; Labarque, G.; Pensaert, M. Serological Profiles after Consecutive Experimental Infections of Pigs with European H1N1, H3N2, and H1N2 Swine Influenza Viruses. Viral Immunol. 2006, 19, 373–382. [Google Scholar] [CrossRef]
- Unterweger, C.; Debeerst, S.; Klingler, E.; Auer, A.; Redlberger-Fritz, M.; Stadler, J.; Pesch, S.; Lillie-Jaschniski, K.; Ladinig, A. Herausforderungen bei der Influenzadiagnostik in einem Schweinebetrieb—Ein Fallbericht. Tierärztl. Prax. Ausg. G Großtiere Nutztiere 2021, 49, 425–431. [Google Scholar] [CrossRef]
- Goecke, N.B.; Krog, J.S.; Hjulsager, C.K.; Skovgaard, K.; Harder, T.C.; Breum, S.Ø.; Larsen, L.E. Subtyping of Swine Influenza Viruses Using a High-Throughput Real-Time PCR Platform. Front. Cell. Infect. Microbiol. 2018, 8, 165. [Google Scholar] [CrossRef]
- Ryt-Hansen, P.; Nielsen, H.G.; Sørensen, S.S.; Larsen, I.; Kristensen, C.S.; Larsen, L.E. The Role of Gilts in Transmission Dynamics of Swine Influenza Virus and Impacts of Vaccination Strategies and Quarantine Management. Porc. Health Manag. 2022, 8, 19. [Google Scholar] [CrossRef]
- Sarli, G.; D’Annunzio, G.; Gobbo, F.; Benazzi, C.; Ostanello, F. The Role of Pathology in the Diagnosis of Swine Respiratory Disease. Vet. Sci. 2021, 8, 256. [Google Scholar] [CrossRef]
- Hemmink, J.D.; Morgan, S.B.; Aramouni, M.; Everett, H.; Salguero, F.J.; Canini, L.; Porter, E.; Chase-Topping, M.; Beck, K.; Loughlin, R.M.; et al. Distinct Immune Responses and Virus Shedding in Pigs Following Aerosol, Intra-Nasal and Contact Infection with Pandemic Swine Influenza A Virus, A(H1N1)09. Vet. Res. 2016, 47, 103. [Google Scholar] [CrossRef]
- Pomorska-Mól, M.; Dors, A.; Kwit, K.; Kowalczyk, A.; Stasiak, E.; Pejsak, Z. Kinetics of Single and Dual Infection of Pigs with Swine Influenza Virus and Actinobacillus Pleuropneumoniae. Vet. Microbiol. 2017, 201, 113–120. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Schelkopf, A.; Allerson, M.; Morrison, R.; Culhane, M.; Perez, A.; Torremorell, M. Breed-to-Wean Farm Factors Associated with Influenza A Virus Infection in Piglets at Weaning. Prev. Vet. Med. 2018, 161, 33–40. