Molecular Diagnosis of Cetacean Morbillivirus in Beaked Whales Stranded in the Canary Islands (1999–2017)
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Nucleotide Identity
3.2. Phylogenetic Analyses
3.3. Histopathology, Immunohistochemistry
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Bressem, M.F.; Duignan, P.J.; Banyard, A.; Barbieri, M.; Colegrove, K.M.; de Guise, S.; di Guardo, G.; Dobson, A.; Domingo, M.; Fauquier, D.; et al. Cetacean Morbillivirus: Current Knowledge and Future Directions. Viruses 2014, 6, 5145–5181. [Google Scholar] [CrossRef] [PubMed]
- Domingo, M.; Visa, J.; Pumarola, M.; Marco, A.J.; Ferrer, L.; Rabanal, R.; Kennedy, S. Pathologic and Immunocytochemical Studies of Morbillivirus Infection in Striped Dolphins (Stenella coeruleoalba). Vet. Pathol. 1992, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, S. Morbillivirus Infections in Aquatic Mammals. J. Comp. Pathol. 1998, 119, 201–225. [Google Scholar] [CrossRef]
- Di Guardo, G.; Marruchella, G.; Agrimi, U.; Kennedy, S. Morbillivirus Infections in Aquatic Mammals: A Brief Overview. J. Vet. Med. Ser. A Physiol. Pathol. Clin. Med. 2005, 52, 88–93. [Google Scholar] [CrossRef]
- De Vries, R.D.; Paul Duprex, W.; De Swart, R.L. Morbillivirus Infections: An Introduction. Viruses 2015, 7, 699–706. [Google Scholar] [CrossRef] [Green Version]
- Gulland, F.M.D.; Dierauf, L.A.; Whitman, K.L. Section IV: Infectious diseases. In CRC Handbook of Marine Mammal Medicine, 3rd ed.; Gulland, F.M.D., Dierauf, L.A., Whitman, K.L., Eds.; CRC Press: New York, NY, USA, 2018; pp. 331–365. ISBN 9781498796873. [Google Scholar] [CrossRef]
- Visser, I.K.G.; Van Bressem, M.F.; De Swart, R.L.; Van de Bildt, M.W.G.; Vos, H.W.; Van der Heijden, R.W.J.; Saliki, J.T.; Orvell, C.; Kitching, P.; Kuiken, T.; et al. Characterization of Morbilliviruses Isolated from Dolphins and Porpoises in Europe. J. Gen. Virol. 1993, 74, 631–641. [Google Scholar] [CrossRef] [Green Version]
- Taubenberger, J.K.; Tsai, M.M.; Atkin, T.J.; Fanning, T.G.; Krafft, A.E.; Moeller, R.B.; Kodsi, S.E.; Mense, M.G.; Lipscomb, T.P. Molecular Genetic Evidence of a Novel Morbillivirus in a Long-Finned Pilot Whale (Globicephalus melas). Emerg. Infect. Dis. 2000, 6, 42–45. [Google Scholar] [CrossRef]
- West, K.L.; Sanchez, S.; Rotstein, D.; Robertson, K.M.; Dennison, S.; Levine, G.; Davis, N.; Schofield, D.; Potter, C.W.; Jensen, B. A Longman’s Beaked Whale (Indopacetus pacificus) Strands in Maui, Hawaii, with First Case of Morbillivirus in the Central Pacific. Mar. Mamm. Sci. 2013, 29, 767–776. [Google Scholar] [CrossRef]
- Groch, K.R.; Colosio, A.C.; Marcondes, M.C.C.; Zucca, D.; Díaz-Delgado, J.; Niemeyer, C.; Marigo, J.; Brandão, P.E.; Fernández, A.; Catão-Dias, J.L. Novel Cetacean Morbillivirus in Guiana Dolphin, Brazil. Emerg. Infect. Dis. 2014, 20, 511–513. [Google Scholar] [CrossRef] [Green Version]
- Stephens, N.; Duignan, P.J.; Wang, J.; Bingham, J.; Finn, H.; Bejder, L.; Patterson, I.A.P.; Holyoake, C. Cetacean Morbillivirus in Coastal Indo-Pacific Bottlenose Dolphins, Western Australia. Emerg. Infect. Dis. 2014, 20, 666–670. [Google Scholar] [CrossRef]
- West, K.L.; Silva-Krott, I.; Landrau-Giovannetti, N.; Rotstein, D.; Saliki, J.; Raverty, S.; Nielsen, O.; Popov, V.L.; Davis, N.; Walker, W.A.; et al. Novel Cetacean Morbillivirus in a Rare Fraser’s Dolphin (Lagenodelphis hosei) Stranding from Maui, Hawai‘i. Sci. Rep. 2021, 11, 15986. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; West, K.L.; Levine, G.; Sanchez, S.; Jensen, B.A. Initial Characterization of Novel Beaked Whale Morbillivirus in Hawaiian Cetaceans. Dis. Aquat. Organ. 2016, 117, 215–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrigue, C.; Oremus, M.; Dodémont, R.; Bustamante, P.; Kwiatek, O.; Libeau, G.; Lockyer, C.; Vivier, J.C.; Dalebout, M.L. A Mass Stranding of Seven Longman’s Beaked Whales (Indopacetus pacificus) in New Caledonia, South Pacific. Mar. Mamm. Sci. 2016, 32, 884–910. [Google Scholar] [CrossRef]
- Centelleghe, C.; Beffagna, G.; Palmisano, G.; Franzo, G.; Casalone, C.; Pautasso, A.; Giorda, F.; Di Nocera, F.; Iaccarino, D.; Santoro, M.; et al. Dolphin Morbillivirus in a Cuvier’s Beaked Whale (Ziphius cavirostris), Italy. Front. Microbiol. 2017, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.P.; Lopez-Jurado, L.F. Whales and the Military. Nature 1991, 351, 448. [Google Scholar] [CrossRef]
- Frantzis, A. Does Acoustic Testing Strand Whales? Nature 1998, 392, 29. [Google Scholar] [CrossRef]
- Jepson, P.; Arbelo, M.; Deaville, R.; Patterson, I.A.P.; Castro, P.; Baker, J.R.; Degollada, E.; Ross, H.M.; Herráez, P.; Pocknell, A.M.; et al. Gas-Bubble Lesions in Stranded Cetaceans. Was Sonar Responsible for a Spate of Whale Deaths after an Atlantic Military Exercise? Nature 2002, 425, 575576. [Google Scholar] [CrossRef]
- Fernández, A.; Edwards, J.F.; Rodríguez, F.; Espinosa de los Monteros, A.; Herráez, P.; Castro, P.; Jaber, J.R.; Martín, V.; Arbelo, M. “Gas and Fat Embolic Syndrome” Involving a Mass Stranding of Beaked Whales (Family Ziphiidae) Exposed to Anthropogenic Sonar Signals. Vet. Pathol. 2005, 42, 446–457. [Google Scholar] [CrossRef]
