Oxidative Stress-Mediated Antibacterial Activity of the Total Flavonoid Extracted from the Agrimonia pilosa Ledeb. in Methicillin-Resistant Staphylococcusaureus (MRSA)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Cultivation
2.2. Preparation of the Total Flavonoid
2.3. MIC Determination
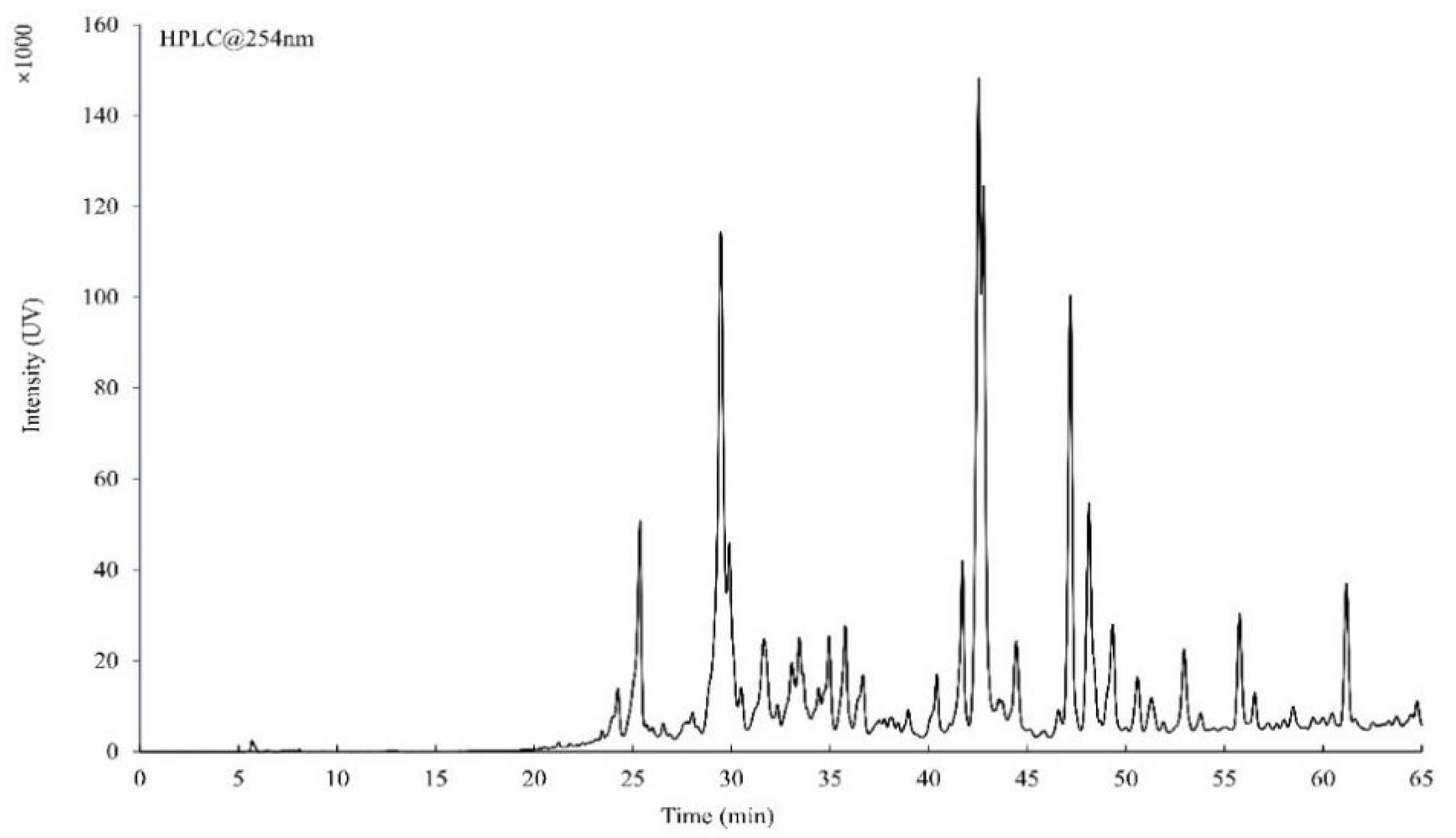
2.4. HPLC and HRMS Analysis of the Total Flavonoid
2.5. Determination of Time–Kill Curves
2.6. The Level of Intracellular H2O2
2.7. The Level of Intracellular ROS Analysis
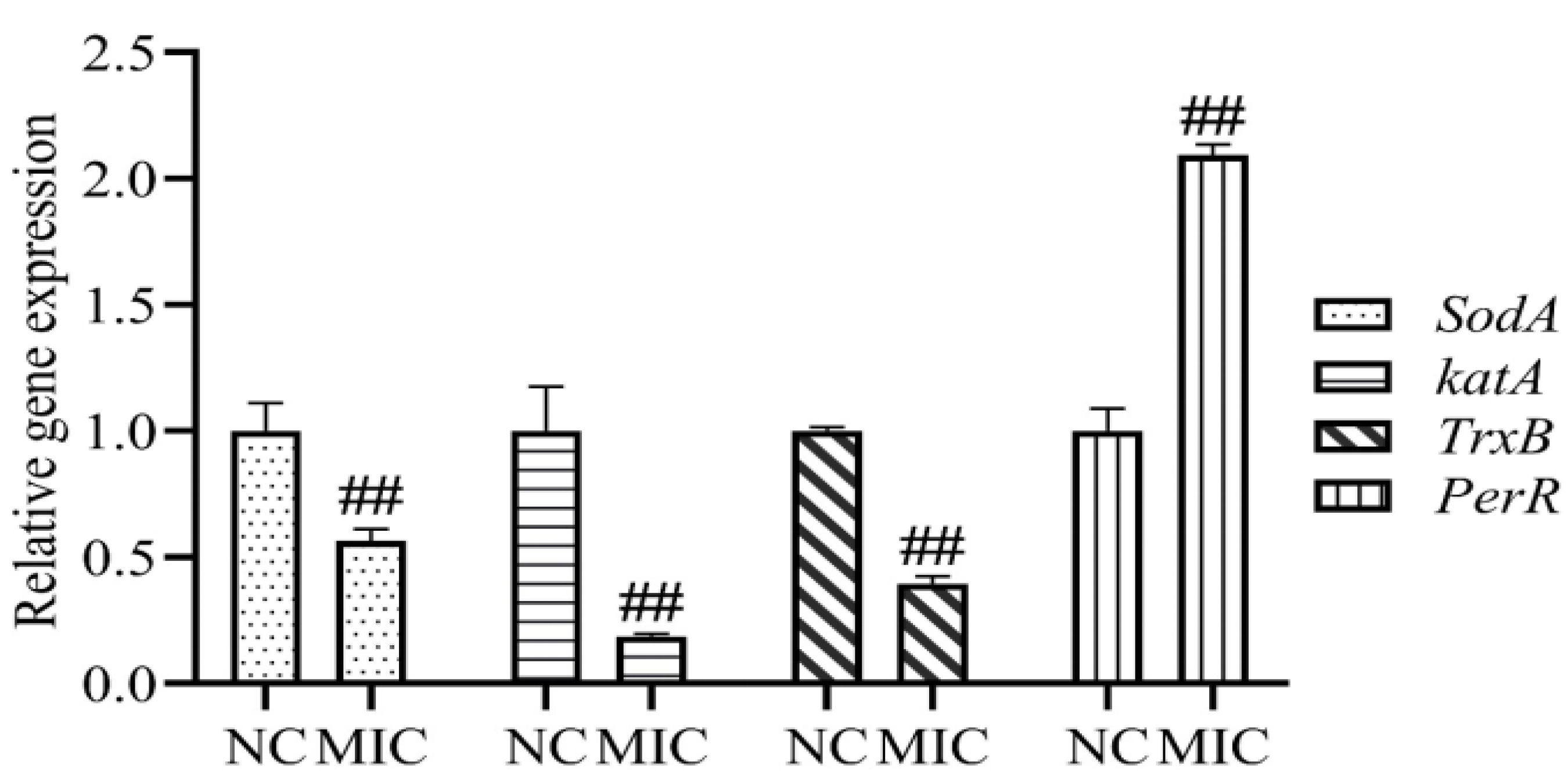
2.8. Oxidative Stress-Related Genes Analysis
