miR-708-5p Regulates Myoblast Proliferation and Differentiation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Transfection
| mimics | Forward: | 5′-AAGGAGCUUACAAUCUAGCUGGG-3′ |
| Reverse: | 5′-CAGCUAGAUUGUAAGCUCCUUUU-3′ | |
| nc | Forward: | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Reverse: | 5′-ACGUGACACGUUCGGAGAATT-3′ | |
| Inhibitor: | 5′-CCCAGCUAGAUUGUAAGCUCCUU-3′ | |
| Inhibitor nc: | 5′-CAGUACUUUUGUGUAGUACAA-3′ | |
2.3. Real-Time Quantitative PCR
2.4. Cell Proliferation Assays
2.5. Flow Cytometry
2.6. Immunofluorescence Analysis
2.7. Target Genes Prediction
2.8. The PPI and miRNA–mRNA Network Construction
2.9. Statistical Analyses
3. Results
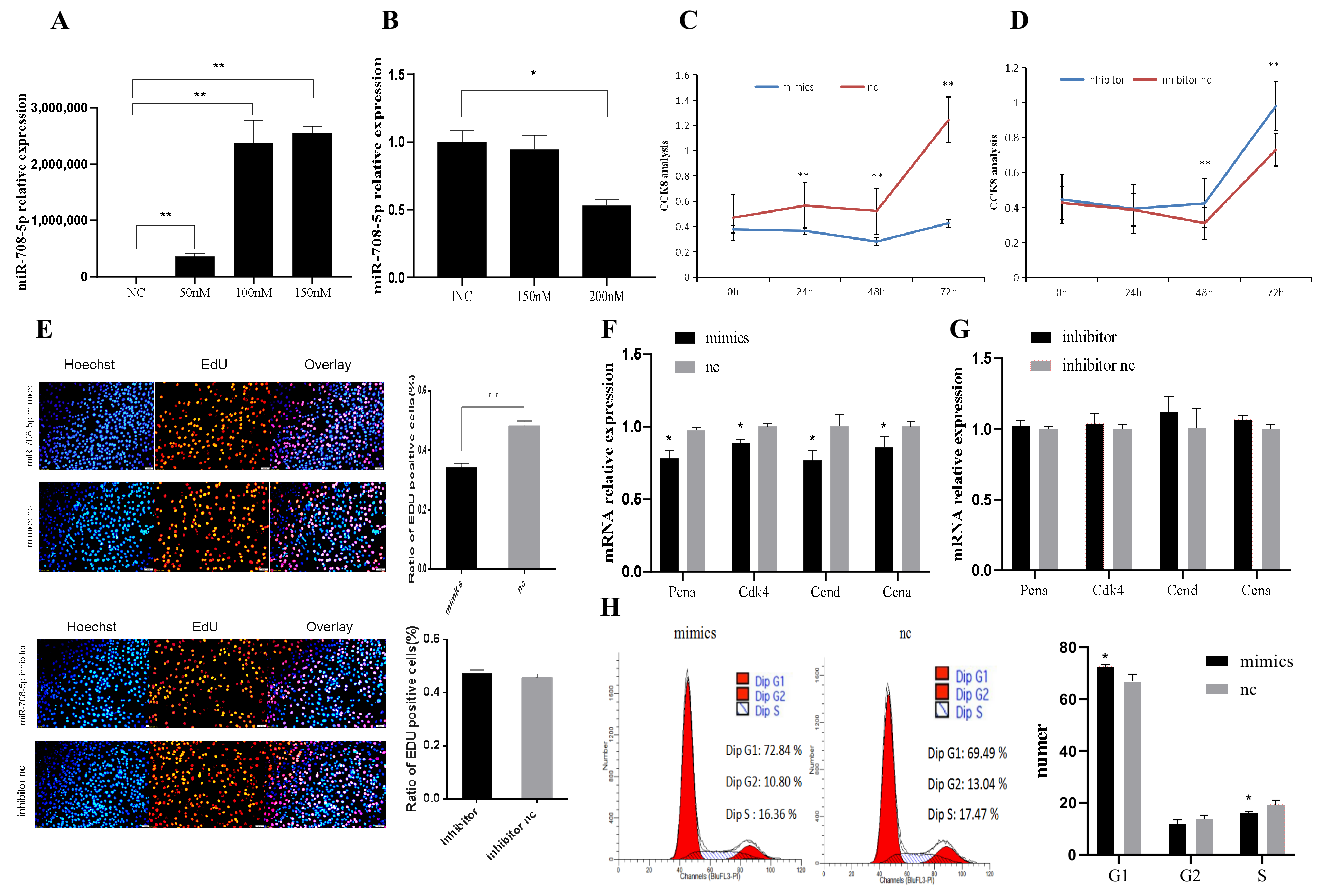
3.1. miR-708-5p Inhibits C2C12 Cells Proliferation
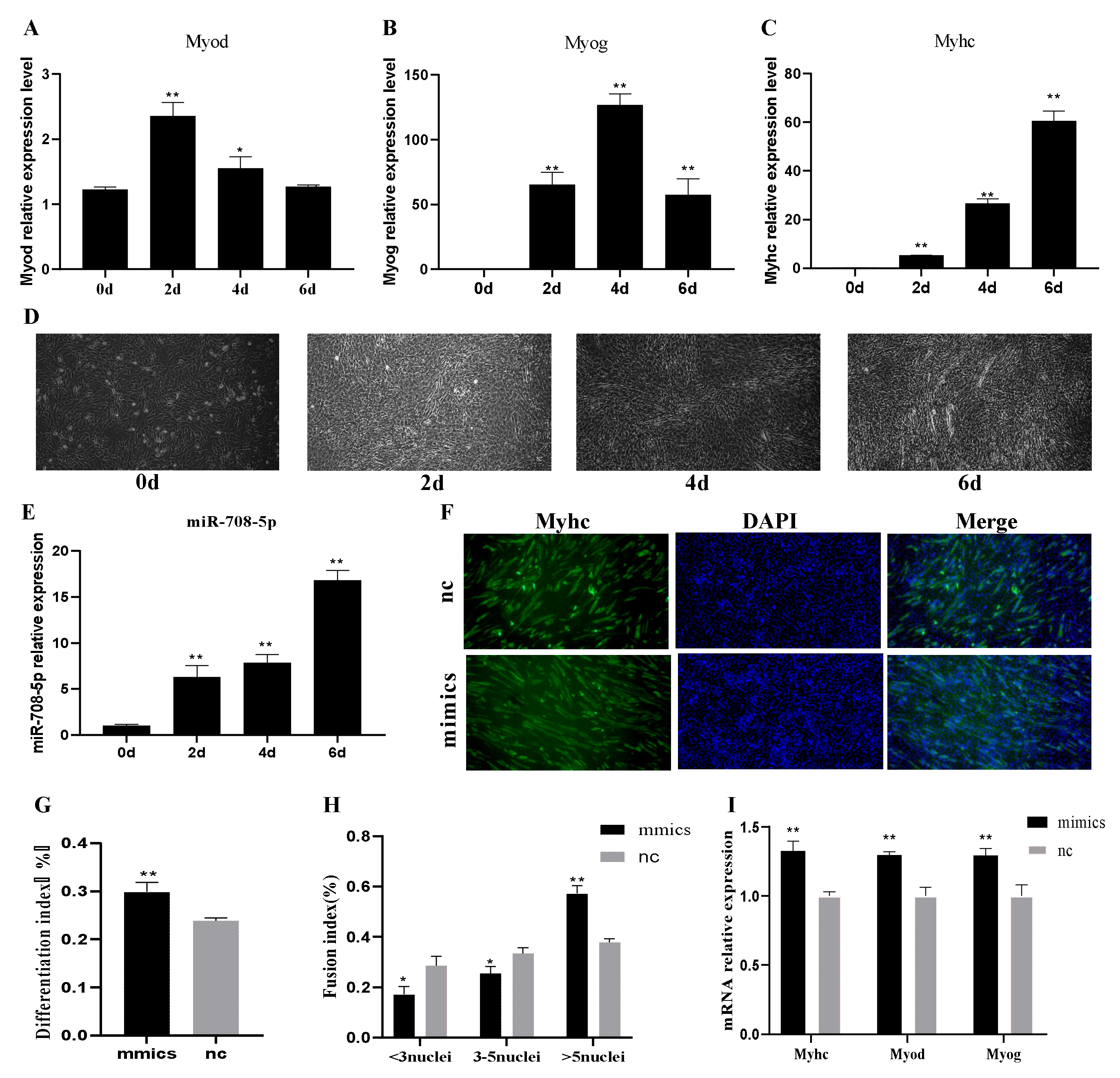
3.2. miR-708-5p Promotes C2C12 Cells Differentiation
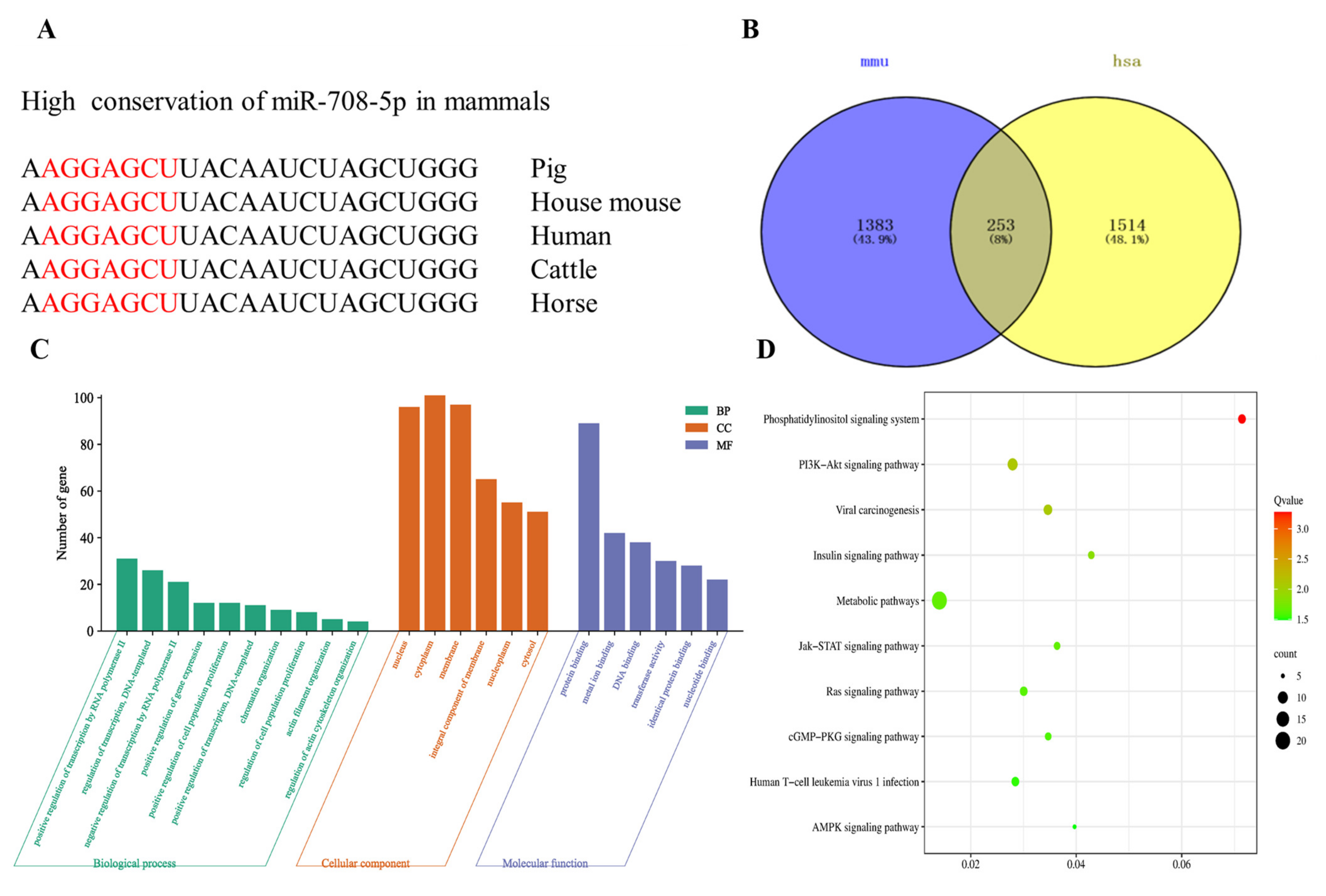
3.3. Target Gene Prediction of miR-708-5p
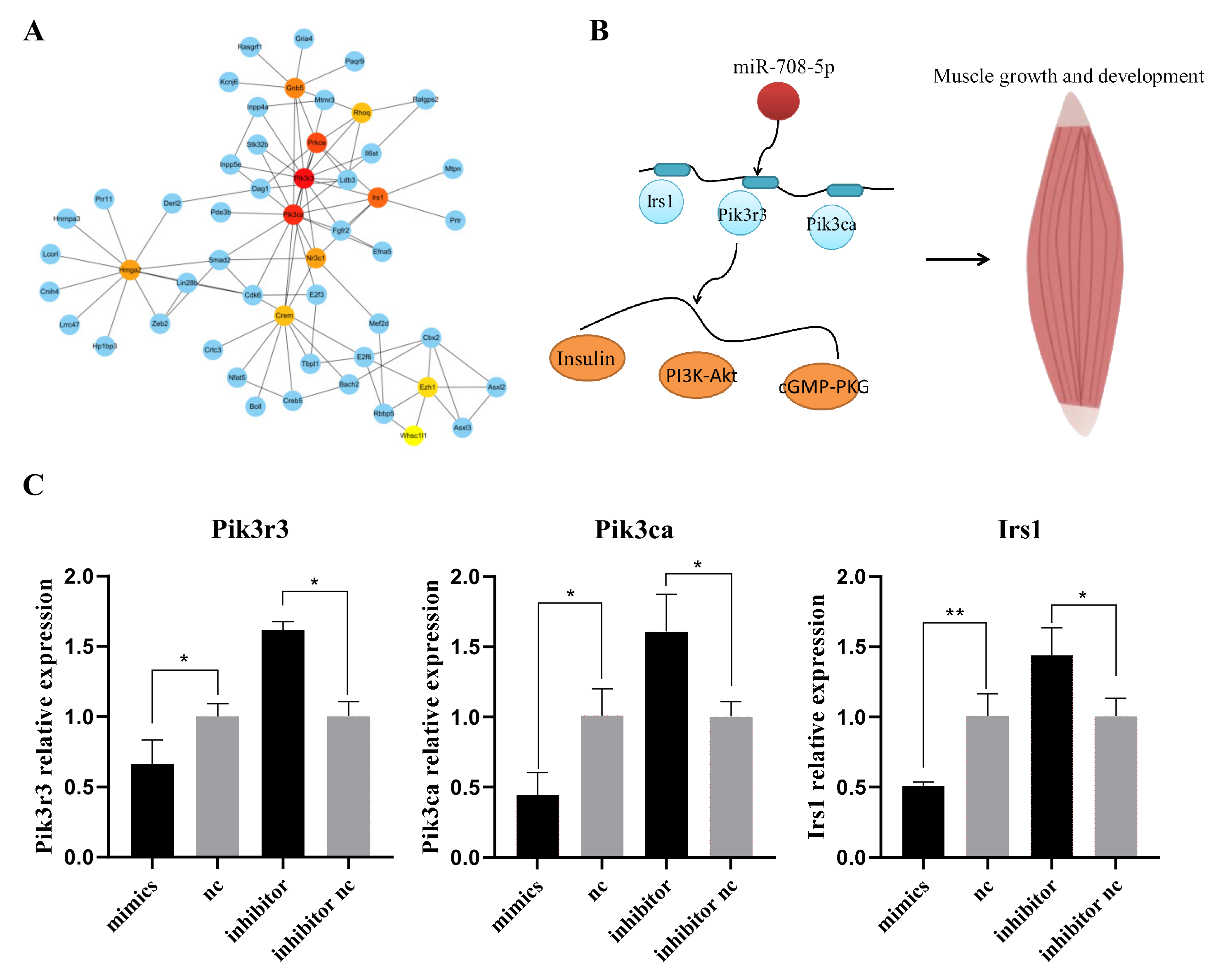
3.4. The PPI and miRNA–mRNA Network Construction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ge, G.; Yang, D.; Tan, Y.; Chen, Y.; Jiang, D.; Jiang, A.; Li, Q.; Liu, Y.; Zhong, Z.; Li, X.; et al. miR-10b-5p regulates C2C12 myoblasts proliferation and differentiation. Biosci. Biotechnol. Biochem. 