- Cox, T.M.; Ragen, T.J.; Read, A.J.; Vos, E.; Baird, R.W.; Balcomb, K.; Barlow, J.; Caldwell, J.; Cranford, T.; Crum, L.; et al. Understanding the Impacts of Anthropogenic Sound on Beaked Whales. J. Cetacean Res. Manag. 2006, 7, 177–187. [Google Scholar]
- Fernández, A.; Sierra, E.; Martín, V.; Méndez, M.; Sacchinni, S.; Bernaldo de Quirós, Y.; Andrada, M.; Rivero, M.; Quesada, O.; Tejedor, M.; et al. Last “Atypical” Beaked Whales Mass Stranding in the Canary Islands (July, 2004). J. Mar. Sci. Res. Dev. 2012, 2, 4–6. [Google Scholar] [CrossRef]
- Arbelo, M.; De Los Monteros, A.E.; Herráez, P.; Andrada, M.; Sierra, E.; Rodríguez, F.; Jepson, P.D.; Fernández, A. Pathology and Causes of Death of Stranded Cetaceans in the Canary Islands (1999–2005). Dis. Aquat. Organ. 2013, 103, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Bernaldo De Quirós, Y.; Fernandez, A.; Baird, R.W.; Brownell, R.L.; Aguilar De Soto, N.; Allen, D.; Arbelo, M.; Arregui, M.; Costidis, A.; Fahlman, A.; et al. Advances in Research on the Impacts of Anti-Submarine Sonar on Beaked Whales. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, A.; Arbelo, M.; Martín, V. No Mass Strandings since Sonar Ban. Nature 2013, 497, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Delgado, J.; Fernández, A.; Sierra, E.; Sacchini, S.; Andrada, M.; Vela, A.I.; Quesada-Canales, O.; Paz, Y.; Zucca, D.; Groch, K.; et al. Pathologic Findings and Causes of Death of Stranded Cetaceans in the Canary Islands (2006–2012). PLoS ONE 2018, 13, e0204444. [Google Scholar] [CrossRef] [Green Version]
- Sierra, E.; Fernández, A.; Espinosa De Los Monteros, A.; Arbelo, M.; Díaz-Delgado, J.; Andrada, M.; Herráez, P. Histopathological Muscle Findings May Be Essential for a Definitive Diagnosis of Suspected Sharp Trauma Associated with Ship Strikes in Stranded Cetaceans. PLoS ONE 2014, 9, e88780. [Google Scholar] [CrossRef]
- Puig-Lozano, R.; Fernández, A.; Sierra, E.; Saavedra, P.; Suárez-Santana, C.M.; De la Fuente, J.; Díaz-Delgado, J.; Godinho, A.; García-Álvarez, N.; Zucca, D.; et al. Retrospective Study of Fishery Interactions in Stranded Cetaceans, Canary Islands. Front. Vet. Sci. 2020, 7, 567258. [Google Scholar] [CrossRef]
- Puig-Lozano, R.; Fernández, A.; Saavedra, P.; Tejedor, M.; Sierra, E.; De la Fuente, J.; Xuriach, A.; Díaz-Delgado, J.; Rivero, M.A.; Andrada, M.; et al. Retrospective Study of Traumatic Intra-Interspecific Interactions in Stranded Cetaceans, Canary Islands. Front. Vet. Sci. 2020, 7, 107. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Delgado, J.; Fernández, A.; Xuriach, A.; Sierra, E.; Bernaldo de Quirós, Y.; Mompeo, B.; Pérez, L.; Andrada, M.; Marigo, J.; Catão-Dias, J.L.; et al. Verminous Arteritis Due to Crassicauda Sp. in Cuvier’s Beaked Whales (Ziphius cavirostris). Vet. Pathol. 2016, 53, 1233–1240. [Google Scholar] [CrossRef] [Green Version]
- Foster, G.; Whatmore, A.M.; Dagleish, M.P.; Baily, J.L.; Deaville, R.; Davison, N.J.; Koylass, M.S.; Perrett, L.L.; Stubberfield, E.J.; Reid, R.J.; et al. Isolation of Brucella Ceti from a Long-Finned Pilot Whale (Globicephala melas) and a Sowerby’s Beaked Whale (Mesoploden bidens). J. Wildl. Dis. 2015, 51, 868–871. [Google Scholar] [CrossRef]
- Davison, N.J.; Brownlow, A.; Doeschate, M.T.; Dale, E.J.; Foster, G.; Muchowski, J.; Perrett, L.L.; Rocchi, M.; Whatmore, A.M.; Dagleish, M.P. Neurobrucellosis Due to Brucella Ceti ST26 in Three Sowerby’s Beaked Whales (Mesoplodon bidens). J. Comp. Pathol. 2021, 182, 1–8. [Google Scholar] [CrossRef]
- Saliki, J.T.; Cooper, E.J.; Rotstein, D.S.; Caseltine, S.L.; Pabst, D.A.; McLellan, W.A.; Govett, P.; Harms, C.; Smolarek, K.A.; Romero, C.H.J. Novel Gammaherpesvirus Associated with Genital Lesions in a Blainville’s Beaked Whale (Mesoplodon densirostris). J. Wildl. Dis. 2006, 42, 142–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbelo, M.; Sierra, E.; Esperón, F.; Watanabe, T.T.N.; Bellière, E.N.; Espinosa de Los Monteros, A.; Fernández, A. Herpesvirus Infection with Severe Lymphoid Necrosis Affecting a Beaked Whale Stranded in the Canary Islands. Dis. Aquat. Organ. 2010, 89, 261–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbelo, M.; Bellière, E.N.; Sierra, E.; Sacchinni, S.; Esperón, F.; Andrada, M.; Rivero, M.; Diaz-Delgado, J.; Fernández, A. Herpes Virus Infection Associated with Interstitial Nephritis in a Beaked Whale (Mesoplodon densirostris). BMC Vet. Res. 2012, 8, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felipe-Jiménez, I.; Fernández, A.; Andrada, M.; Arbelo, M.; Segura-Göthlin, S.; Colom-Rivero, A.; Sierra, E. Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands. Animals 2021, 11, 1923. [Google Scholar] [CrossRef]
- Vela, A.I.; Fernandez, A.; Sánchez-Porro, C.; Sierra, E.; Mendez, M.; Arbelo, M.; Ventosa, A.; Domínguez, L.; Fernández-Garayzábal, J.F. Flavobacterium Ceti Sp. Nov., Isolated from Beaked Whales (Ziphius cavirostris). Int. J. Syst. Evol. Microbiol. 2007, 57, 2604–2608. [Google Scholar] [CrossRef] [Green Version]
- Canarias Conservación Cetacean Research Society. “Familia Ziphiidae”. Canarias Conservación Cetacean Research Society (2018). Available online: https://www.canariasconservacion.org/Zifios-Ziphiidae.htm (accessed on 29 June 2021).
- Esri; Garmin; GEBCO; NOAA; NGDC. ArcMap, Version: ArcGIS Desktop 10.8.1; Esri INC: Redlands, CA, USA, 2021.
