Characterization of Intestinal Microbiota in Lambs with Different Susceptibility to Escherichia coli F17
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Escherichia coli F17 Preparation
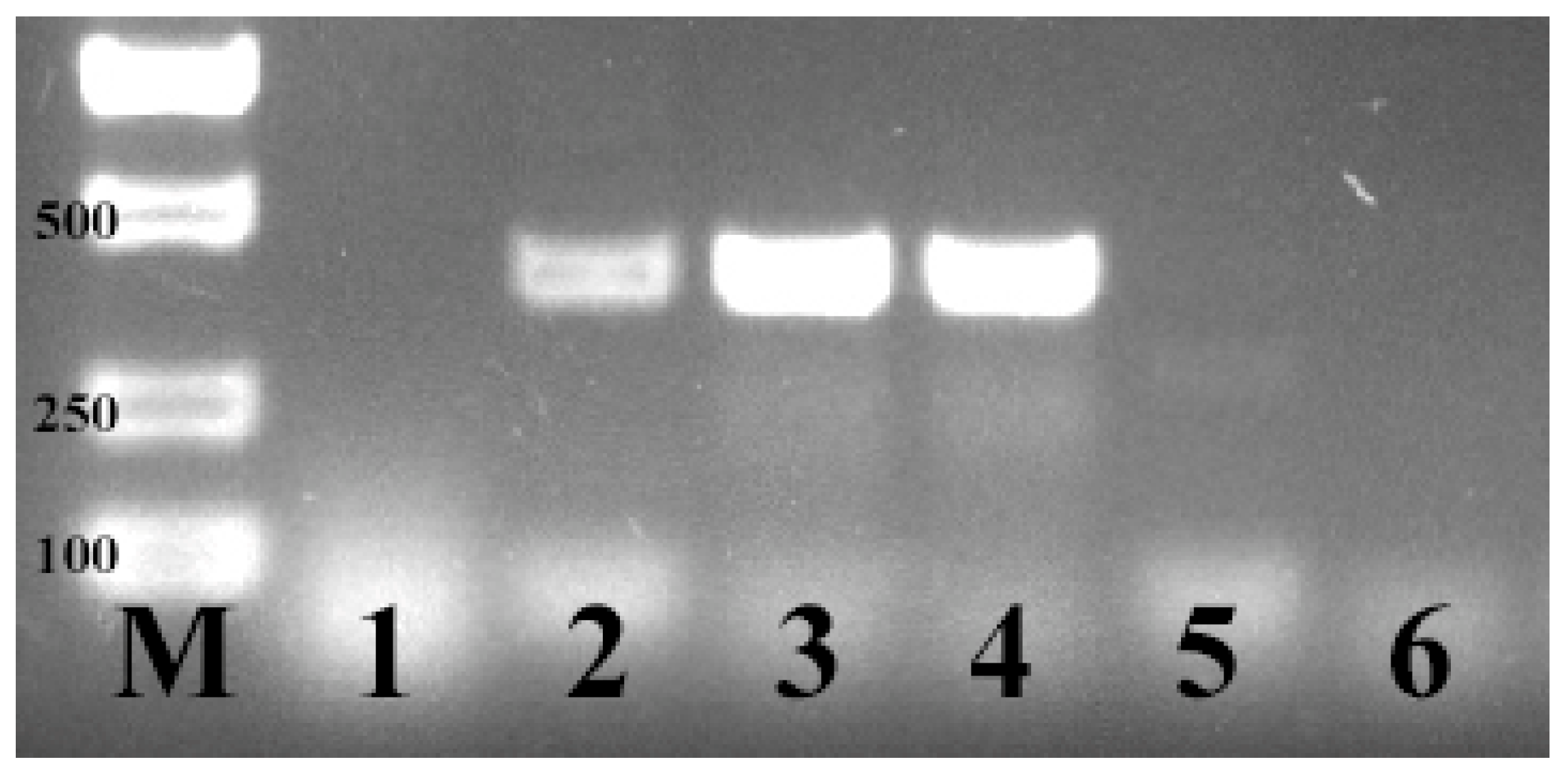
2.2. PCR Conditions for E. coli F17 Identification
2.3. Animals and Challenge Experimental Design
2.4. Jejunal Bacterial Community Sequencing
2.5. 16s rRNA Data Analysis Process
2.6. Statistical Analyses
3. Results
3.1. Identification of E. coli F17
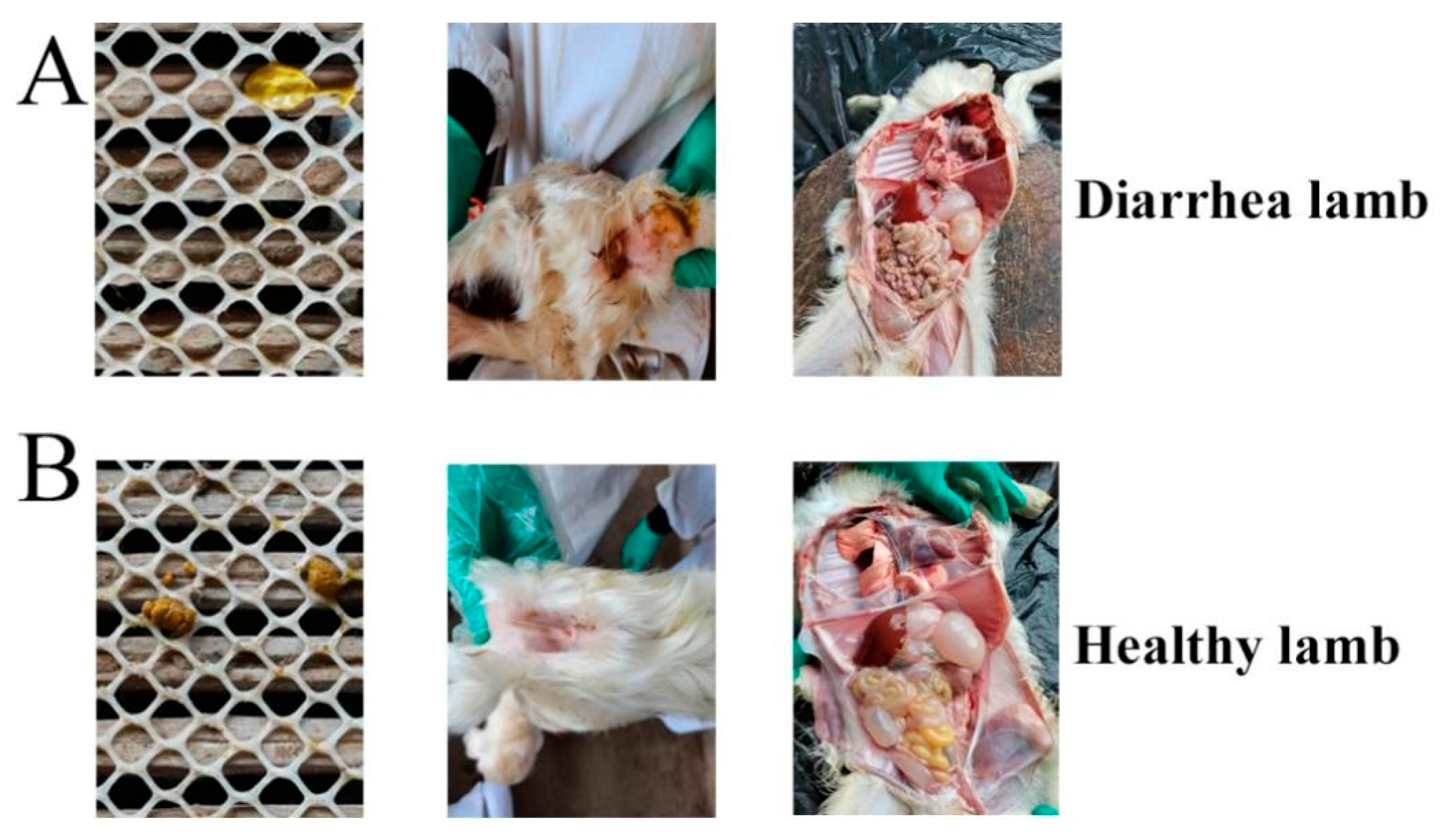
3.2. Pathogenicity Record of E. coli F17 Challenge Experiment
3.3. Identification of Hu Sheep Sensitive and Resistant to the E. coli F17 Strain
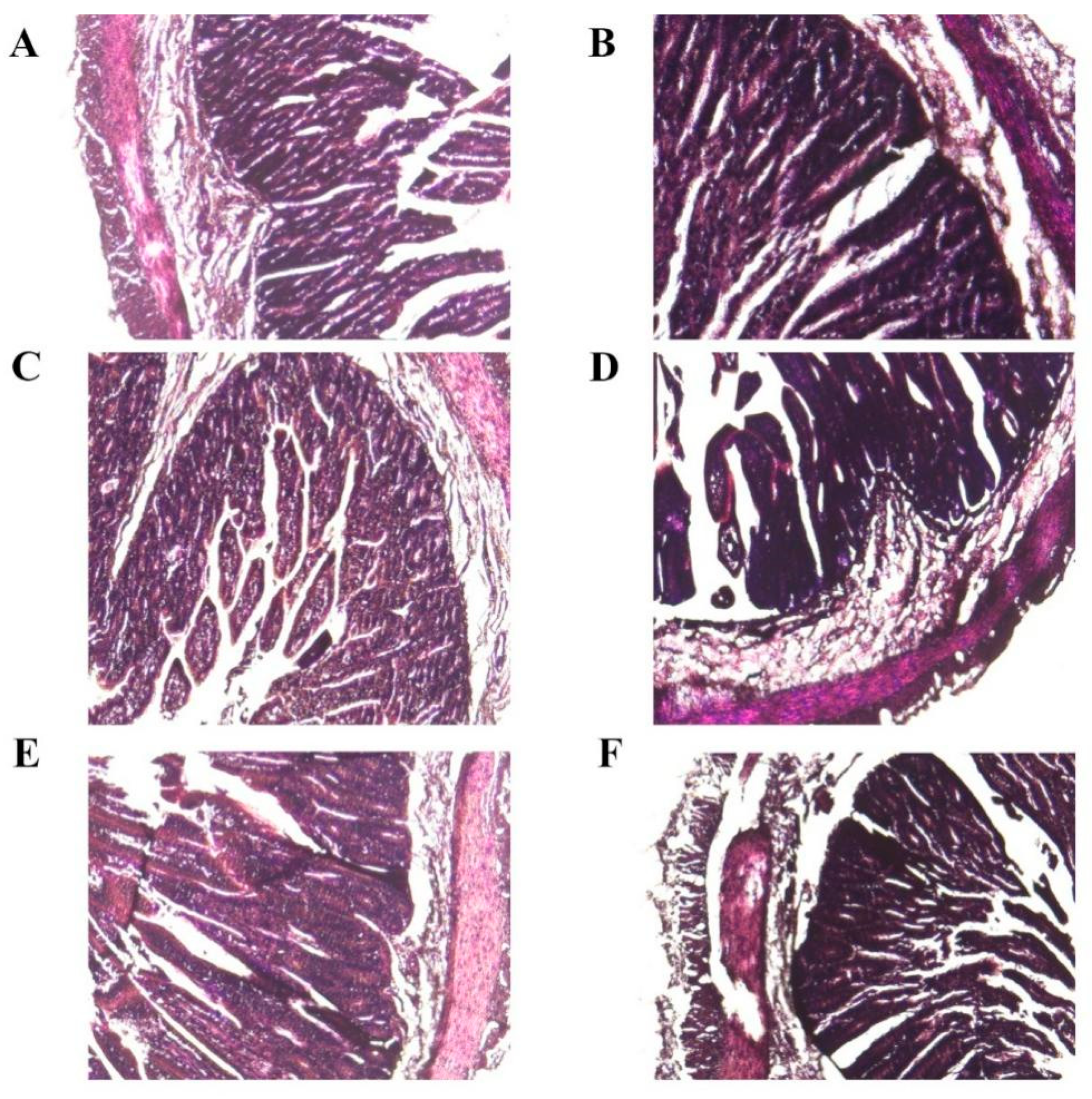
3.4. Histopathological Examination Experiment
3.5. Bacteria Plate Counting Result
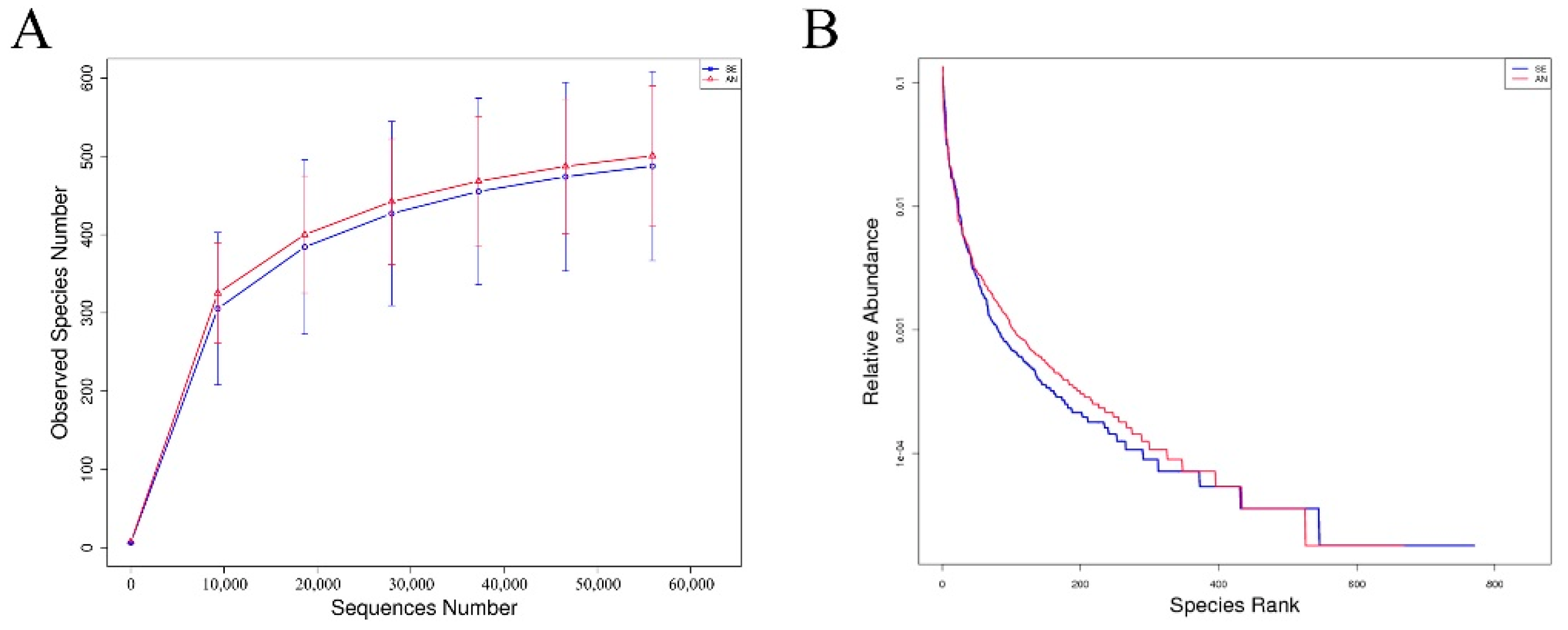
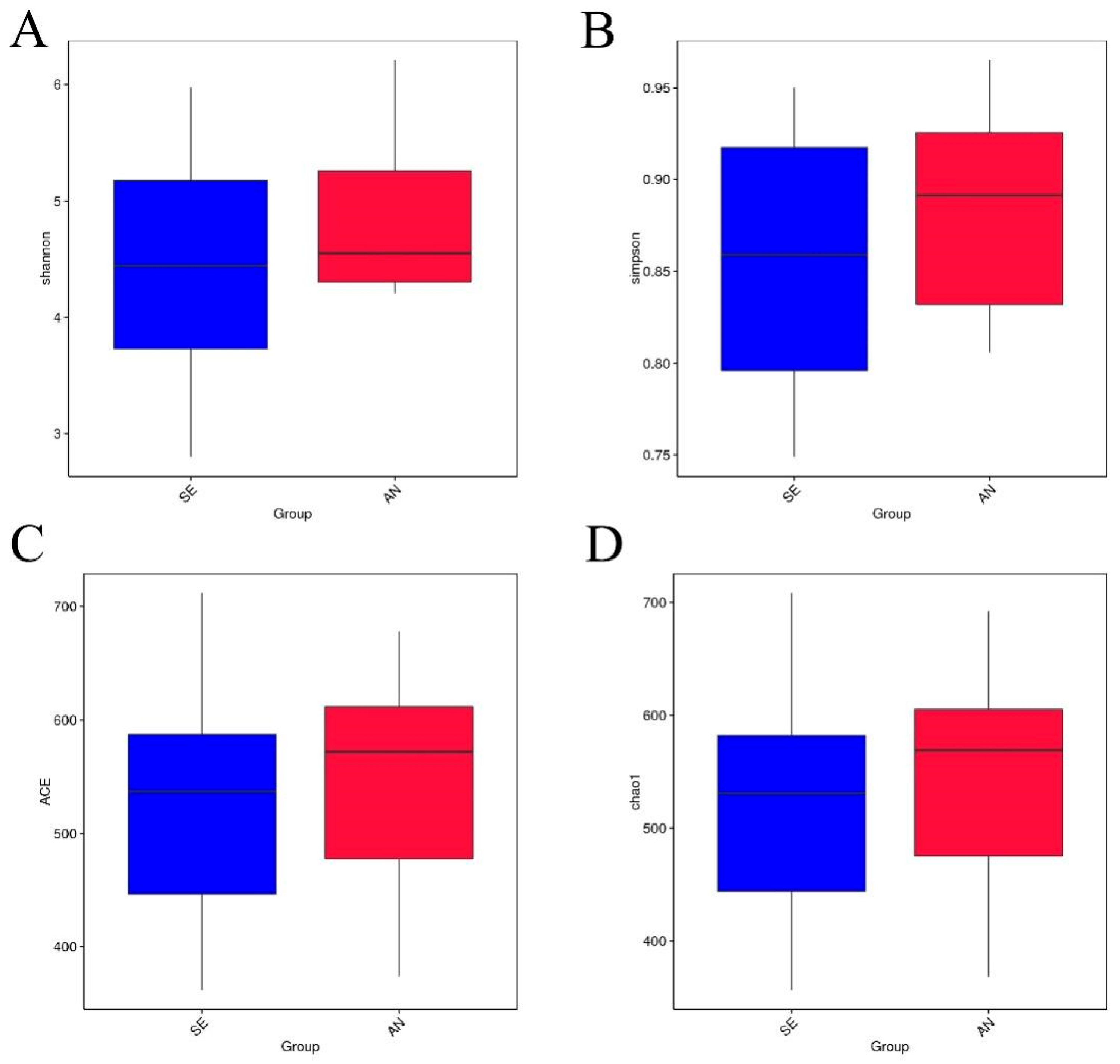
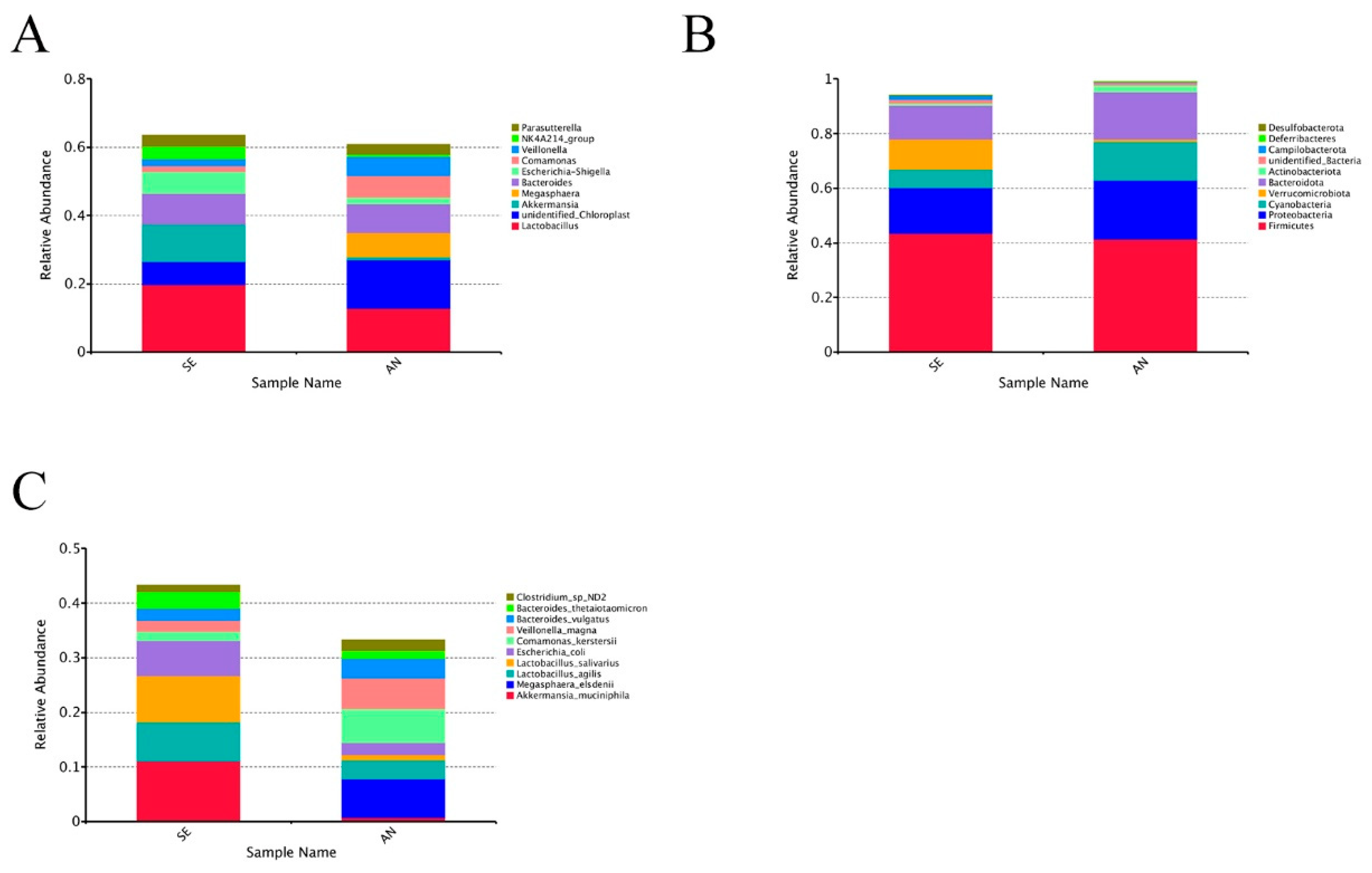
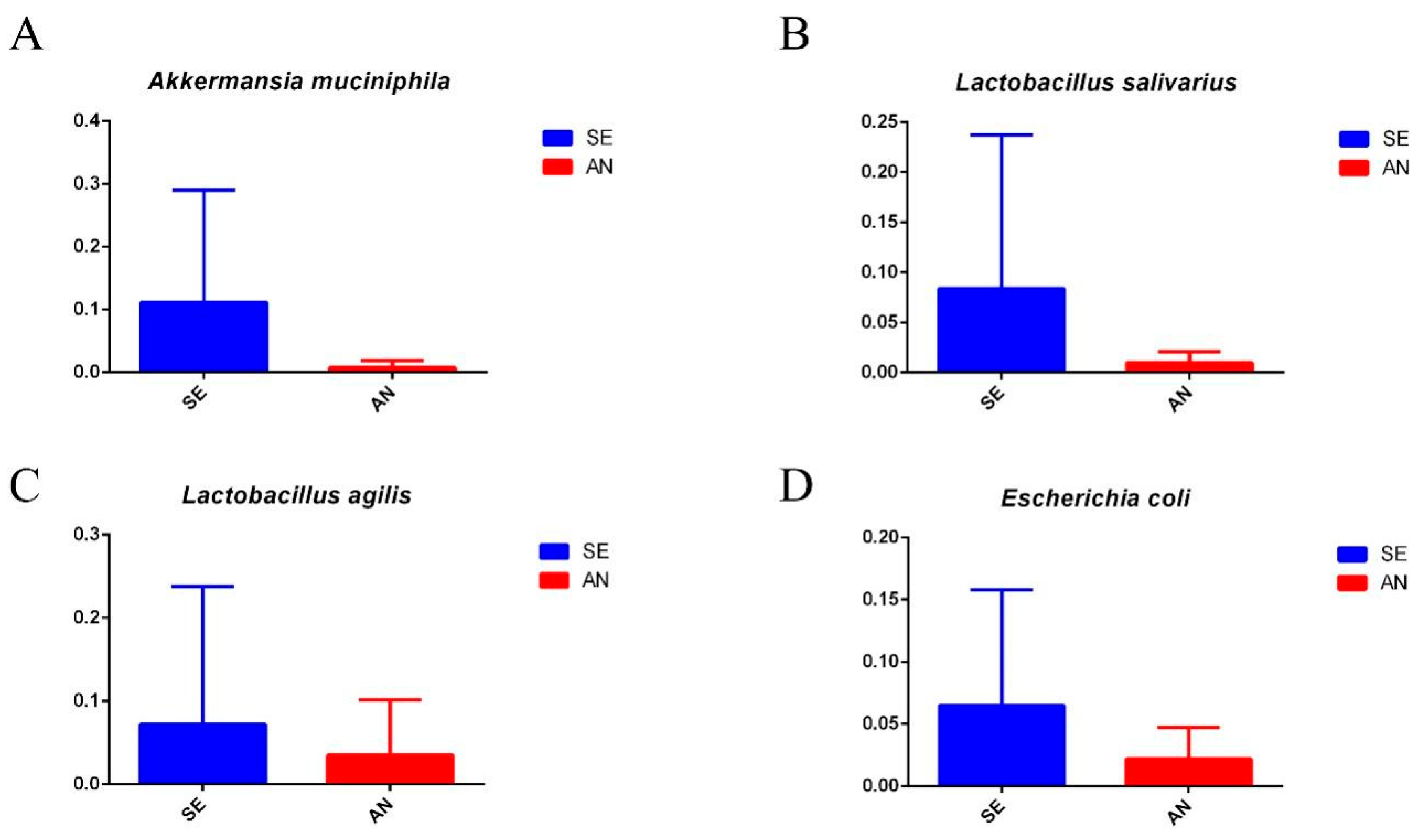
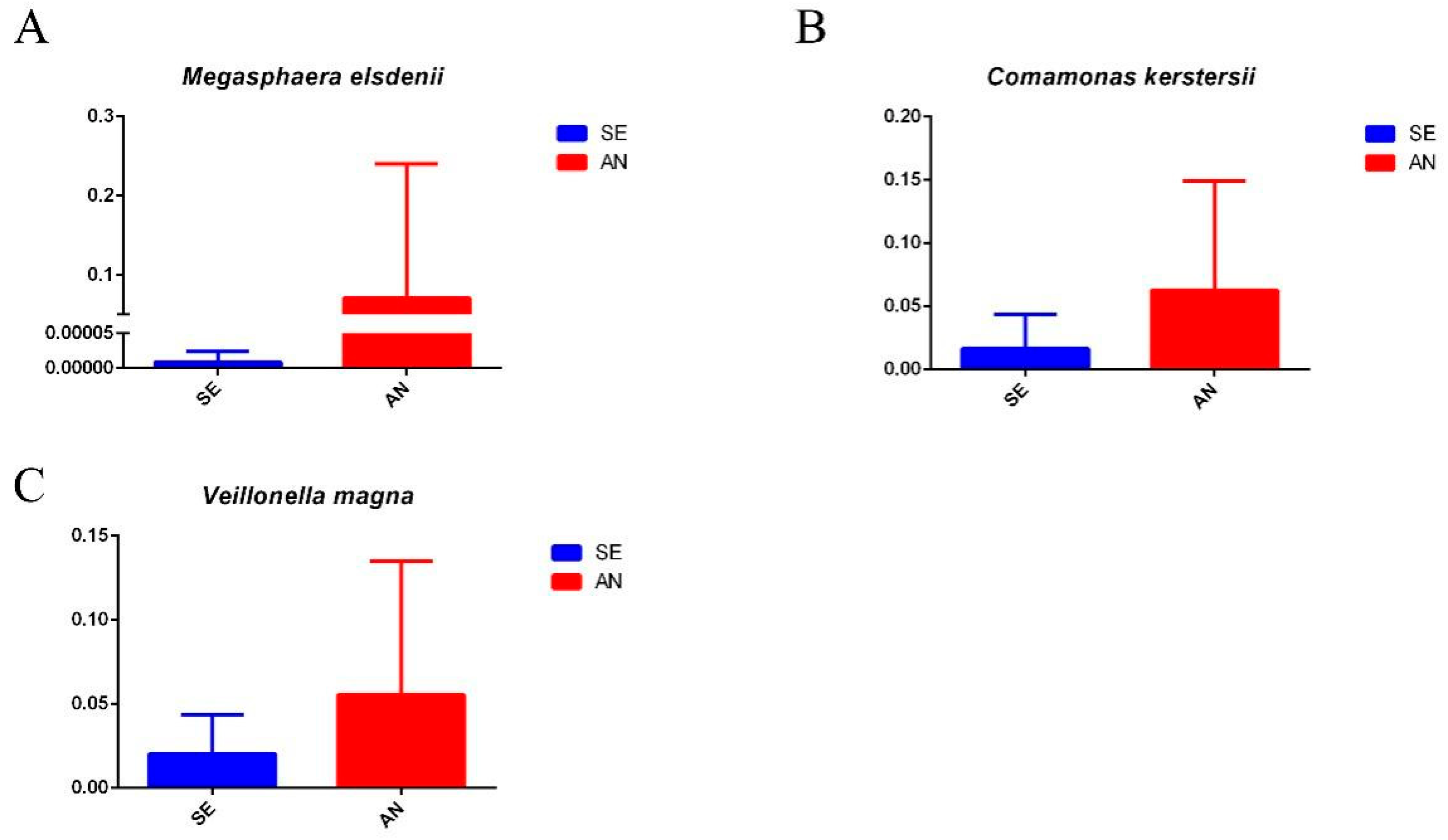
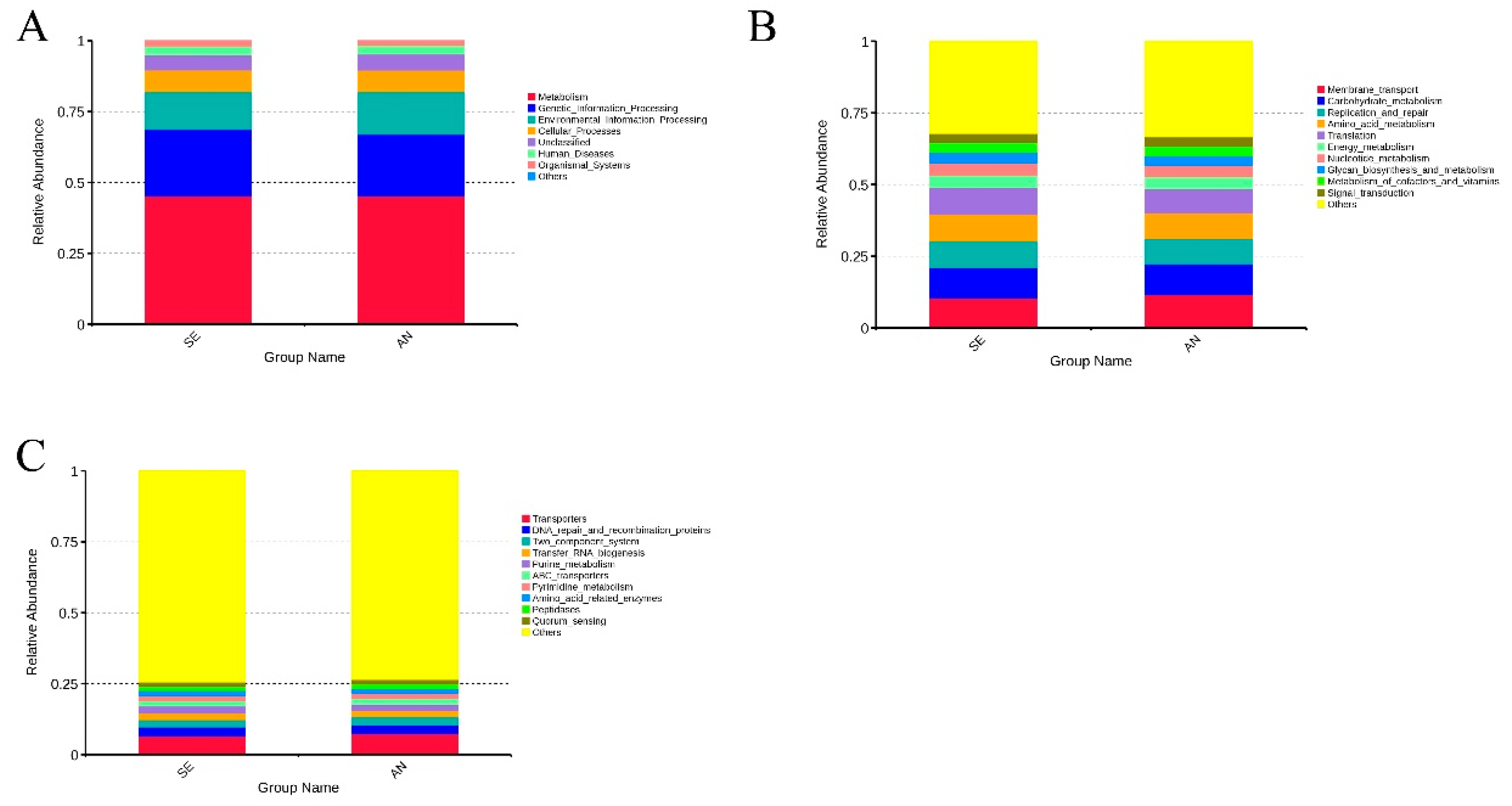
3.6. Jejunal Bacterial Communities Profile of Lambs with Different Susceptibility to Escherichia coli F17
4. Discussion
4.1. Effect of E. coli F17 on Lambs
4.2. Intestinal Microbiota in Lambs with Different Susceptibility to E. coli F17
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Fratamico, P.M.; DebRoy, C.; Liu, Y.; Needleman, D.S.; Baranzoni, G.M.; Feng, P. Advances in Molecular Serotyping and Subtyping of Escherichia coli. Front. Microbiol. 2016, 7, 644. [Google Scholar] [CrossRef] [PubMed]
- Viidu, D.A.; Motus, K. Implementation of a pre-calving vaccination programme against rotavirus, coronavirus and enterotoxigenic Escherichia coli (F5) and association with dairy calf survival. BMC Vet. Res. 2022, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Cid, D.; Ruiz Santa Quiteria, J.A.; de la Fuente, R. F17 fimbriae in Escherichia coli from lambs and kids. Vet. Rec. 1993, 132, 251. [Google Scholar] [CrossRef] [PubMed]