2019, 83, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Rehfeldt, C.; Kuhn, G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis. J. Anim. Sci. 2006, 84 (Suppl. 13), E113–E123. [Google Scholar] [CrossRef] [PubMed]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How muscle structure and composition influence meat and flesh quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G. Muscle development in the embryo and hatchling. Poult Sci. 2007, 86, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Lee, J.H.; Kang, N.; Kim, K.N.; Jeon, Y.J. The Effects of marine algal polyphenols, phlorotannins, on skeletal muscle growth in C2C12 muscle cells via Smad and IGF-1 signaling pathways. Mar. Drugs 2021, 19, 266. [Google Scholar] [CrossRef]
- Relaix, F.; Bencze, M.; Borok, M.J.; Der Vartanian, A.; Gattazzo, F.; Mademtzoglou, D.; Perez-Diaz, S.; Prola, A.; Reyes-Fernandez, P.C.; Rotini, A.; et al. Perspectives on skeletal muscle stem cells. Nat. Commun. 2021, 12, 692. [Google Scholar] [CrossRef]
- Zhang, Y.; Yun, Z.; Gong, L.; Qu, H.; Duan, X.; Jiang, Y.; Zhu, H. Comparison of miRNA evolution and function in plants and animals. microRNA 2018, 7, 4–10. [Google Scholar] [CrossRef]
- Zhao, Y.; Cong, L.; Lukiw, W.J. Plant and animal microRNAs (miRNAs) and their potential for inter-kingdom communication. Cell. Mol. Neurobiol. 2018, 38, 133–140. [Google Scholar] [CrossRef]