- Jsseldijk, L.L.; Brownlow, A.C.; Mazzariol, S. European Best Practice on Cetacean Post Mortem Investigation and Tissue Sampling. In Proceedings of the Joint ASCOBANS/ACCOBAMS 2019, 25th Meeting of the Advisory Committee, Stralsund, Germany, 17–19 September 2019. [Google Scholar] [CrossRef]
- Kuiken, T.; García Hartmann, M. Cetacean pathology: Dissection techniques and tissue sampling. In ECS Newsletter 17—Special Issue, Proceedings of the First ECS Workshop on Cetacean Pathology, Leiden, The Netherlands, 13–14 September 1991; European Cetacean Society: Leiden, The Netherlands, 1991; pp. 1–39. [Google Scholar]
- Groch, K.R.; Díaz-Delgado, J.; Santos-Neto, E.B.; Ikeda, J.M.P.; Carvalho, R.R.; Oliveira, R.B.; Guari, E.B.; Flach, L.; Sierra, E.; Godinho, A.I.; et al. The Pathology of Cetacean Morbillivirus Infection and Comorbidities in Guiana Dolphins During an Unusual Mortality Event (Brazil, 2017–2018). Vet. Pathol. 2020, 57, 845–857. [Google Scholar] [CrossRef]
- Barrett, T.; Visser, I.K.G.; Mamaev, L.; Goatley, L.; Van Bressem, M.F.; Osterhaus, A.D.M.E. Dolphin and Porpoise Morbilliviruses Are Genetically Distinct from Phocine Distemper Virus. Virology 1993, 193, 1010–1012. [Google Scholar] [CrossRef]
- Reidarson, T.H.; McBain, J.; House, C.; King, D.P.; Stott, J.L.; Krafft, A.; Taubenberger, J.K.; Heyning, J.; Lipscomb, T.P. Morbillivirus Infection in Stranded Common Dolphins from the Pacific Ocean. J. Wildl. Dis. 1998, 34, 771–776. [Google Scholar] [CrossRef]
- Bellière, E.N.; Esperón, F.; Fernández, A.; Arbelo, M.; Muñoz, M.J.; Sánchez-Vizcaíno, J.M. Phylogenetic Analysis of a New Cetacean Morbillivirus from a Short-Finned Pilot Whale Stranded in the Canary Islands. Res. Vet. Sci. 2011, 90, 324–328. [Google Scholar] [CrossRef]
- Sacristán, C.; Carballo, M.; Muñoz, M.J.; Bellière, E.N.; Neves, E.; Nogal, V.; Esperón, F. Diagnosis of Cetacean Morbillivirus: A Sensitive One Step Real Time RT Fast-PCR Method Based on SYBR® Green. J. Virol. Methods 2015, 226, 25–30. [Google Scholar] [CrossRef]
- Groch, K.R.; Taniwaki, S.A.; Favero, C.M.; Brandão, P.E.; Díaz-Delgado, J.; Fernández, A.; Catão-Dias, J.L.; Sierra, E. A Novel Real-Time PCR to Detect Cetacean Morbillivirus in Atlantic Cetaceans. J. Virol. Methods 2020, 285, 113964. [Google Scholar] [CrossRef] [PubMed]
- Gröne, A.; Weckmann, M.T.; Capen, C.C.; Rosol, T.J. Canine Glyceraldehyde-3-Phosphate Dehydrogenase Complementary DNA: Polymerase Chain Reaction Amplification, Cloning, Partial Sequence Analysis, and Use as Loading Control in Ribonuclease Protection Assays. Am. J. Vet. Res. 1996, 57, 254–257. [Google Scholar] [PubMed]
- Kemper, C.M.; Tomo, I.; Bingham, J.; Bastianello, S.S.; Wang, J.; Gibbs, S.E.; Woolford, L.; Dickason, C.; Kelly, D. Morbillivirus-Associated Unusual Mortality Event in South Australian Bottlenose Dolphins Is Largest Reported for the Southern Hemisphere. R. Soc. Open Sci. 2016, 3, 160838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandevanter, D.R.; Warrener, P.; Bennett, L.; Schultz, E.R.; Coulter, S.; Garber, R.L.; Rose, T.M. Detection and Analysis of Diverse Herpesviral Species by Consensus Primer PCR. J. Clin. Microbiol. 1996, 34, 1666–1671. [Google Scholar] [CrossRef] [Green Version]
- BLAST. BLAST: Basic Local Alignment Search Tool. 2021. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 November 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zemtsova, G.E.; Montgomery, M.; Levin, M.L. Relative Sensitivity of Conventional and Real-Time PCR Assays for Detection of SFG Rickettsia in Blood and Tissue Samples from Laboratory Animals. PLoS ONE 2015, 10, e0116658. [Google Scholar] [CrossRef]
- Lipscomb, T.P.; Mense, M.G.; Habecker, P.L.; Taubenberger, J.K.; Schoelkopf, R. Morbilliviral Dermatitis in Seals. Vet. Pathol. 2001, 38, 724–726. [Google Scholar] [CrossRef] [Green Version]
- Dagleish, M.P.; Perri, A.; Maley, M.; Ballingall, K.T.; Baily, J.L.; Davison, N.J.; Brownlow, A.C.; Rocchi, M.S. Novel Dermatitis and Relative Viral Nucleic Acid Tissue Loads in a Fin Whale (Balaenoptera physalus) with Systemic Cetacean Morbillivirus Infection. J. Comp. Pathol. 2021, 183, 57–62. [Google Scholar] [CrossRef]
- Keck, N.; Kwiatek, O.; Dhermain, F.; Dupraz, F.; Boulet, H.; Danes, C.; Laprie, C.; Perrin, A.; Godenir, J.; Micout, L.; et al. Resurgence of Morbillivirus Infection in Mediterranean Dolphins off the French Coast. Vet. Rec. 2010, 166, 654–655. [Google Scholar] [CrossRef] [Green Version]