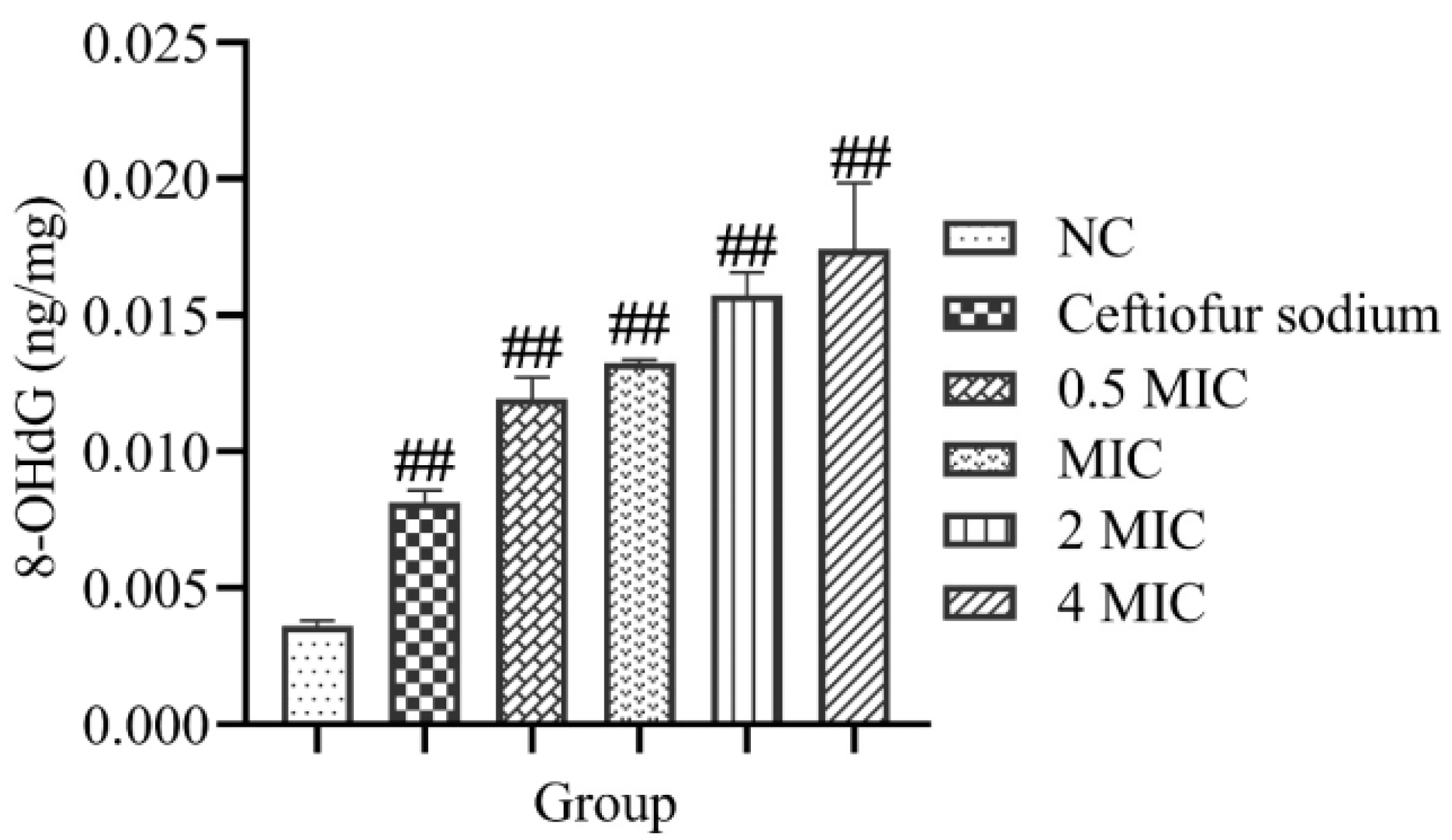
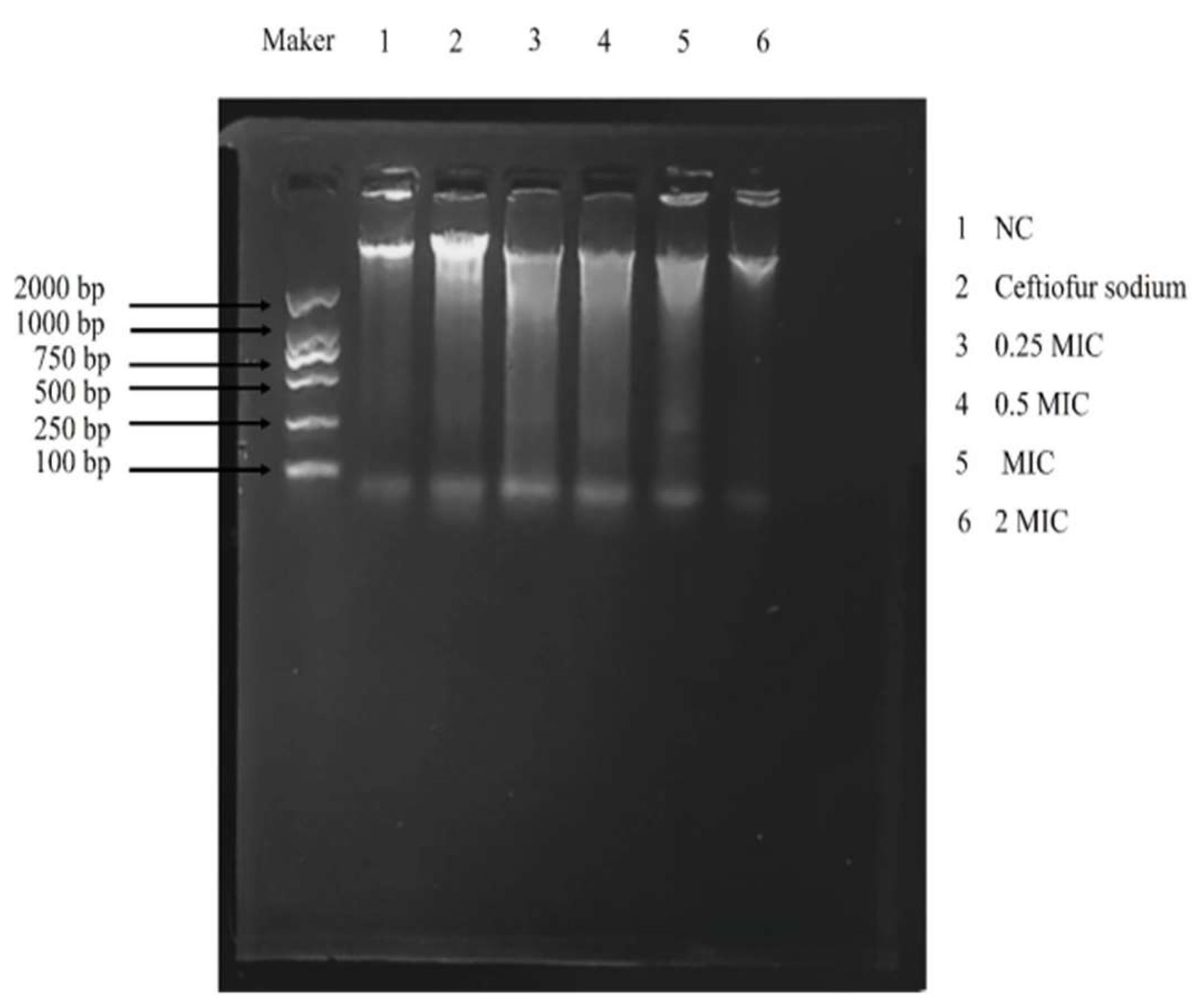
2.9. 8-Hydroxy-2′-Deoxyguanosine (8-OHdG) and DNA Gel Electrophoresis
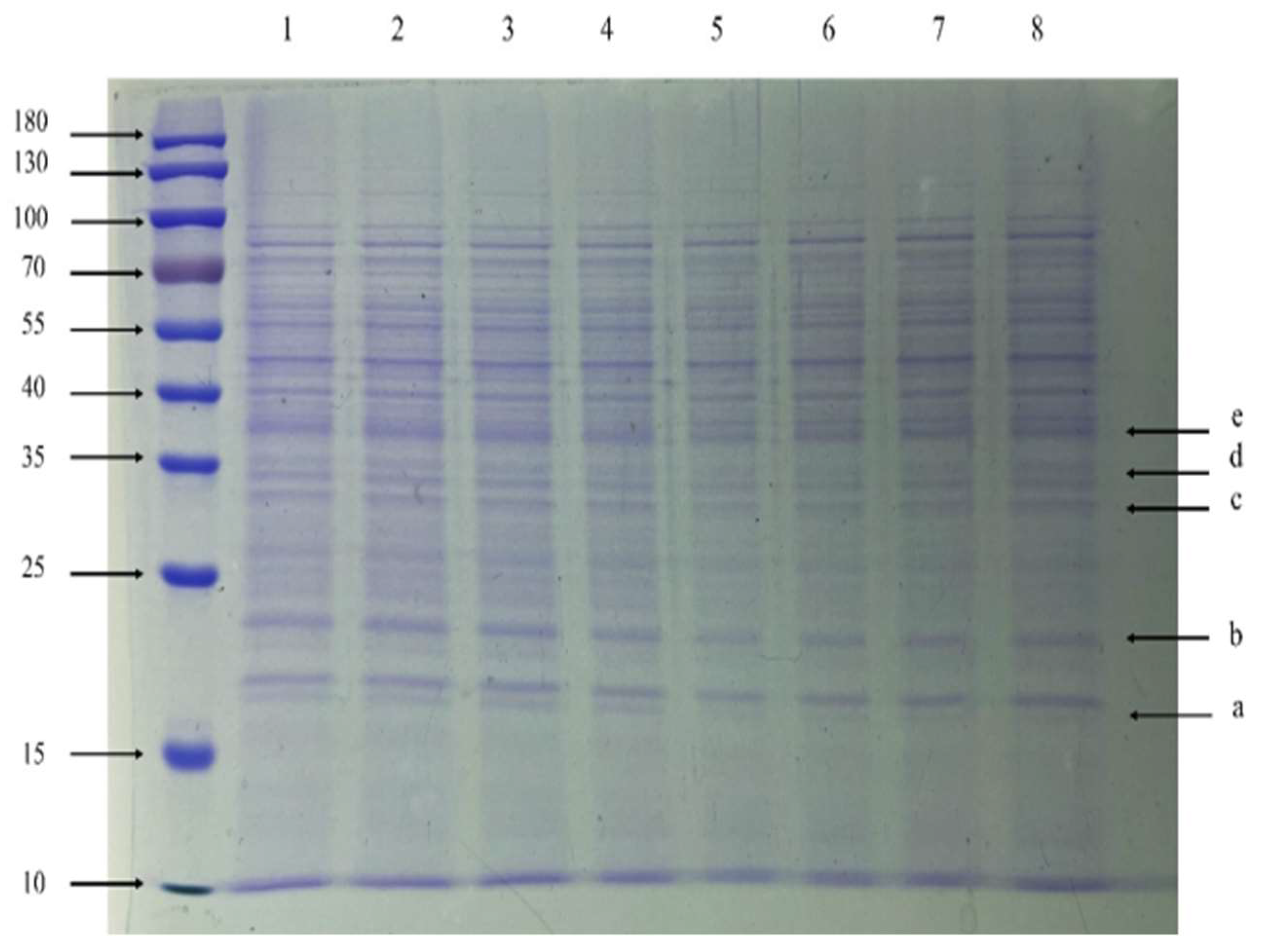
2.10. SDS-PAGE and NanoLC-ESI-MS/MS Analysis
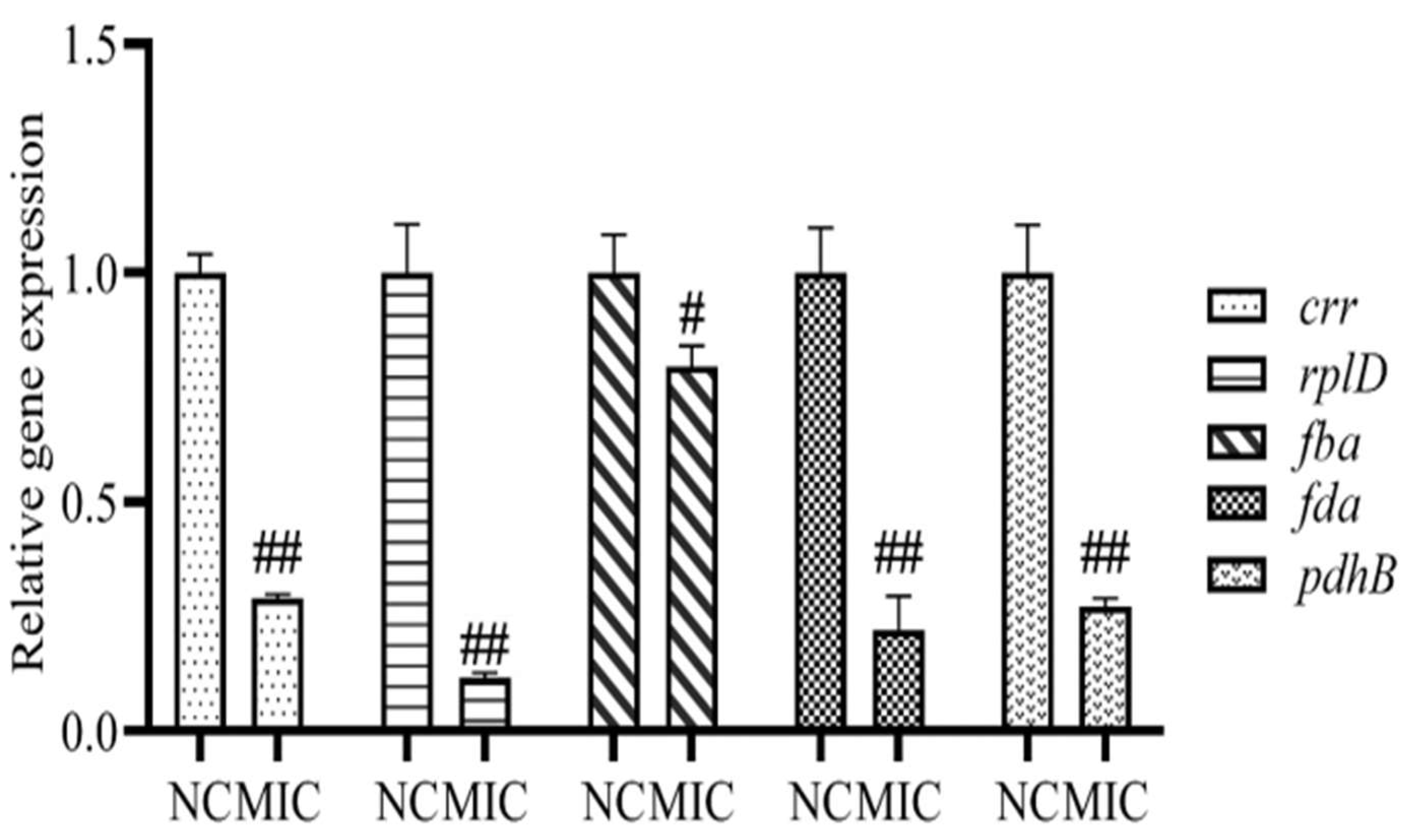