- Vogeli, P.; Bertschinger, H.U.; Stamm, M.; Stricker, C.; Hagger, C.; Fries, R.; Rapacz, J.; Stranzinger, G. Genes specifying receptors for F18 fimbriated Escherichia coli, causing oedema disease and postweaning diarrhoea in pigs, map to chromosome 6. Anim. Genet. 1996, 27, 321–328. [Google Scholar]
- Duan, Q.; Wu, W.; Pang, S.; Pan, Z.; Zhang, W.; Zhu, G. Coimmunization with Two Enterotoxigenic Escherichia coli (ETEC) Fimbrial Multiepitope Fusion Antigens Induces the Production of Neutralizing Antibodies against Five ETEC Fimbriae (F4, F5, F6, F18, and F41). Appl. Environ. Microbiol. 2020, 86, e00217-20. [Google Scholar] [CrossRef]
- Ryu, J.H.; Kim, S.; Park, J.; Choi, K.S. Characterization of virulence genes in Escherichia coli strains isolated from pre-weaned calves in the Republic of Korea. Acta Vet. Scand. 2020, 62, 45. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Ivanov, P.A.; Senchenkova, S.N.; Naumenko, O.I.; Ovchinnikova, O.O.; Shashkov, A.S.; Golomidova, A.K.; Babenko, V.V.; Kulikov, E.E.; Letarov, A.V. Structure and gene cluster of the O antigen of Escherichia coli F17, a candidate for a new O-serogroup. Int. J. Biol. Macromol. 2019, 124, 389–395. [Google Scholar] [CrossRef]
- Bihannic, M.; Ghanbarpour, R.; Auvray, F.; Cavalie, L.; Chatre, P.; Boury, M.; Brugere, H.; Madec, J.Y.; Oswald, E. Identification and detection of three new F17 fimbrial variants in Escherichia coli strains isolated from cattle. Vet. Res. 2014, 45, 76. [Google Scholar] [CrossRef]
- Contrepois, M.; Bertin, Y.; Pohl, P.; Picard, B.; Girardeau, J.P. A study of relationships among F17 a producing enterotoxigenic and non-enterotoxigenic Escherichia coli strains isolated from diarrheic calves. Vet. Microbiol. 1998, 64, 75–81. [Google Scholar] [CrossRef]