- Bellière, E.N.; Esperón, F.; Arbelo, M.; Muñoz, M.J.; Fernández, A.; Sánchez-Vizcaíno, J.M. Presence of Herpesvirus in Striped Dolphins Stranded during the Cetacean Morbillivirus Epizootic along the Mediterranean Spanish Coast in 2007. Arch. Virol. 2010, 155, 1307–1311. [Google Scholar] [CrossRef]
- Casalone, C.; Mazzariol, S.; Pautasso, A.; Di Guardo, G.; Di Nocera, F.; Lucifora, G.; Ligios, C.; Franco, A.; Fichi, G.; Cocumelli, C.; et al. Cetacean Strandings in Italy: An Unusual Mortality Event along the Tyrrhenian Sea Coast in 2013. Dis. Aquat. Organ. 2014, 109, 81–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, K.L.; Levine, G.; Jacob, J.; Jensen, B.; Sanchez, S.; Colegrove, K.; Rotstein, D. Coinfection and Vertical Transmission of Brucella and Morbillivirus in a Neonatal Sperm Whale (Physeter macrocephalus) in Hawaii, USA. J. Wildl. Dis. 2015, 51, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Delgado, J.; Groch, K.R.; Sierra, E.; Sacchini, S.; Zucca, D.; Quesada-Canales, Ó.; Arbelo, M.; Fernández, A.; Santos, E.; Ikeda, J.; et al. Comparative Histopathologic and Viral Immunohistochemical Studies on CeMV Infection among Western Mediterranean, Northeast-Central, and Southwestern Atlantic Cetaceans. PLoS ONE 2019, 14, e0213363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groch, K.R.; Santos-Neto, E.B.; Díaz-Delgado, J.; Ikeda, J.M.P.; Carvalho, R.R.; Oliveira, R.B.; Guari, E.B.; Bisi, T.L.; Azevedo, A.F.; Lailson-Brito, J.; et al. Guiana Dolphin Unusual Mortality Event and Link to Cetacean Morbillivirus, Brazil. Emerg. Infect. Dis. 2018, 24, 1349–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierra, E.; Sánchez, S.; Saliki, J.T.; Blas-Machado, U.; Arbelo, M.; Zucca, D.; Fernández, A. Retrospective Study of Etiologic Agents Associated with Nonsuppurative Meningoencephalitis in Stranded Cetaceans in the Canary Islands. J. Clin. Microbiol. 2014, 52, 2390–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierra, E.; Zucca, D.; Arbelo, M.; García-álvarez, N.; Andrada, M.; Déniz, S.; Fernández, A. Fatal Systemic Morbillivirus Infection in Bottlenose Dolphin, Canary Islands, Spain. Emerg. Infect. Dis. 2014, 20, 269–271. [Google Scholar] [CrossRef]
- Sierra, E.; Fernández, A.; Suárez-Santana, C.; Xuriach, A.; Zucca, D.; Bernaldo De Quirós, Y.; García-Álvarez, N.; De La Fuente, J.; Sacchini, S.; Andrada, M.; et al. Morbillivirus and Pilot Whale Deaths, Canary Islands, Spain, 2015. Emerg. Infect. Dis. 2016, 22, 740–742. [Google Scholar] [CrossRef]
- Bento, M.C.R. de M.; Eira, C.I.C.S.; Vingada, J.V.; Marçalo, A.L.; Ferreira, M.C.T.; Fernandez, A.L.; Tavares, L.M.M.; Duarte, A.I. New Insight into Dolphin Morbillivirus Phylogeny and Epidemiology in the Northeast Atlantic: Opportunistic Study in Cetaceans Stranded along the Portuguese and Galician Coasts. BMC Vet. Res. 2016, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Bossart, G.D.; Reif, J.S.; Schaefer, A.M.; Goldstein, J.; Fair, P.A.; Saliki, J.T. Morbillivirus Infection in Free-Ranging Atlantic Bottlenose Dolphins (Tursiops truncatus) from the Southeastern United States: Seroepidemiologic and Pathologic Evidence of Subclinical Infection. Vet. Microbiol. 2010, 143, 160–166. [Google Scholar] [CrossRef]
- Sierra, E.; Fernández, A.; Zucca, D.; Câmara, N.; Felipe-Jiménez, I.; Suárez-Santana, C.; Bernaldo de Quirós, Y.; Díaz-Delgado, J.; Arbelo, M. Morbillivirus Infection in Risso’s Dolphin Grampus griseus: A Phylogenetic and Pathological Study of Cases from the Canary Islands. Dis. Aquat. Organ. 2018, 129, 165–174. [Google Scholar] [CrossRef]
- Fauquier, D.A.; Litz, J.; Sanchez, S.; Colegrove, K.; Schwacke, L.H.; Hart, L.; Saliki, J.; Smith, C.; Goldstein, T.; Bowen-Stevens, S.; et al. Evaluation of Morbillivirus Exposure in Cetaceans from the Northern Gulf of Mexico 2010–2014. Endanger. Species Res. 2017, 33, 211–220. [Google Scholar] [CrossRef] [Green Version]
- Mazzariol, S.; Centelleghe, C.; Beffagna, G.; Povinelli, M.; Terracciano, G.; Cocumelli, C.; Pintore, A.; Denurra, D.; Casalone, C.; Pautasso, A.; et al. Mediterranean Fin Whales (Balaenoptera physalus) Threatened by Dolphin Morbillivirus. Emerg Infect Dis. 2016, 22, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Profeta, F.; Di Francesco, C.E.; Marsilio, F.; Mignone, W.; Di Nocera, F.; De Carlo, E.; Lucifora, G.; Pietroluongo, G.; Baffoni, M.; Cocumelli, C.; et al. Retrospective Seroepidemiological Investigations against Morbillivirus, Toxoplasma gondii and Brucella Spp. in Cetaceans Stranded along the Italian Coastline (1998–2014). Res. Vet. Sci. 2015, 101, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Serrano, L.; Simeone, C.A.; Colegrove, K.M.; Duignan, P.J.; Goldstein, T.; Gulland, F.M.D. Cetacean Morbillivirus in Odontocetes Stranded along the Central California Coast, USA, 2000–2015. J. Wildl. Dis. 2017, 53, 386–392. [Google Scholar] [CrossRef]