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Allerson, M.; Culhane, M.; Morrison, R.; Davies, P.; Perez, A.; Torremorell, M. Effect of Influenza A Virus Sow Vaccination on Infection in Pigs at Weaning: A Prospective Longitudinal Study. Transbound. Emerg. Dis. 2021, 68, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Marthaler, D.; Culhane, M.; Sreevatsan, S.; Alkhamis, M.; Torremorell, M. Complete Genome Sequencing of Influenza A Viruses within Swine Farrow-to-Wean Farms Reveals the Emergence, Persistence, and Subsidence of Diverse Viral Genotypes. J. Virol. 2017, 91, e00745-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, J.J.H.; Torremorell, M.; Craft, M.E. Mathematical Modeling of Influenza A Virus Dynamics within Swine Farms and the Effects of Vaccination. PLoS ONE 2014, 9, e106177. [Google Scholar] [CrossRef]
- Rose, N.; Hervé, S.; Eveno, E.; Barbier, N.; Eono, F.; Dorenlor, V.; Andraud, M.; Camsusou, C.; Madec, F.; Simon, G. Dynamics of Influenza A Virus Infections in Permanently Infected Pig Farms: Evidence of Recurrent Infections, Circulation of Several Swine Influenza Viruses and Reassortment Events. Vet. Res. 2013, 44, 72. [Google Scholar] [CrossRef] [Green Version]
- Nirmala, J.; Perez, A.; Culhane, M.R.; Allerson, M.W.; Sreevatsan, S.; Torremorell, M. Genetic Variability of Influenza A Virus in Pigs at Weaning in Midwestern United States Swine Farms. Transbound. Emerg. Dis. 2021, 68, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.B.; Poljak, Z.; Friendship, R.; Nagy, É.; Wideman, G.; Grgić, H. Assessment of Exposure to Influenza A Viruses in Pigs between Weaning and Market Age. Vet. Res. 2021, 52, 60. [Google Scholar] [CrossRef] [PubMed]
- Allerson, M.; Deen, J.; Detmer, S.E.; Gramer, M.R.; Joo, H.S.; Romagosa, A.; Torremorell, M. The Impact of Maternally Derived Immunity on Influenza A Virus Transmission in Neonatal Pig Populations. Vaccine 2013, 31, 500–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cador, C.; Hervé, S.; Andraud, M.; Gorin, S.; Paboeuf, F.; Barbier, N.; Quéguiner, S.; Deblanc, C.; Simon, G.; Rose, N. Maternally-Derived Antibodies Do Not Prevent Transmission of Swine Influenza A Virus between Pigs. Vet. Res. 2016, 47, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido-Mantilla, J.; Sanhueza, J.; Alvarez, J.; Culhane, M.R.; Davies, P.; Allerson, M.W.; Torremorell, M. Impact of Nurse Sows on Influenza A Virus Transmission in Pigs under Field Conditions. Prev. Vet. Med. 2021, 188, 105257. [Google Scholar] [CrossRef]