2.11. The mRNA Expression Analysis of Differential Proteins
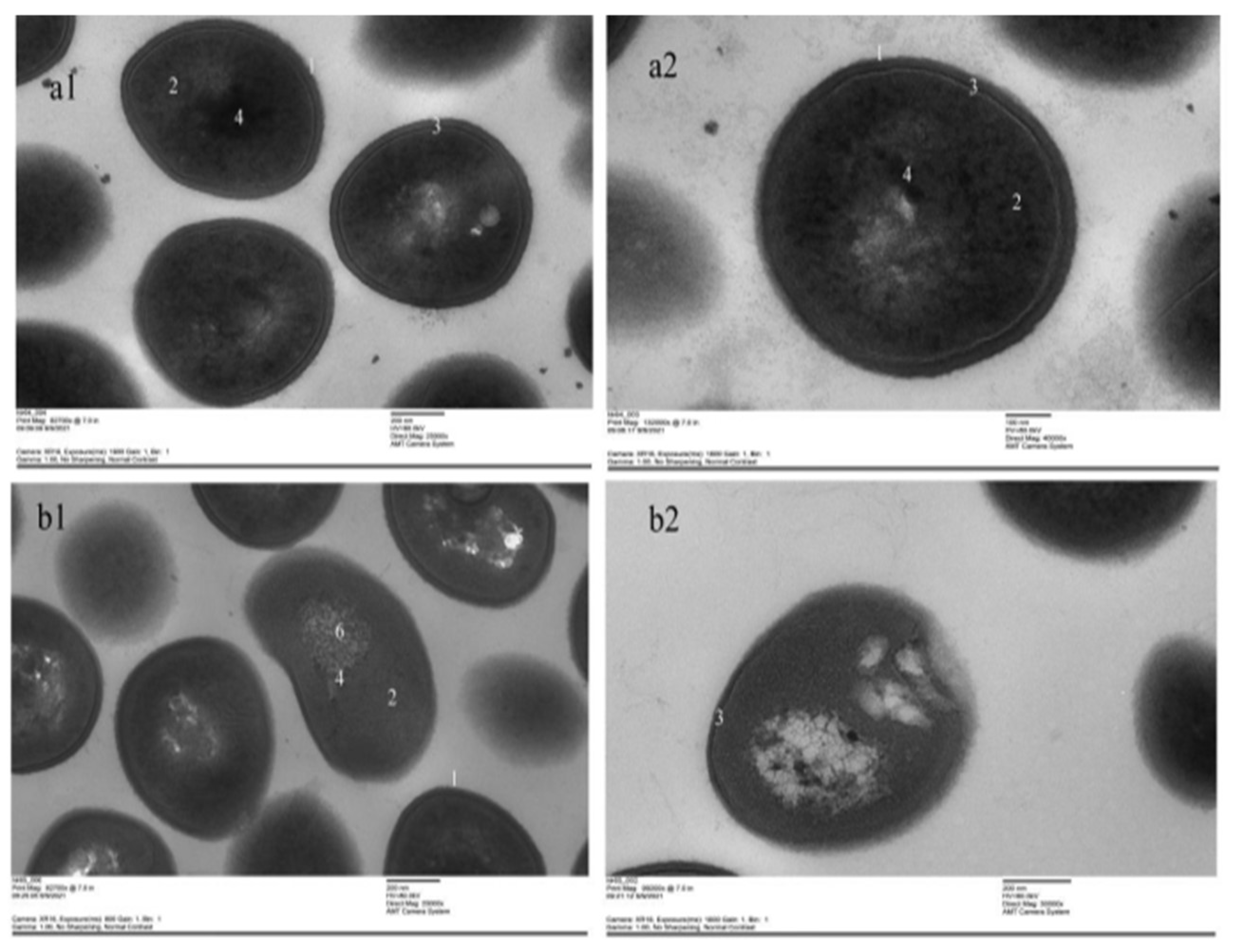
2.12. Transmission Electron Microscope (TEM) Observation
2.13. Statistical Analysis
3. Results
3.1. HPLC and HRMS Analysis of the Total Flavonoid Extracted from the A. pilosa Ledeb.
3.2. The Time–Kill Curves
3.3. The Level of Intracellular H2O2
3.4. The Level of Intracellular ROS
3.5. Oxidative Stress-Related Genes
3.6. 8-OHdG and DNA Gel Electrophoresis
- Y = 0.0276X + 0.0245 (R2 = 0.9862)
- Y is the concentration of 8-OHdG (ng/mg); X is the OD value of sample.