- Yang, L.; Qi, Q.; Wang, J.; Song, C.; Wang, Y.; Chen, X.; Chen, H.; Zhang, C.; Hu, L.; Fang, X. MiR-452 regulates C2C12 myoblast proliferation and differentiation via targeting ANGPT1. Front. Genet. 2021, 12, 640807. [Google Scholar] [CrossRef]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef]
- Wu, J.; Fan, W.; Ma, L.; Geng, X. miR-708-5p promotes fibroblast-like synoviocytes’ cell apoptosis and ameliorates rheumatoid arthritis by the inhibition of Wnt3a/beta-catenin pathway. Drug Des. Dev. Ther. 2018, 12, 3439–3447. [Google Scholar] [CrossRef]
- Wu, N.; Liu, G.B.; Zhang, Y.M.; Wang, Y.; Zeng, H.T.; Xiang, H. MiR-708-5p/Pit-1 axis mediates high phosphate-induced calcification in vascular smooth muscle cells via Wnt8b/beta-catenin pathway. Kaohsiung J. Med. Sci. 2022, 38, 653–661. [Google Scholar] [CrossRef]
- Monteleone, N.J.; Lutz, C.S. miR-708-5p: A microRNA with emerging roles in cancer. Oncotarget 2017, 8, 71292–71316. [Google Scholar] [CrossRef]
- Wang, R.; Feng, Y.; Xu, H.; Huang, H.; Zhao, S.; Wang, Y.; Li, H.; Cao, J.; Xu, G.; Huang, S. Synergistic effects of miR-708-5p and miR-708-3p accelerate the progression of osteoporosis. J. Int. Med. Res. 2020, 48, 1220777567. [Google Scholar] [CrossRef]
- Shen, H.; Liu, T.; Fu, L.; Zhao, S.; Fan, B.; Cao, J.; Li, X. Identification of microRNAs involved in dexamethasone-induced muscle atrophy. Mol. Cell. Biochem. 2013, 381, 105–113. [Google Scholar] [CrossRef]