- Meiners, C.; Loesken, S.; Doehring, S.; Starick, E.; Pesch, S.; Maas, A.; Noe, T.; Beer, M.; Harder, T.; Grosse Beilage, E. Field Study on Swine Influenza Virus (SIV) Infection in Weaner Pigs and Sows. Tierärztl. Prax. Ausg. G Großtiere Nutztiere 2014, 42, 351–359. [Google Scholar] [CrossRef] [Green Version]
- White, L.A.; Torremorell, M.; Craft, M.E. Influenza A Virus in Swine Breeding Herds: Combination of Vaccination and Biosecurity Practices Can Reduce Likelihood of Endemic Piglet Reservoir. Prev. Vet. Med. 2017, 138, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Grebe, K.M.; Yewdell, J.W.; Bennink, J.R. Heterosubtypic Immunity to Influenza A Virus: Where Do We Stand? Microbes Infect. 2008, 10, 1024–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poljak, Z.; Carman, S.; McEwen, B. Assessment of Seasonality of Influenza in Swine Using Field Submissions to a Diagnostic Laboratory in Ontario between 2007 and 2012. Influenza Other Respir. Viruses 2014, 8, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Grøntvedt, C.A.; Er, C.; Gjerset, B.; Hauge, A.G.; Brun, E.; Jørgensen, A.; Lium, B.; Framstad, T. Influenza A(H1N1)Pdm09 Virus Infection in Norwegian Swine Herds 2009/10: The Risk of Human to Swine Transmission. Prev. Vet. Med. 2013, 110, 429–434. [Google Scholar] [CrossRef] [PubMed]

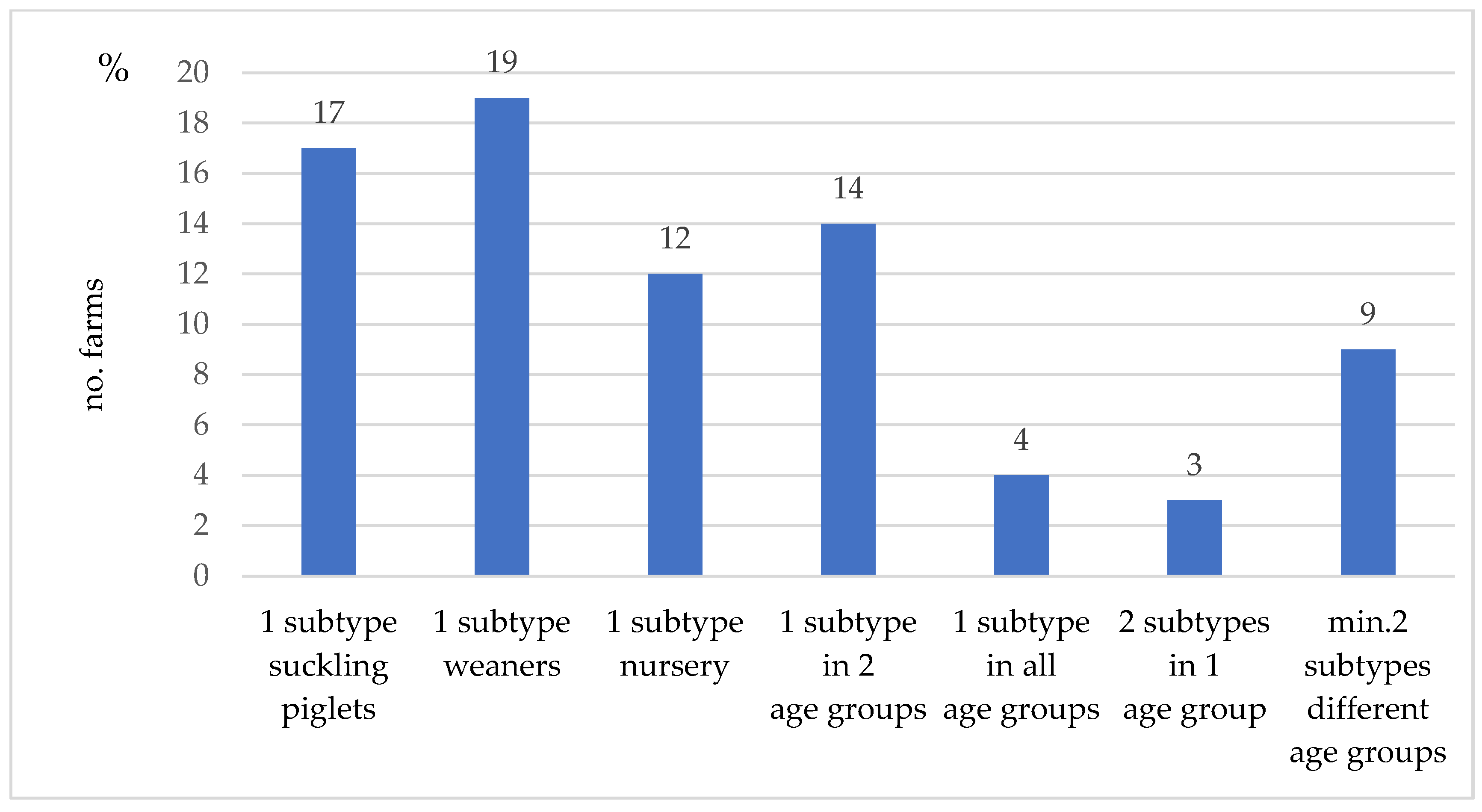
| Country | No. of Farms | No. of Positive Farms | Suckling Piglets | Weaners | Nursery |
|---|---|---|---|---|---|
| (1–4 w.o.a.) | (4–6 w.o.a) | (7–9 w.o.a) | |||
| No. of Positive Farms | No. of Positive Farms | No. of Positive Farms | |||
| Others * | 3 | 2 | 2 | 1 | 1 |
| Denmark | 26 | 23 | 12 | 17 | 12 |
| France | 10 | 6 | 2 | 4 | 3 |
| Germany | 20 | 17 | 11 | 13 | 10 |
| Ireland | 6 | 5 | 4 | 4 | 3 |
| Italy | 19 | 12 | 6 | 6 | 8 |
| Netherlands | 5 | 4 | 3 | 4 | 3 |
| Poland | 7 | 7 | 3 | 6 | 5 |
| Spain | 17 | 12 | 8 | 3 | 3 |
| UK | 18 | 15 | 6 | 10 | 14 |
| Total | 131 | 103 | 57 | 68 | 62 |
| Suckling Piglets | Weaners | Nursery | |
|---|---|---|---|
| (1–4 w.o.a.) | (4–6 w.o.a.) | (7–8 w.o.a) | |
| Country (No.) Farms | no. samples/ | no. samples/ | no. samples/ |
| no. positive | no. positive | no. positive | |