- Siuce, J.; Maturrano, L.; Wheeler, J.C.; Rosadio, R. Diarrheagenic Escherichia coli isolates from neonatal alpacas mainly display F17 fimbriae adhesion gene. Trop. Anim. Health Prod. 2020, 52, 3917–3921. [Google Scholar] [CrossRef]
- Dezfulian, H.; Batisson, I.; Fairbrother, J.M.; Lau, P.C.; Nassar, A.; Szatmari, G.; Harel, J. Presence and characterization of extraintestinal pathogenic Escherichia coli virulence genes in F165-positive E. coli strains isolated from diseased calves and pigs. J. Clin. Microbiol. 2003, 41, 1375–1385. [Google Scholar] [CrossRef]
- Zhang, Y.; Si, X.; Yang, L.; Wang, H.; Sun, Y.; Liu, N. Association between intestinal microbiota and inflammatory bowel disease. Anim. Model. Exp. Med. 2022, 5, 311–322. [Google Scholar] [CrossRef]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef]
- Sabatino, A.; Regolisti, G.; Cosola, C.; Gesualdo, L.; Fiaccadori, E. Intestinal Microbiota in Type 2 Diabetes and Chronic Kidney Disease. Curr. Diab. Rep. 2017, 17, 16. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Li, X.Q.; Zhang, W.; Zhou, D.; Liu, H.Y.; Wang, J.F. Dose-dependent effects of Lactobacillus rhamnosus on serum interleukin-17 production and intestinal T-cell responses in pigs challenged with Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 1787–1798. [Google Scholar] [CrossRef]
- Higginson, E.E.; Sayeed, M.A.; Pereira Dias, J.; Shetty, V.; Ballal, M.; Srivastava, S.K.; Willis, I.; Qadri, F.; Dougan, G.; Mutreja, A. Microbiome Profiling of Enterotoxigenic Escherichia coli (ETEC) Carriers Highlights Signature Differences between Symptomatic and Asymptomatic Individuals. mBio 2022, 13, e0015722. [Google Scholar] [CrossRef]
- Bertin, Y.; Martin, C.; Oswald, E.; Girardeau, J.P. Rapid and specific detection of F17-related pilin and adhesin genes in diarrheic and septicemic Escherichia coli strains by multiplex PCR. J. Clin. Microbiol. 1996, 34, 2921–2928. [Google Scholar] [CrossRef]