- Groch, K.R.; Groch, K.R.; Kolesnikovas, C.K.M.; de Castilho, P.V.; Moreira, L.M.P.; Barros, C.R.M.B.; Morais, C.R.; Renault-Braga, E.P.; Sierra, E.; Fernandez, A.; et al. Cetacean Morbillivirus in Southern Right Whales, Brazil. Transbound. Emerg. Dis. 2019, 66, 606–610. [Google Scholar] [CrossRef] [Green Version]
- Stone, B.M.; Blyde, D.J.; Saliki, J.T.; Morton, J.M. Morbillivirus Infection in Live Stranded, Injured, Trapped, and Captive Cetaceans in Southeastern Queensland and Northern New South Wales, Australia. J. Wildl. Dis. 2012, 48, 47–55. [Google Scholar] [CrossRef]
- Domingo, M.; Vilafranca, M.; Visa, J.; Prats, N.; Trudgett, A.; Visser, I. Evidence for Chronic Morbillivirus Infection in the Mediterranean Striped Dolphin (Stenella coeruleoalba). Vet. Microbiol. 1995, 44, 229–239. [Google Scholar] [CrossRef]
- Fernández, A.; Espero, F.; Herraéz, P.; Espinosa de Los Monteros, A.; Clavel, C.; Bernabé, A.; Sánchez-Vizcaino, J.M.; Verborgh, P.; DeStephanis, R.; Toledano, F.; et al. Morbillivirus and Pilot Whale Deaths, Mediterranean Sea. Emerg. Infect. Dis. 2008, 14, 792–794. [Google Scholar] [CrossRef]
- Jo, W.K.; Kruppa, J.; Habierski, A.; van de Bildt, M.; Mazzariol, S.; Di Guardo, G.; Siebert, U.; Kuiken, T.; Jung, K.; Osterhaus, A.; et al. Evolutionary Evidence for Multi-Host Transmission of Cetacean Morbillivirus. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Raga, J.; Banyard, A.; Domingo, M.; Corteyn, M.; Fernández, M.; Aznar, F.; Barrett, T. Dolphin Morbillivirus Epizootic Resurgence, Mediterranean Sea. Emerg. Infect. Dis. 2008, 14, 471–473. [Google Scholar] [CrossRef]
- Soto, S.; Marco, A.; Domingo, M.; González, R.; Alegre, F.; González, B.; Medina, P.; Raga, J.A. Epizootic of Dolphin Morbillivirus on the Catalonian Mediterranean Coast in 2007. Vet. Rec. 2011, 169, 102. [Google Scholar] [CrossRef] [PubMed]
- Van Bressem, M.F.; Raga, J.; Di Guardo, G.; Jepson, P.; Duignan, P.; Siebert, U.; Barrett, T.; Santos, M.; Moreno, I.; Siciliano, S.; et al. Emerging Infectious Diseases in Cetaceans Worldwide and the Possible Role of Environmental Stressors. Dis. Aquat. Organ. 2009, 86, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, G.; Di Francesco, C.E.; Eleni, C.; Cocumelli, C.; Scholl, F.; Casalone, C.; Peletto, S.; Mignone, W.; Tittarelli, C.; Di Nocera, F.; et al. Morbillivirus Infection in Cetaceans Stranded along the Italian Coastline: Pathological, Immunohistochemical and Biomolecular Findings. Res. Vet. Sci. 2013, 94, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Guerri, C.; Melero, M.; Esperón, F.; Bellière, E.N.; Arbelo, M.; Crespo, J.L.; Sierra, E.; García-Párraga, D.; Sánchez-Vizcaíno, J.M. Unusual Striped Dolphin Mass Mortality Episode Related to Cetacean Morbillivirus in the Spanish Mediterranean Sea. BMC Vet. Res. 2013, 9, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzariol, S.; Centelleghe, C.; Provvido, A.D.; Di Renzo, L.; Cardeti, G.; Cersini, A.; Fichi, G.; Petrella, A.; Esmeralda, C.; Francesco, D.; et al. Dolphin Morbillivirus Associated with the Mass Stranding of Sperm Whales, Italy. Emerg. Infect. Dis. 2017, 23, 144–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pautasso, A.; Iulini, B.; Grattarola, C.; Giorda, F.; Goria, M.; Peletto, S.; Masoero, L.; Mignone, W.; Varello, K.; Petrella, A.; et al. Novel Dolphin Morbillivirus (DMV) Outbreak among Mediterranean Striped Dolphins Stenella coeruleoalba in Italian Waters. Dis. Aquat. Organ. 2018, 132, 215–220. [Google Scholar] [CrossRef]
- Rubio-Guerri, C.; Jiménez, M.Á.; Melero, M.; Díaz-Delgado, J.; Sierra, E.; Arbelo, M.; Bellière, E.N.; Crespo-Picazo, J.L.; García-Párraga, D.; Esperón, F.; et al. Genetic Heterogeneity of Dolphin Morbilliviruses Detected in the Spanish Mediterranean in Inter-Epizootic Period. BMC Vet. Res. 2018, 14, 248. [Google Scholar] [CrossRef]
- Mira, F.; Rubio-Guerri, C.; Purpari, G.; Puleio, R.; Caracappa, G.; Gucciardi, F.; Russotto, L.; Loria, G.R.; Guercio, A. Circulation of a Novel Strain of Dolphin Morbillivirus (DMV) in Stranded Cetaceans in the Mediterranean Sea. Sci. Rep. 2019, 9, 9792. [Google Scholar] [CrossRef]
- Cerutti, F.; Giorda, F.; Grattarola, C.; Mignone, W.; Beltramo, C.; Keck, N.; Lorusso, A.; Di Francesco, G.; Di Renzo, L.; Di Guardo, G.; et al. Specific Capture and Whole-Genome Phylogeography of Dolphin Morbillivirus. Sci. Rep. 2020, 10, 20831. [Google Scholar] [CrossRef]