- Deng, S.; Zhao, Q.; Zhou, X.; Zhang, L.; Bao, L.; Zhen, L.; Zhang, Y.; Fan, H.; Liu, Z.; Yu, Z. Neonatal heart-enriched miR-708 promotes differentiation of cardiac progenitor cells in rats. Int. J. Mol. Sci. 2016, 17, 875. [Google Scholar] [CrossRef]
- Guilbaud, M.; Gentil, C.; Peccate, C.; Gargaun, E.; Holtzmann, I.; Gruszczynski, C.; Falcone, S.; Mamchaoui, K.; Yaou, R.B.; Leturcq, F.; et al. miR-708-5p and miR-34c-5p are involved in nNOS regulation in dystrophic context. Skelet Muscle 2018, 8, 15. [Google Scholar] [CrossRef]
- Zhao, Z.; Qin, X. MicroRNA-708 targeting ZNF549 regulates colon adenocarcinoma development through PI3K/AKt pathway. Sci. Rep. 2020, 10, 16729. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, C.; Zhang, L.; Zhang, Y.; Fu, F.; Chen, Y.; Zhou, J. MicroRNA-708 suppresses cell proliferation and enhances chemosensitivity of cervical cancer cells to cDDP by negatively targeting Timeless. OncoTargets Ther. 2020, 13, 225–235. [Google Scholar] [CrossRef]
- Sun, S.; Hang, T.; Zhang, B.; Zhu, L.; Wu, Y.; Lv, X.; Huang, Q.; Yao, H. miRNA-708 functions as a tumor suppressor in colorectal cancer by targeting ZEB1 through Akt/mTOR signaling pathway. Am. J. Transl. Res. 2019, 11, 5338–5356. [Google Scholar]