- Jin, C.; Bao, J.; Wang, Y.; Chen, W.; Wu, T.; Wang, L.; Lv, X.; Gao, W.; Wang, B.; Zhu, G.; et al. Changes in long non-coding RNA expression profiles related to the antagonistic effects of Escherichia coli F17 on lamb spleens. Sci. Rep. 2018, 8, 16514. [Google Scholar] [CrossRef]
- Wu, Z.C.; Liu, Y.; Dong, W.H.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. CD14 in the TLRs signaling pathway is associated with the resistance to E. coli F18 in Chinese domestic weaned piglets. Sci. Rep.-Uk 2016, 6, 24611. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Roewer, L.; Kayser, M.; Dieltjes, P.; Nagy, M.; Bakker, E.; Krawczak, M.; de Knijff, P. Analysis of molecular variance (AMOVA) of Y-chromosome-specific microsatellites in two closely related human populations. Hum. Mol. Genet. 1996, 5, 1029–1033. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Asshauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Ren, W.; Wang, P.; Yan, J.; Liu, G.; Zeng, B.; Hussain, T.; Peng, C.; Yin, J.; Li, T.; Wei, H.; et al. Melatonin alleviates weanling stress in mice: Involvement of intestinal microbiota. J. Pineal Res. 2018, 64, e12448. [Google Scholar] [CrossRef]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal Enterotoxigenic Escherichia coli. EcoSal Plus 2016, 7, 10. [Google Scholar] [CrossRef]
- Burgess, M.N.; Bywater, R.J.; Cowley, C.M.; Mullan, N.A.; Newsome, P.M. Biological evaluation of a methanol-soluble, heat-stable Escherichia coli enterotoxin in infant mice, pigs, rabbits, and calves. Infect. Immun. 1978, 21, 526–531. [Google Scholar] [CrossRef]
- Barry, J.; Bokkers, E.A.M.; Berry, D.P.; de Boer, I.J.M.; McClure, J.; Kennedy, E. Associations between colostrum management, passive immunity, calf-related hygiene practices, and rates of mortality in preweaning dairy calves. J. Dairy Sci. 2019, 102, 10266–10276. [Google Scholar] [CrossRef]