- Duignan, P.J.; House, C.; Geraci, J.R.; Early, G.; Copland, H.G.; Walsh, M.T.; Bossart, G.D.; Cray, C.; Sadove, S.; Aubin, D.J.S.; et al. Morbillivirus Infection in Two Species of Pilot Whale (Globicephala Sp.) From the Western Atlantic. Mar Mamm. Sci. 1995, 11, 150–162. [Google Scholar] [CrossRef]
- Macleod, C.; Perrin, W.; Pitman, R.; Barlow, J.; Ballance, L.; D’Amico, A.; Gerrodette, T.; Joyce, G.; Mullin, K.; Palka, D. Known and Inferred Distributions of Beaked Whale Species (Cetacea: Ziphiidae). J. Cetacean Res. Manag. 2005, 7, 271. [Google Scholar]
- Blixenkrone-Møller, M.; Bolt, G.; Jensen, T.D.; Harder, T.; Svansson, V. Comparative Analysis of the Attachment Protein Gene (H) of Dolphin Morbillivirus. Virus Res. 1996, 40, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Jo, W.K.; Grilo, M.L.; Wohlsein, P.; Andersen-Ranberg, E.U.; Hansen, M.S.; Kinze, C.C.; Hjulsager, C.K.; Olsen, M.T.; Lehnert, K.; Prenger-Berninghoff, E.; et al. Dolphin Morbillivirus in a Fin Whale (Balaenoptera physalus) in Denmark, 2016. J. Wildl. Dis. 2017, 53, 921–924. [Google Scholar] [CrossRef] [PubMed]
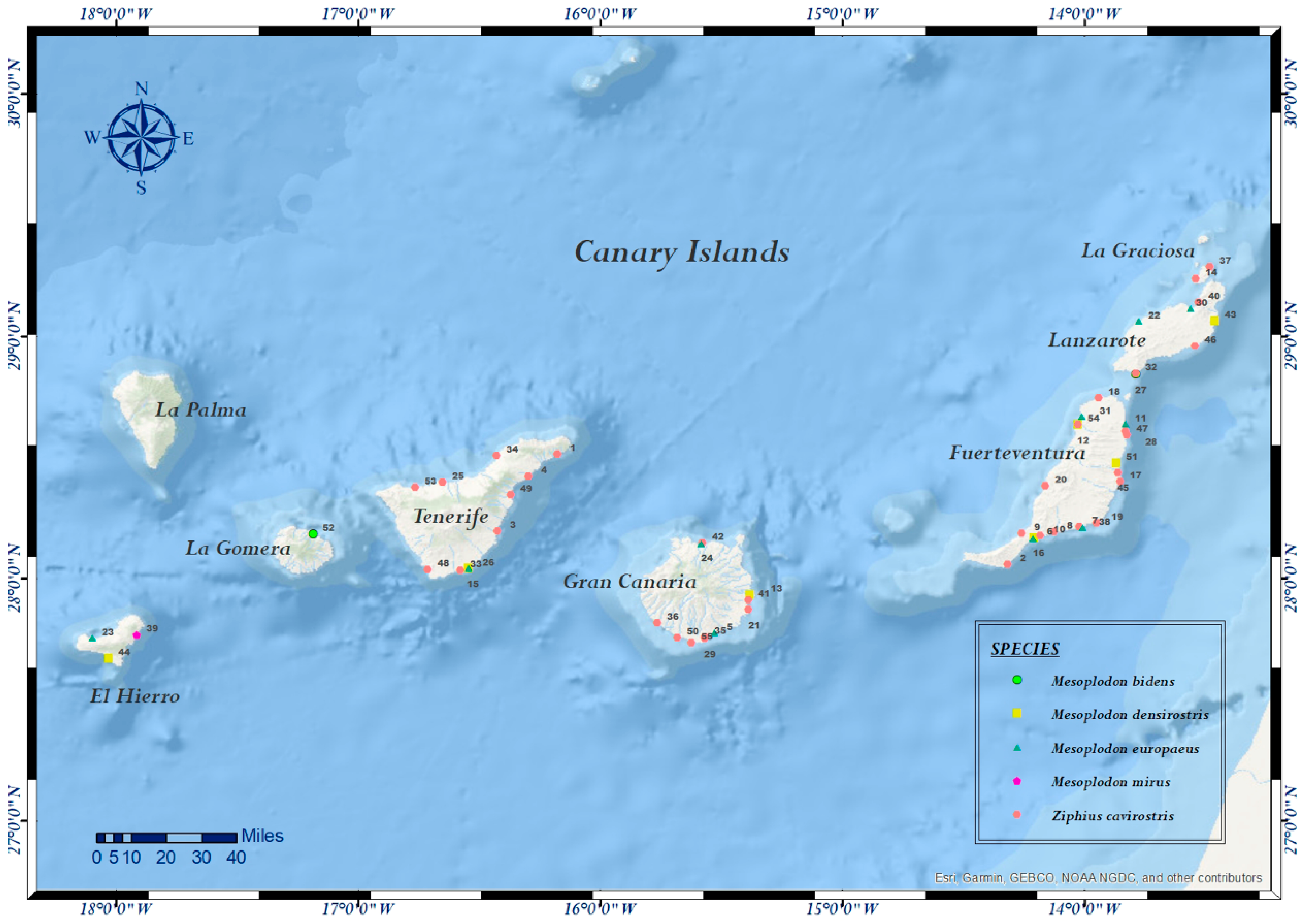
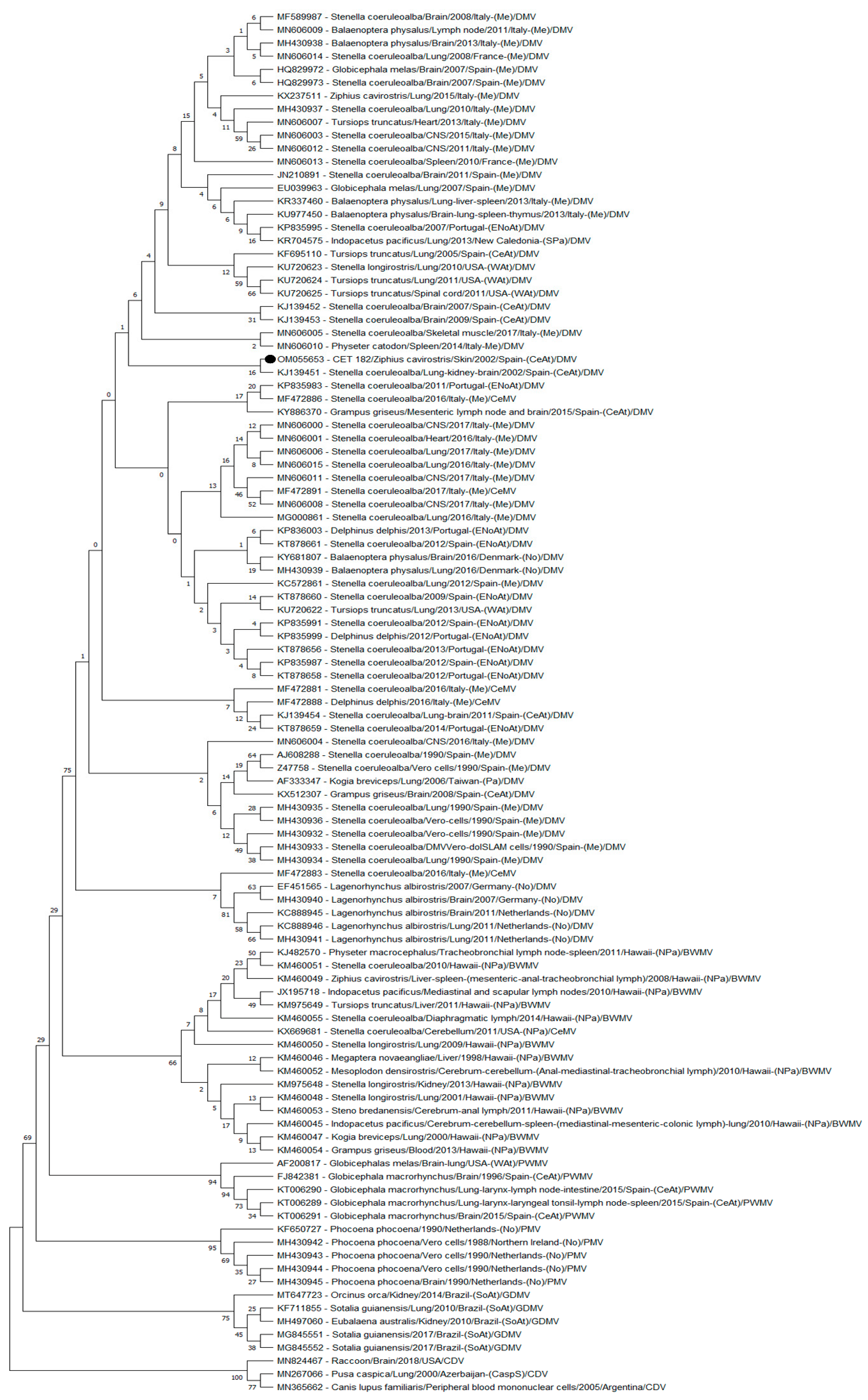

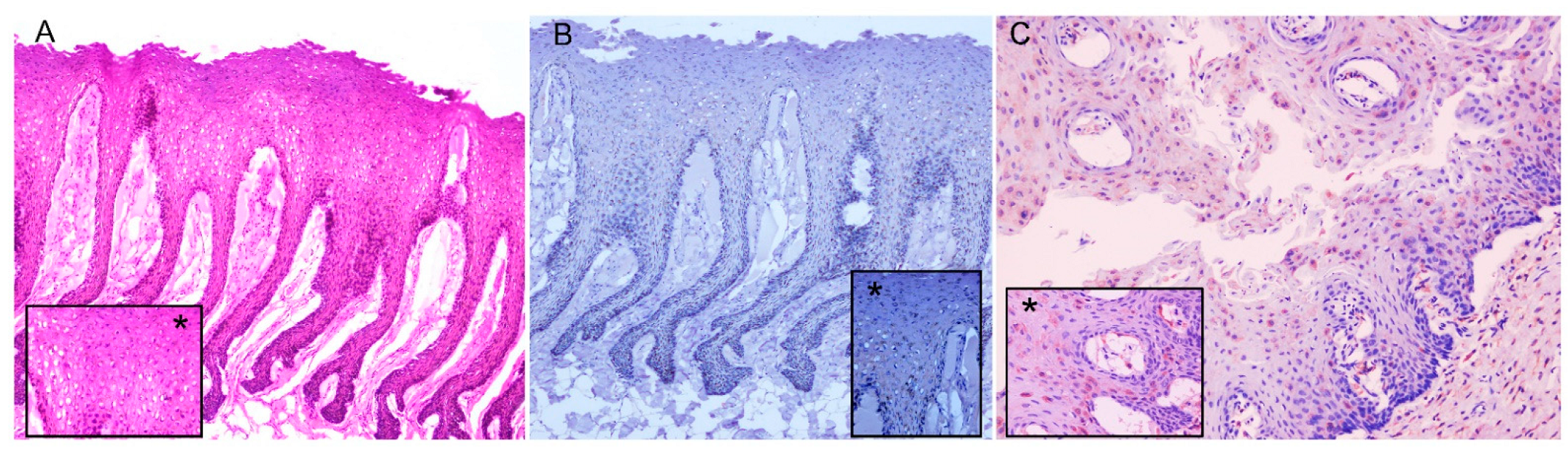
| Case Nº | ID Code | Species | Sex | Age | SD | SL | Coordinates | SS | DC | NS | Tested Samples | PCR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CET 86 | Z. c. | F | A | 27/11/1999 | TNF | 28.513067382968398, –16.176165295752952 | A | 3 | G | Skin, lung, liver, kidney | C, F |
| 2 | CET 103 | Z. c. | M | J | 19/04/2000 | FTV | 28.056538132454694, –14.317684331100672 | D | 3 | G | Lung, liver, kidney, brain | C, F |
| 3 | CET 108 | Z. c. | F | A | 10/06/2000 | TNF | 28.195070013507404,–16.42181088596981 | D | 3 | G | Skin, skeletal muscle, lung, liver, kidney | C, F |
| 4 | CET 113 | Z. c. | F | S | 16/07/2000 | TNF | 28.420528319456402, –16.295411555275532 | D | 3 | P | Skin, skeletal muscle | C, F, P |
| 5 | CET 134 | M. e. | F | C | 28/06/2001 | GC | 27.77761716736704, –15.52674193489951 | A | 1 | P | Skin, lung, liver, kidney, brain | C, F, P |
| 6 | CET 180 | M.d. | F | A | 24/09/2002 | FTV | 28.167568704666728, –14.206111760665271 | A | 2 | NE | Skin | C, F, P |
| 7 | CET 181 | Z. c. | M | S | 24/09/2002 | FTV | 28.211852124234827, –14.019063839127336 | A | 2 | NE | Skin, skeletal muscle, lung, mediastinal and mesenteric lymph node, liver, kidney, brain, spleen | C, F |
| 8 | CET 182 | Z. c. | M | S | 24/09/2002 | FTV | 28.19146832713967, –14.122994013994195 | D | 2 | NE | Skin, skeletal muscle, lung, liver, mesenteric lymph node, kidney, brain, spleen | C, F, P |
| 9 | CET 183 | Z. c. | M | S | 24/09/2002 | FTV | 28.186564312428384, –14.257397680615142 | D | 2 | NE | Skin, skeletal muscle, lung, liver, mesenteric lymph node, kidney, brain | C, F, P |
| 10 | CET 184 | Z. c. | M | S | 24/09/2002 | FTV | 28.175274186060737, –14.183223454614003 | D | 2 | NE | Skin, skeletal muscle, lung, liver, mediastinal and mesenteric lymph node, kidney, brain, spleen, thyroid | C, F, P |
| 11 | CET 185 | M. e. | F | A | 24/09/2002 | FTV | 28.638713939591398, –13.830348964499422 | D | 2 | NE | Skin, skeletal muscle, lung, liver, mesenteric lymph node, kidney, brain, spleen | C, F, P |
| 12 | CET 189 | Z. c. | F | A | 27/09/2002 | FTV | 28.63498925367787, –14.026753961358423 | D | 4 | NE | Skin, lung, liver, kidney | C, F, P |
| 13 | CET 213 | M. d. | F | A | 28/06/2003 | GC | 27.933119723146724, –15.37973786995963 | A | 1 | P | Skin, skeletal muscle, lung, liver, kidney, brain | C, F |
| 14 | CET 236 | Z. c. | F | C | 21/03/2004 | LGr | 29.23649579711158, –13.538613817871068 | D | 3 | G | Skin, skeletal muscle, lung, liver, kidney, brain | C, F |
| 15 | CET 243 | M. d. | M | A | 18/04/2004 | TNF | 28.040710105511895, –16.5426071324846 | A | 1 | P | Skin, skeletal muscle, lung, liver, kidney | C, F |
| 16 | CET 259 | M. e. | F | J | 21/06/2004 | FTV | 28.164953252754323, –14.212321787452456 | D | 2 | P | Skin, skeletal muscle, lung, liver, kidney, brain | C, F, P |
| 17 | CET 264 | Z. c. | F | ND | 23/07/2004 | FTV | 28.40069477695754, –13.852448567548068 | D | 4 | G | Liver, skeletal muscle, lung, kidney | C, F |
| 18 | CET 265 | Z. c. | M | A | 24/07/2004 | FTV | 28.744473643139884, –13.940991510227075 | D | 4 | G | Skin, skeletal muscle, lung, liver, kidney | C, F |
| 19 | CET 294 | Z. c. | F | A | 18/04/2005 | FTV | 28.228156055155715, –13.949995729432887 | D | 4 | G | Skin, skeletal muscle, lung, liver, spleen | C, F |