- Sui, C.; Liu, D.; Hu, Y.; Zhang, L. MicroRNA-708-5p affects proliferation and invasion of osteosarcoma cells by targeting URGCP. Exp. Ther. Med. 2019, 17, 2235–2241. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, T.; Wu, L.; Xu, H.L.; Liu, R.M. Molecular mechanisms of MCM3AP-AS1 targeted the regulation of miR-708-5p on cell proliferation and apoptosis in gastric cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2452–2461. [Google Scholar] [PubMed]
- Salant, G.M.; Tat, K.L.; Goodrich, J.A.; Kugel, J.F. miR-206 knockout shows it is critical for myogenesis and directly regulates newly identified target mRNAs. RNA Biol. 2020, 17, 956–965. [Google Scholar] [PubMed]
- Xu, M.; Chen, X.; Chen, D.; Yu, B.; Li, M.; He, J.; Huang, Z. Regulation of skeletal myogenesis by microRNAs. J. Cell Physiol. 2020, 235, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, J.; Qi, Q.; Yang, L.; Wang, Y.; Zhang, C.; Hu, L.; Chen, H.; Fang, X. miR-487b-3p suppresses the proliferation and differentiation of myoblasts by targeting IRS1 in skeletal muscle myogenesis. Int. J. Biol. Sci. 2018, 14, 760–774. [Google Scholar] [CrossRef]
- Morgan, J.E.; Moore, S.E.; Walsh, F.S.; Partridge, T.A. Formation of skeletal muscle in vivo from the mouse C2 cell line. J. Cell Sci. 1992, 102 Pt 4, 779–787. [Google Scholar]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Filipkowski, R.K.; Rylski, M.; Wilczynski, G.M.; Konopacki, F.A.; Jaworski, J.; Ciemerych, M.A.; Sicinski, P.; Kaczmarek, L. The critical role of cyclin D2 in adult neurogenesis. J. Cell Biol. 2004, 167, 209–213. [Google Scholar] [CrossRef]