- Zhang, W.; Heng, J.; Kim, S.W.; Chen, F.; Deng, Z.; Zhang, S.; Guan, W. Dietary enzymatically-treated Artemisia annua L. supplementation could alleviate oxidative injury and improve reproductive performance of sows reared under high ambient temperature. J. Therm. Biol. 2020, 94, 102751. [Google Scholar] [CrossRef]
- Rodrigues, L.M.; Neto, T.; Garbossa, C.A.P.; Martins, C.; Garcez, D.; Alves, L.K.S.; de Abreu, M.L.T.; Ferreira, R.A.; Cantarelli, V.S. Benzoic Acid Combined with Essential Oils Can Be an Alternative to the Use of Antibiotic Growth Promoters for Piglets Challenged with E. coli F4. Animals 2020, 10, 1978. [Google Scholar] [CrossRef]
- Choi, J.; Wang, L.; Liu, S.; Lu, P.; Zhao, X.; Liu, H.; Lahaye, L.; Santin, E.; Liu, S.; Nyachoti, M.; et al. Effects of a microencapsulated formula of organic acids and essential oils on nutrient absorption, immunity, gut barrier function, and abundance of enterotoxigenic Escherichia coli F4 in weaned piglets challenged with E. coli F4. J. Anim. Sci. 2020, 98, skaa259. [Google Scholar] [CrossRef]
- Lopez-Colom, P.; Castillejos, L.; Rodriguez-Sorrento, A.; Puyalto, M.; Mallo, J.J.; Martin-Orue, S.M. Impact of in-feed sodium butyrate or sodium heptanoate protected with medium-chain fatty acids on gut health in weaned piglets challenged with Escherichia coli F4+. Arch. Anim. Nutr. 2020, 74, 271–295. [Google Scholar] [CrossRef]
- Mathew, A.G.; Upchurch, W.G.; Chattin, S.E. Incidence of antibiotic resistance in fecal Escherichia coli isolated from commercial swine farms. J. Anim. Sci. 1998, 76, 429–434. [Google Scholar] [CrossRef]
- Rhouma, M.; Braley, C.; Theriault, W.; Thibodeau, A.; Quessy, S.; Fravalo, P. Evolution of Pig Fecal Microbiota Composition and Diversity in Response to Enterotoxigenic Escherichia coli Infection and Colistin Treatment in Weaned Piglets. Microorganisms 2021, 9, 1459. [Google Scholar] [CrossRef]