| 20 | CET 304 | Z. c. | F | C | 13/07/2005 | FTV | 28.38034460360221, –14.161693075458741 | D | 2 | P | Skin, skeletal muscle, lung, liver, kidney | C, F |
| 21 | CET 322 | Z. c. | M | A | 17/02/2006 | GC | 27.870618611481834, –15.38656422608156 | D | 4 | I | Skin, lung, liver, kidney | C, F |
| 22 | CET 333 | M. e. | F | S | 28/03/2006 | EH | 29.06227107414346, –13.774638588905537 | A | 2 | G | Skin, skeletal muscle, lung, thymus, liver, mesenteric lymph node, kidney, brain, spleen | C, F |
| 23 | CET 334 | M. e. | F | S | 28/03/2006 | EH | 27.755578877787087, –18.09553359589475 | A | 2 | G | Skin, skeletal muscle, lung, liver, kidney, brain, spleen | C, F |
| 24 | CET 338 | M. e. | F | J | 06/04/2006 | GC | 28.14436864892595, –15.581793297520823 | D | 2 | P | Skin, skeletal muscle, lung, liver, blood, mesenteric lymph node, kidney, brain, spleen | C, F |
| 25 | CET 352 | Z. c. | ND | J | 06/07/2006 | TNF | 28.396221323032027,–16.648897697794496 | D | 3 | P | Skin, lung, liver, kidney, brain, spleen | P |
| 26 | CET 354 | M. e. | M | C | 28/07/2006 | TNF | 28.040710105511895, –16.5426071324846 | D | 4 | M | Skin, lung, liver, kidney, spleen | P |
| 27 | CET 379 | M. b. | M | A | 16/04/2007 | LZ | 28.842800242252817, –13.788144917745738 | D | 2 | E | Skin, lung, liver, kidney, brain, spleen | P |
| 28 | CET 471 | Z. c. | F | S | 06/11/2008 | FTV | 28.607731990351333, –13.8297794478201 | D | 2 | G | Lung, kidney, brain, spleen | P |
| 29 | CET 503 | Z. c. | F | A | 21/09/2009 | GC | 27.750297975375215, –15.568322782893084 | D | 4 | I | Skin, lung, kidney, liver | P |
| 30 | CET 510 | M. e. | M | A | 14/12/2009 | LZ | 29.11583432185095, –13.560196813234894 | D | 2 | E | Skin, lung, liver, mesenteric lymph node, kidney, brain | P |
| 31 | CET 547 | M. e. | M | A | 29/08/2010 | FTV | 28.667401632579352, –14.011239799734405 | D | 4 | M | Skin, lung, liver, mesenteric lymph node, kidney, brain | P |
| 32 | CET 576 | Z. c. | F | A | 16/05/2011 | LZ | 28.84666379719735, –13.788277756973725 | D | 2 | G | Lung, kidney, brain, spleen | F, P |
| 33 | CET 579 | Z. c. | M | S | 13/06/2011 | TNF | 28.031868453191404, –16.575344550886005 | D | 4 | M | Skin, lung, mesenteric lymph node, liver, kidney, brain, spleen | P |
| 34 | CET 591 | Z. c. | F | A | 01/11/2011 | TNF | 28.506326419217828, –16.425276036108105 | D | 4 | I | Skin, lung, prescapular lymph node, liver, kidney, brain, spleen | P |
| 35 | CET 593 | Z. c. | M | A | 18/11/2011 | GC | 27.75629856377708, –15.567180176416969 | D | 4 | G | Skin, lung, prescapular lymph node, liver, kidney | P |
| 36 | CET 620 | Z. c. | M | A | 20/05/2012 | GC | 27.817154801714093, –15.764000859185387 | D | 4 | NE | Skin, lung, liver, kidney, spleen | P |
| 37 | CET 624 | Z. c. | F | A | 13/07/2012 | LGr | 29.286035258319366, –13.481903003501081 | D | 3 | G | Skin, lung, liver, mesenteric lymph node, kidney, brain | F, P |
| 38 | CET 631 | M. e. | M | A | 21/10/2012 | FTV | 28.210771928688324, –14.009197485266776 | D | 4 | G | Skin, lung, penis, palate, esophagus, brain | C, P |
| 39 | CET 636 | M. m. | M | S | 30/11/2012 | EH | 27.765920776532404,–17.910161998928636 | D | 2 | M | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen | P |
| 40 | CET 645 | Z. c. | M | J | 09/02/2013 | LZ | 29.138139438489326, –13.528173725646699 | D | 4 | M | Skin, liver, brain | F, P |
| 41 | CET 680 | Z. c. | F | N | 02/07/2013 | GC | 27.91040435902637,–15.386896660597422 | D | 4 | M | Skin, lung, liver, intestine, mesenteric lymph node, kidney, brain, spleen | P |
| 42 | CET 688 | Z. c. | F | A | 18/11/2013 | GC | 28.14483414396133, –15.57739880000554 | D | 4 | I | Skin, brain | P |
| 43 | CET 695 | M. d. | F | A | 12/07/2014 | LZ | 29.064020665490503, –13.460099910240649 | D | 4 | M | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen | P |
| 44 | CET 711 | M. d. | M | S | 03/04/2014 | EH | 27.66937590944239, –18.027604222360484 | D | 5 | P | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen | P |
| 45 | CET 712 | Z. c. | F | S | 28/04/2014 | FTV | 28.43722859059096, –13.862160820030729 | D | 4 | G | Skin, prescapular lymph node, spleen | P |
| 46 | CET 719 | Z. c. | F | A | 06/06/2014 | LZ | 28.958846751180907, –13.542006915443775 | D | 3 | M | Skin, lung, liver, mesenteric lymph node, kidney, spleen | P |
| 47 | CET 720 | Z. c. | ND | S | 10/06/2014 | FTV | 28.59159662129131,–13.82462012207941 | D | 4 | I | Skin, lung, mesenteric lymph node, liver, kidney, brain | P |