- Zhao, X.; Gu, H.; Wang, L.; Zhang, P.; Du, J.; Shen, L.; Jiang, D.; Wang, J.; Li, X.; Zhang, S.; et al. MicroRNA-23a-5p mediates the proliferation and differentiation of C2C12 myoblasts. Mol. Med. Rep. 2020, 22, 3705–3714. [Google Scholar] [CrossRef]
- Zhang, J.; Hua, C.; Zhang, Y.; Wei, P.; Tu, Y.; Wei, T. KAP1-associated transcriptional inhibitory complex regulates C2C12 myoblasts differentiation and mitochondrial biogenesis via miR-133a repression. Cell Death Dis. 2020, 11, 732. [Google Scholar] [CrossRef]
- Du, C.; Jin, Y.Q.; Qi, J.J.; Ji, Z.X.; Li, S.Y.; An, G.S.; Jia, H.T.; Ni, J.H. Effects of myogenin on expression of late muscle genes through MyoD-dependent chromatin remodeling ability of myogenin. Mol. Cells. 2012, 34, 133–142. [Google Scholar] [CrossRef]
- Li, B.; Xie, S.; Cai, C.; Qian, L.; Jiang, S.; Ma, D.; Xiao, G.; Gao, T.; Yang, J.; Cui, W. MicroRNA-95 promotes myogenic differentiation by down-regulation of aminoacyl-tRNA synthase complex-interacting multifunctional protein 2. Oncotarget 2017, 8, 111356–111368. [Google Scholar] [CrossRef]