- Bin, P.; Tang, Z.; Liu, S.; Chen, S.; Xia, Y.; Liu, J.; Wu, H.; Zhu, G. Intestinal microbiota mediates Enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet. Res. 2018, 14, 385. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shang, Z.; Liu, X.; Qiao, Y.; Wang, K.; Qiao, J. Clostridium butyricum Alleviates Enterotoxigenic Escherichia coli K88-Induced Oxidative Damage Through Regulating the p62-Keap1-Nrf2 Signaling Pathway and Remodeling the Cecal Microbial Community. Front. Immunol. 2021, 12, 771826. [Google Scholar] [CrossRef] [PubMed]
- Roussel, C.; De Paepe, K.; Galia, W.; De Bodt, J.; Chalancon, S.; Leriche, F.; Ballet, N.; Denis, S.; Alric, M.; Van de Wiele, T.; et al. Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 2020, 18, 141. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Hutchings, M.R.; Hutchings, K.E.K.; Gally, D.L.; Houdijk, J.G.M. Changes in the Ileal, but Not Fecal, Microbiome in Response to Increased Dietary Protein Level and Enterotoxigenic Escherichia coli Exposure in Pigs. Appl. Environ. Microbiol. 2019, 85, e01252-19. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.O.; Chaban, B.; Harding, J.C.; Hill, J.E. Characterization of the fecal microbiota of pigs before and after inoculation with “Brachyspira hampsonii”. PLoS ONE 2014, 9, e106399. [Google Scholar] [CrossRef]
- Pop, M.; Walker, A.W.; Paulson, J.; Lindsay, B.; Antonio, M.; Hossain, M.A.; Oundo, J.; Tamboura, B.; Mai, V.; Astrovskaya, I.; et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014, 15, R76. [Google Scholar] [CrossRef]
- Gorkiewicz, G.; Thallinger, G.G.; Trajanoski, S.; Lackner, S.; Stocker, G.; Hinterleitner, T.; Gully, C.; Hogenauer, C. Alterations in the colonic microbiota in response to osmotic diarrhea. PLoS ONE 2013, 8, e55817. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef]