3.7. SDS-PAGE and NanoLC-ESI-MS/MS Analysis
3.8. mRNA Expression of Differential Proteins
3.9. TEM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parisi, A.; Caruso, M.; Normanno, G.; Latorre, L.; Miccolupo, A.; Fraccalvieri, R.; Intini, F.; Manginelli, T.; Santagada, G. MRSA in swine, farmers and abattoir workers in Southern Italy. Food Microbiol. 2019, 82, 287–293. [Google Scholar] [CrossRef]
- Dayan, G.H.; Mohamed, N.; Scully, I.L.; Cooper, D.; Begier, E.; Eiden, J.; Jansen, K.U.; Gurtman, A.; Anderson, A.S. Staphylococcus aureus: The current state of disease, pathophysiology and strategies for prevention. Expert Rev. Vaccines 2016, 15, 1373–1392. [Google Scholar] [CrossRef]
- Peton, V.; Le Loir, Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014, 21, 602–615. [Google Scholar] [CrossRef]
- Barber, M. Methicillin-resistant Staphylococci. J. Clin. Pathol. 1961, 14, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Magro, G.; Rebolini, M.; Beretta, D.; Piccinini, R. Methicillin-resistant Staphylococcus aureus CC22-MRSA-IV as an agent of dairy cow intramammary infections. Vet. Microbiol. 2018, 227, 29–33. [Google Scholar] [CrossRef]
- Benrabia, I.; Hamdi, T.M.; Shehata, A.A.; Neubauer, H.; Wareth, G. Methicillin-Resistant Staphylococcus aureus (MRSA) in Poultry Species in Algeria: Long-Term Study on Prevalence and Antimicrobial Resistance. Vet. Sci. 2020, 7, 54. [Google Scholar] [CrossRef]
- Baba, K.; Ishihara, K.; Ozawa, M.; Tamura, Y.; Asai, T. Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int. J. Antimicrob. Agents 2010, 36, 352–354. [Google Scholar] [CrossRef]
- Jones, T.F.; Kellum, M.E.; Porter, S.S.; Bell, M.; Schaffner, W. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2002, 8, 82–84. [Google Scholar] [CrossRef]
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef]
- Dhama, K.; Tiwari, R.; Chakraborty, S.; Saminathan, M.; Kumar, A.; Karthik, K.; Wani, M.Y.; Amarpal; Singh, S.V.; Rahal, A. Evidence Based Antibacterial Potentials of Medicinal Plants and Herbs Countering Bacterial Pathogens Especially in the Era of Emerging Drug Resistance: An Integrated Update. Int. J. Pharmacol. 2014, 10, 1–43. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M. Methicillin-resistant Staphylococcus aureus among animals: Current overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Song, S.; Wang, J.; Zhang, Q.; Qiu, F.; Zhao, F. Tiliroside, the major component of Agrimonia pilosa Ledeb ethanol extract, inhibits MAPK/JNK/p38-mediated inflammation in lipopolysaccharide-activated RAW 264.7 macrophages. Exp. Ther. Med. 2016, 12, 499–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.Y.; Yu, Q.M.; Kong, H.J.; Lee, J.Y.; Yang, K.M.; Seo, J.S. Antioxidant and Anti-Inflammatory Activities of Agrimonia pilosa Ledeb. Extract. Evid. Based Complement. Alternat. Med. 2020, 2020, 8571207. [Google Scholar] [CrossRef] [PubMed]
- Yamaki, M.; Kashihara, M.; Ishiguro, K.; Takagi, S. Antimicrobial Principles of Xian he cao (Agrimonia pilosa). Planta Med. 1989, 55, 169–170. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Wang, H.W.; Cai, X. Optimization of the extraction of total flavonoids from Scutellaria baicalensis Georgi using the response surface methodology. J. Food Sci Tech. Mys. 2015, 52, 2336–2343. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.D.; Nahar, L.; Kumarasamy, Y. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 2007, 42, 321–324. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Zhang, H.; Sun, W.; Li, Z.; Zhang, F.; Zhang, H.; Chen, F.; Zhang, H.; An, J.; et al. Antimicrobial mechanism of strictinin isomers extracted from the root of Rosa roxburghii Tratt (Ci Li Gen). J. Ethnopharmacol. 2020, 250, 112498. [Google Scholar] [CrossRef]
- Esmadi, F.T.; Khabour, O.F.; Abbas, K.; Mohammad, A.E.; Obeidat, R.T.; Mfady, D. Synthesis, characterization and biological activity of some unsymmetrical Schiff base transition metal complexes. Drug Chem. Toxicol. 2016, 39, 41–47. [Google Scholar] [CrossRef]
- Dhawan, A.; Bajpayee, M.; Parmar, D. Comet assay: A reliable tool for the assessment of DNA damage in different models. Cell Biol. Toxicol. 2009, 25, 5–32. [Google Scholar] [CrossRef] [PubMed]
- Clement, C.C.; Aphkhazava, D.; Nieves, E.; Callaway, M.; Olszewski, W.; Rotzschke, O.; Santambrogio, L. Protein expression profiles of human lymph and plasma mapped by 2D-DIGE and 1D SDS–PAGE coupled with nanoLC–ESI–MS/MS bottom-up proteomics. J. Proteom. 2013, 78, 172–187. [Google Scholar] [CrossRef] [Green Version]