| Others (n = 3) * | 5/2 | 5/1 | 5/1 |
| Denmark (n = 26) | 52/17 | 52/31 | 51/19 |
| France (n = 10) | 20/4 | 19/7 | 20/3 |
| Germany (n = 20) | 41/16 | 36/18 | 36/16 |
| Ireland (n = 6) | 12/5 | 12/7 | 12/5 |
| Italy (n = 19) | 42/8 | 34/6 | 34/10 |
| Netherlands (n = 5) | 10/6 | 10/7 | 10/5 |
| Poland (n = 7) | 7/3 | 8/6 | 7/5 |
| Spain (n = 17) | 34/16 | 34/5 | 34/4 |
| UK (n = 18) | 36/9 | 36/18 | 36/25 |
| Total: 12 Countries | 259/86 a,b | 246/106 b | 245/93 a |
| Subtype | Suckling Piglets | Weaners | Nursery | Total |
|---|---|---|---|---|
| (1–4 w.o.a.) | (4–6 w.o.a.) | (7–9 w.o.a.) | ||
| H1avN1 | 4 | 8 | 2 | 14 |
| H1avN2 | 11 | 13 | 5 | 29 |
| H1avNx | 2 | 2 | 1 | 5 |
| H1huN2 | 5 | 4 | 14 | 23 |
| H1huN1 | 4 | 4 | 0 | 8 |
| H1huNx | 1 | 1 | 2 | 4 |
| H3N2 | 2 | 1 | 1 | 4 |
| H3Nx | 1 | 1 | 2 | 4 |
| H1pdmN1 | 9 | 11 | 8 | 28 |
| H1pdmN2 | 5 | 2 | 3 | 10 |
| H1pdmNx | 5 | 11 | 3 | 19 |
| total | 49 | 58 | 41 | 148 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lillie-Jaschniski, K.; Lisgara, M.; Pileri, E.; Jardin, A.; Velazquez, E.; Köchling, M.; Albin, M.; Casanovas, C.; Skampardonis, V.; Stadler, J. A New Sampling Approach for the Detection of Swine Influenza a Virus on European Sow Farms. Vet. Sci. 2022, 9, 338. https://doi.org/10.3390/vetsci9070338
Lillie-Jaschniski K, Lisgara M, Pileri E, Jardin A, Velazquez E, Köchling M, Albin M, Casanovas C, Skampardonis V, Stadler J. A New Sampling Approach for the Detection of Swine Influenza a Virus on European Sow Farms. Veterinary Sciences. 2022; 9(7):338. https://doi.org/10.3390/vetsci9070338
Chicago/Turabian StyleLillie-Jaschniski, Kathrin, Marina Lisgara, Emanuela Pileri, Agnes Jardin, Eduardo Velazquez, Monika Köchling, Michael Albin, Carlos Casanovas, Vassilis Skampardonis, and Julia Stadler. 2022. "A New Sampling Approach for the Detection of Swine Influenza a Virus on European Sow Farms" Veterinary Sciences 9, no. 7: 338. https://doi.org/10.3390/vetsci9070338
APA StyleLillie-Jaschniski, K., Lisgara, M., Pileri, E., Jardin, A., Velazquez, E., Köchling, M., Albin, M., Casanovas, C., Skampardonis, V., & Stadler, J. (2022). A New Sampling Approach for the Detection of Swine Influenza a Virus on European Sow Farms. Veterinary Sciences, 9(7), 338. https://doi.org/10.3390/vetsci9070338