- Duarte, M.E.; Tyus, J.; Kim, S.W. Synbiotic Effects of Enzyme and Probiotics on Intestinal Health and Growth of Newly Weaned Pigs Challenged With Enterotoxigenic F18+ Escherichia coli. Front. Vet. Sci. 2020, 7, 573. [Google Scholar] [CrossRef]
- Huang, C.; Ming, D.; Wang, W.; Wang, Z.; Hu, Y.; Ma, X.; Wang, F. Pyrroloquinoline Quinone Alleviates Jejunal Mucosal Barrier Function Damage and Regulates Colonic Microbiota in Piglets Challenged With Enterotoxigenic Escherichia coli. Front. Microbiol. 2020, 11, 1754. [Google Scholar] [CrossRef]
- Almuzara, M.; Cittadini, R.; Estraviz, M.L.; Ellis, A.; Vay, C. First report of Comamonas kerstersii causing urinary tract infection. New Microbes New Infect 2018, 24, 4–7. [Google Scholar] [CrossRef]
- Liu, J.; Merritt, J.; Qi, F. Genetic transformation of Veillonella parvula. FEMS Microbiol. Lett. 2011, 322, 138–144. [Google Scholar] [CrossRef]


| Group | No. of Lambs | Treatment | Dose (CFU) |
|---|---|---|---|
| High-dose challenge group | 20 | E. coli F17 | 5 × 109 |
| Low-dose challenge group | 20 | E. coli F17 | 5 × 108 |
| Positive control group | 5 | E. coli F17 | 1 × 1010 |
| Negative control group | 5 | LB medium | - |
| Type | Form |
|---|---|
| Type 1 | Separate hard lumps, like nuts |
| Type 2 | Sausage-shaped but lumpy |
| Type 3 | Like a sausage or snake but with cracks on its surface |
| Type 4 | Soft blobs with clear-cut edges |
| Type 5 | Like a sausage or snake, smooth and soft. |
| Type 6 | Fluffy pieces with ragged edges, a mushy stool |
| Type 7 | Watery, no solid pieces |
| Group | No. of Challenged Lambs | Prevalence | Death Rate |
|---|---|---|---|
| Low-dose challenge group | 20 | 14/20, 75% | 2/20, 10% |
| High-dose challenge group | 20 | 18/20, 90% | 7/20, 35% |