| 48 | CET 770 | Z. c. | M | S | 28/07/2015 | TNF | 28.03511014479708, –16.709505678909764 | D | 3 | G | Skin, lung, liver, intestine, mesenteric lymph node, brain, spleen | P |
| 49 | CET 771 | Z. c. | F | A | 05/08/2015 | TNF | 28.3461443622214, –16.368973513719958 | D | 2 | G | Skin, lung, liver, intestine, mesenteric lymph node, kidney, brain, spleen | P |
| 50 | CET 818 | Z. c. | M | S | 16/08/2016 | GC | 27.756233743172498,–15.67996962889147 | D | 4 | I | Lung, intestine, mesenteric lymph node, kidney, brain, spleen | P |
| 51 | CET 824 | M. d. | F | A | 11/11/2016 | FTV | 28.4768803133933, –13.867047314550653 | D | 2 | E | Skin, prescapular lymph node, liver, kidney, brain, spleen | P |
| 52 | CET 827 | M. b. | F | A | 07/12/2016 | LG | 28.182564817917488, –17.184569356359297 | A | 4 | I | Skin, lung, liver, mesenteric lymph node, intestine, brain, spleen | P |
| 53 | CET 833 | Z. c. | ND | ND | 13/02/2017 | TNF | 28.375804156993976, –16.762765447612193 | D | 4 | I | Skin, lung, liver, mesenteric lymph node, kidney, brain | P |
| 54 | CET 852 | M. d. | F | A | 05/05/2017 | FTV | 28.635507152083715, –14.026812972482025 | D | 2 | P | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen | P |
| 55 | CET 855 | Z. c. | M | A | 22/05/2017 | GC | 27.734397352882862, –15.622884576151774 | D | 3 | I | Skin, lung, liver, intestine, mesenteric lymph node, kidney, brain, spleen | P |
| Region | Period | Species | Diagnostic Test | % Prevalence | References |
|---|---|---|---|---|---|
| Portuguese and Galician coasts (North-East Atlantic Ocean) | 2004–2015 | Stenella coeruleoalba, Delphinus delphis | PCR | 5.7% (16/279) | [64] |
| Southeastern coast of USA (North-West Atlantic Ocean) | 2003–2007 | Tursiops truncatus | Serology | 9.8% (12/122) | [65] |
| The Canary Islands (Central East Atlantic Ocean) | 2003–2015 | Grampus griseus | PCR | 16.6% (2/12) | [66] |
| Northern Gulf of Mexico and USA (West Atlantic Ocean) | 2010–2014 | Tursiops truncatus | PCR | 9.9% (14/142) stranded cetaceans; 1% (1/83) free-ranging live | [67] |
| Serology | 29% (5/7) of live stranded 23% (23/102) of live free-ranging | ||||
| Histopathology + PCR | 6.6% (9/136) | ||||
| Italy (Mediterranean Sea) | 2006–2014 | Balaenoptera physalus | PCR Serology IHC | 21.74% (5/23) | [68] |
| Hawaii (Pacific Ocean) | 1997–2014 | Megaptera novaeangliae, Kogia breviceps, Stenella longirostris, Ziphius cavirostris, Stenella frontalis, Indopacetus pacificus, Stenella coeruleoalba, Mesoplodon densirostris, Tursiops truncatus, Physeter macrocephalus, Steno bredanensis, Grampus griseus | PCR | 24% (15/62) | [13] |
| Italy (Mediterranean Sea) | 1998–2014 | Stenella coeruleoalba, Tursiops truncatus, Balaenoptera physalus and Globicephala melas | Serology | 32.8% (23/70) | [69] |
| Central California coast (Pacific Ocean) | 2000–2015 | Phocoena phocoena, Delphinus capensis, Lagenorhynchus obliquidens, Stenella coeruleoalba, Grampus griseus | PCR (1/11) | 36.36% (4/11) | [70] |
| Serology (3/11) | |||||
| Brazil (South Atlantic Ocean) | 2010–2017 | Eubalaena australis | PCR | 37.5% (3/8) | [71] |
| Australia (Pacific Ocean) | 2005–2011 | Peponocephala electra, Tursiops aduncus, Lagenodelphis hosei, Tursiops truncatus, Balaenoptera edeni | Serology | 48.1% (13/27) | [72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felipe-Jiménez, I.; Fernández, A.; Arbelo, M.; Segura-Göthlin, S.; Colom-Rivero, A.; Suárez-Santana, C.M.; De La Fuente, J.; Sierra, E. Molecular Diagnosis of Cetacean Morbillivirus in Beaked Whales Stranded in the Canary Islands (1999–2017). Vet. Sci. 2022, 9, 121. https://doi.org/10.3390/vetsci9030121
Felipe-Jiménez I, Fernández A, Arbelo M, Segura-Göthlin S, Colom-Rivero A, Suárez-Santana CM, De La Fuente J, Sierra E. Molecular Diagnosis of Cetacean Morbillivirus in Beaked Whales Stranded in the Canary Islands (1999–2017). Veterinary Sciences. 2022; 9(3):121. https://doi.org/10.3390/vetsci9030121
Chicago/Turabian StyleFelipe-Jiménez, Idaira, Antonio Fernández, Manuel Arbelo, Simone Segura-Göthlin, Ana Colom-Rivero, Cristian M. Suárez-Santana, Jesús De La Fuente, and Eva Sierra. 2022. "Molecular Diagnosis of Cetacean Morbillivirus in Beaked Whales Stranded in the Canary Islands (1999–2017)" Veterinary Sciences 9, no. 3: 121. https://doi.org/10.3390/vetsci9030121
APA StyleFelipe-Jiménez, I., Fernández, A., Arbelo, M., Segura-Göthlin, S., Colom-Rivero, A., Suárez-Santana, C. M., De La Fuente, J., & Sierra, E. (2022). Molecular Diagnosis of Cetacean Morbillivirus in Beaked Whales Stranded in the Canary Islands (1999–2017). Veterinary Sciences, 9(3), 121. https://doi.org/10.3390/vetsci9030121