- Grigor’eva, A.; Bardasheva, A.; Tupitsyna, A.; Amirkhanov, N.; Tikunova, N.; Pyshnyi, D.; Ryabchikova, E. Changes in the Ultrastructure of Staphylococcus aureus Treated with Cationic Peptides and Chlorhexidine. Microorganisms 2020, 8, 1991. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, J.; Tan, J.; Liu, X.; Wang, B. Flavonoids from Agrimonia pilosa Ledeb: Free Radical Scavenging and DNA Oxidative Damage Protection Activities and Analysis of Bioactivity-Structure Relationship Based on Molecular and Electronic Structures. Molecules 2017, 22, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Liu, H.X.; Zhuang, Y.L.; Ding, L.Q.; Chen, L.X.; Qiu, F. Studies on isolation and identification of flavonoids in herbs of Agrimonia pilosa. Zhongguo Zhong Yao Za Zhi 2008, 33, 2925–2928. [Google Scholar] [PubMed]
- Donnapee, S.; Li, J.; Yang, X.; Ge, A.H.; Donkor, P.O.; Gao, X.M.; Chang, Y.X. Cuscuta chinensis Lam.: A systematic review on ethnopharmacology, phytochemistry and pharmacology of an important traditional herbal medicine. J. Ethnopharmacol. 2014, 157, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Liancai, Z.; Jun, T.; Bochu, W.; Rui, H.; Yuping, L.; Chao, Z. Antioxidant activities of aqueous extract from Agrimonia pilosa Ledeb and its fractions. Chem. Biodivers. 2009, 6, 1716–1726. [Google Scholar] [CrossRef]
- Kim, S.B.; Hwang, S.H.; Suh, H.W.; Lim, S.S. Phytochemical Analysis of Agrimonia pilosa Ledeb, Its Antioxidant Activity and Aldose Reductase Inhibitory Potential. Int. J. Mol. Sci. 2017, 18, 379. [Google Scholar] [CrossRef]
- Liu, W.J.; Hou, X.Q.; Chen, H.; Liang, J.Y.; Sun, J.B. Chemical constituents from Agrimonia pilosa Ledeb. and their chemotaxonomic significance. Nat. Prod. Res. 2016, 30, 2495–2499. [Google Scholar] [CrossRef]
- Jiang, Q.H.; Ma, J.H.; Wang, Y.; Ding, L.Q.; Chen, L.X.; Qiu, F. Simultaneous determination of nine major constituents in Agrimonia pilosa Ledeb. by HPLC-DAD-ESI-MS/MS. Anal. Methods 2014, 6, 4373–4379. [Google Scholar] [CrossRef]
- Babeshina, L.G.; Kuznetzov, A.A.; Reshetov, Y.E.; Krasnov, E.A. Agrimony pilosa Ledeb. (Rosaceae)-Chemical Composition, Biological Effects and Anatomy. Biosci. Biotechnol. Res. Asia 2014, 11, 65–68. [Google Scholar] [CrossRef]
- Park, M.J.; Kang, Y.-H. Isolation of Isocoumarins and Flavonoids as α-Glucosidase Inhibitors from Agrimonia pilosa L. Molecules 2020, 25, 2572. [Google Scholar] [CrossRef]
- Kateete, D.P.; Bwanga, F.; Seni, J.; Mayanja, R.; Kigozi, E.; Mujuni, B.; Ashaba, F.K.; Baluku, H.; Najjuka, C.F.; Kallander, K.; et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob. Resist. Infect. Control 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Bal, A.M.; David, M.Z.; Garau, J.; Gottlieb, T.; Mazzei, T.; Scaglione, F.; Tattevin, P.; Gould, I.M. Future trends in the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infection: An in-depth review of newer antibiotics active against an enduring pathogen. J. Glob. Antimicrob. Resist. 2017, 10, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Basri, D.F.; Sandra, V. Synergistic Interaction of Methanol Extract from Canarium odontophyllum Miq. Leaf in Combination with Oxacillin against Methicillin-Resistant Staphylococcus aureus (MRSA) ATCC 33591. Int J. Microbiol. 2016, 2016, 5249534. [Google Scholar] [CrossRef] [Green Version]
- Al-Habib, A.; Al-Saleh, E.; Safer, A.M.; Afzal, M. Bactericidal effect of grape seed extract on methicillin-resistant Staphylococcus aureus (MRSA). J. Toxicol. Sci. 2010, 35, 357–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.; Gan, R.Y.; Zhang, D.; Farha, A.K.; Habimana, O.; Mavumengwana, V.; Li, H.B.; Wang, X.H.; Corke, H. Large-Scale Screening of 239 Traditional Chinese Medicinal Plant Extracts for Their Antibacterial Activities against Multidrug-Resistant Staphylococcus aureus and Cytotoxic Activities. Pathogens 2020, 9, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Park, C.; Jang, H.J.; Kim, B.O.; Bae, H.W.; Chung, I.Y.; Kim, E.S.; Cho, Y.H. Antibacterial strategies inspired by the oxidative stress and response networks. J. Microbiol. 2019, 57, 203–212. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Clements, M.O.; Crossley, H.; Ingham, E.; Foster, S.J. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 2001, 69, 3744–3754. [Google Scholar] [CrossRef] [Green Version]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-hydroxy-2’ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2009, 27, 120–139. [Google Scholar] [CrossRef] [Green Version]