| Positive control group | 5 | 5/5, 100% | 3/5, 60% |
| Negative control group | 5 | 0/5, 0% | 0/5, 0% |
| 10 µL Coated Plate | Intestinal Tract | Dilution Multiple | Average Count of Bacterial (CFU/mL) | |||
|---|---|---|---|---|---|---|
| 104 | 105 | 106 | 107 | |||
| sensitive candidates | duodenum | >1000 | >500 | 128 | 13 | 1.29 × 108 |
| jejunum | >1000 | >500 | 176 | 19 | 1.83 × 108 | |
| ileum | >1000 | >500 | 120 | 7 | 0.95 × 108 | |
| resistant candidates | duodenum | 113 | 5 | NG | NG | 8.20 × 105 |
| jejunum | >500 | 170 | 9 | NG | 1.30 × 106 | |
| ileum | 119 | 8 | NG | NG | 9.95 × 105 | |
| positive control group | duodenum | >1000 | >500 | 158 | 8 | 1.19 × 108 |
| jejunum | >1000 | >500 | 241 | 11 | 1.76 × 108 | |
| ileum | >1000 | >500 | 147 | 3 | 0.89 × 108 | |
| negative control group | duodenum | 70 | 6 | NG | NG | 6.50 × 105 |
| jejunum | 64 | NG | NG | NG | 6.40 × 105 | |
| ileum | 336 | 27 | NG | NG | 3.03 × 105 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Chen, W.; Yuan, Z. Characterization of Intestinal Microbiota in Lambs with Different Susceptibility to Escherichia coli F17. Vet. Sci. 2022, 9, 670. https://doi.org/10.3390/vetsci9120670
Sun J, Chen W, Yuan Z. Characterization of Intestinal Microbiota in Lambs with Different Susceptibility to Escherichia coli F17. Veterinary Sciences. 2022; 9(12):670. https://doi.org/10.3390/vetsci9120670
Chicago/Turabian StyleSun, Jingyi, Weihao Chen, and Zehu Yuan. 2022. "Characterization of Intestinal Microbiota in Lambs with Different Susceptibility to Escherichia coli F17" Veterinary Sciences 9, no. 12: 670. https://doi.org/10.3390/vetsci9120670
APA StyleSun, J., Chen, W., & Yuan, Z. (2022). Characterization of Intestinal Microbiota in Lambs with Different Susceptibility to Escherichia coli F17. Veterinary Sciences, 9(12), 670. https://doi.org/10.3390/vetsci9120670