- Choe, M.; Park, Y.H.; Lee, C.R.; Kim, Y.R.; Seok, Y.J. The general PTS component HPr determines the preference for glucose over mannitol. Sci. Rep. 2017, 7, 43431. [Google Scholar] [CrossRef]
- Zebre, A.C.; Ake, F.M.; Ventroux, M.; Koffi-Nevry, R.; Noirot-Gros, M.F.; Deutscher, J.; Milohanic, E. Interaction with enzyme IIBMpo (EIIBMpo) and phosphorylation by phosphorylated EIIBMpo exert antagonistic effects on the transcriptional activator ManR of Listeria monocytogenes. J. Bacteriol. 2015, 197, 1559–1572. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Cai, M.; Zhou, H.; Hou, L.; Peng, H.; He, H. Discovery of efficient inhibitors against pyruvate dehydrogenase complex component E1 with bactericidal activity using computer aided design. Pestic. Biochem. Physiol. 2021, 177, 104894. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Korotchkina, L.G.; Sidhu, S. Interaction of E1 and E3 components with the core proteins of the human pyruvate dehydrogenase complex. J. Mol. Catal. B Enzym. 2009, 61, 2–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, Y.P.; Gurmit, S.; Budhayash, G.; Satendra, S.; Madhu, Y.; Upasana, S.; Brijendra, S. Molecular modeling, dynamics studies and virtual screening of Fructose 1, 6 biphosphate aldolase-II in community acquired- methicillin resistant Staphylococcus aureus (CA-MRSA). Bioinformation 2013, 9, 158–164. [Google Scholar] [CrossRef]
- Callens, M.; Kuntz, D.A.; Opperdoes, F.R. Kinetic properties of fructose bisphosphate aldolase from Trypanosoma brucei compared to aldolase from rabbit muscle and Staphylococcus aureus. Mol. Biochem. Parasitol. 1991, 47, 1–9. [Google Scholar] [CrossRef]
- Zengel, J.M.; Lindahl, L. Ribosomal protein L4 of Escherichia coli: In vitro analysis of L4-mediated attenuation control. Biochimie 1991, 73, 719–727. [Google Scholar] [CrossRef]
- Locke, J.B.; Hilgers, M.; Shaw, K.J. Novel Ribosomal Mutations in Staphylococcus aureus Strains Identified through Selection with the Oxazolidinones Linezolid and Torezolid (TR-700). Antimicrob. Agents Chemother. 2009, 53, 5265–5274. [Google Scholar] [CrossRef] [Green Version]



| Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| 16s | CCAGCAGCCGCGGTAAT | CGCGCTTTACGCCCAATA |
| sodA | AAGCGTGTTCCCATACGTCTAAACC | TTGGTTCAGGTTGGGCTTGGTTAG |
| katA | GCTGCTGAAATTATAGCTACAGAT | TACTTGAATATACATTGTCCATTT |
| TrxB | AAGACGGCAAAGTGGGTTCTGTG | TGGCGCTGTTAATGGCTTCATACC |
| perR | TCCATTCGATGATGTGTTACGTCA | TGTGAACAATGTGGTAAGATCGTTG |
| Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| crr | TCCTTCGCCGTCTAATTGAACTGTG | GCAGGACGTGTTGACAATGTCTTTC |
| rplD | ACTTCTTGGAGTTGGTCCGAATACG | GGAACAGGTCGTGCTCGTCAAG |
| fba | ACTGAACCTAATGCTGGCGCTAATG | ACTGTTGGCGGACAAGAAGATGATG |
| fda | TGTTCCAACTTGTTCACGATATGCG | GCAGTGTATTTGCCTTCTACTTCGC |
| pdhB | TTGATGCGATTGCTGGACAAATTGC | TGTGTGTACGCCACCACCAAATG |
| 16s | CCAGCAGCCGCGGTAAT | CGCGCTTTACGCCCAATA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Cheng, H.; Chen, F.; Song, S.; Zhang, H.; Sun, W.; Bao, X.; Zhang, H.; He, C. Oxidative Stress-Mediated Antibacterial Activity of the Total Flavonoid Extracted from the Agrimonia pilosa Ledeb. in Methicillin-Resistant Staphylococcusaureus (MRSA). Vet. Sci. 2022, 9, 71. https://doi.org/10.3390/vetsci9020071
He L, Cheng H, Chen F, Song S, Zhang H, Sun W, Bao X, Zhang H, He C. Oxidative Stress-Mediated Antibacterial Activity of the Total Flavonoid Extracted from the Agrimonia pilosa Ledeb. in Methicillin-Resistant Staphylococcusaureus (MRSA). Veterinary Sciences. 2022; 9(2):71. https://doi.org/10.3390/vetsci9020071
Chicago/Turabian StyleHe, Liren, Han Cheng, Fuxin Chen, Suquan Song, Hang Zhang, Weidong Sun, Xiaowei Bao, Haibin Zhang, and Chenghua He. 2022. "Oxidative Stress-Mediated Antibacterial Activity of the Total Flavonoid Extracted from the Agrimonia pilosa Ledeb. in Methicillin-Resistant Staphylococcusaureus (MRSA)" Veterinary Sciences 9, no. 2: 71. https://doi.org/10.3390/vetsci9020071
APA StyleHe, L., Cheng, H., Chen, F., Song, S., Zhang, H., Sun, W., Bao, X., Zhang, H., & He, C. (2022). Oxidative Stress-Mediated Antibacterial Activity of the Total Flavonoid Extracted from the Agrimonia pilosa Ledeb. in Methicillin-Resistant Staphylococcusaureus (MRSA). Veterinary Sciences, 9(2), 71. https://doi.org/10.3390/vetsci9020071