- Wang, H.; Nicolay, B.N.; Chick, J.M.; Gao, X.; Geng, Y.; Ren, H.; Gao, H.; Yang, G.; Williams, J.A.; Suski, J.M.; et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017, 546, 426–430. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Yang, Y.; Yang, J.; Qin, B.; Gu, L. miR-217-5p regulates myogenesis in skeletal muscle stem cells by targeting FGFR2. Mol. Med. Rep. 2020, 22, 850–858. [Google Scholar]
- Ogawa, M.; Sakakibara, Y.; Kamemura, K. Requirement of decreased O-GlcNAc glycosylation of Mef2D for its recruitment to the myogenin promoter. Biochem. Biophys. Res. Commun. 2013, 433, 558–562. [Google Scholar] [CrossRef]
- Canaud, G.; Hammill, A.M.; Adams, D.; Vikkula, M.; Keppler-Noreuil, K.M. A review of mechanisms of disease across PIK3CA-related disorders with vascular manifestations. Orphanet. J. Rare Dis. 2021, 16, 306. [Google Scholar] [CrossRef]
- Yuan, L.; Fan, L.; Li, Q.; Cui, W.; Wang, X.; Zhang, Z. Inhibition of miR-181b-5p protects cardiomyocytes against ischemia/reperfusion injury by targeting AKT3 and PI3KR3. J. Cell Biochem. 2019, 120, 19647–19659. [Google Scholar] [CrossRef]
- Eckstein, S.S.; Weigert, C.; Lehmann, R. Divergent Roles of IRS (Insulin Receptor Substrate) 1 and 2 in Liver and Skeletal Muscle. Curr. Med. Chem. 2017, 24, 1827–1852. [Google Scholar] [CrossRef]
| Gene | Primers | Primer Sequence (5′→3′) | Annealing Temperature (°C) |
|---|---|---|---|
| U6 | F | CTCGCTTCGGCAGCACA | 60 |
| R | AACGCTTCACGAATTTGCGT | ||
| β-actin | F | CTTGCGTTTGGCTATCCGTG | 60 |
| R | TCTCCGAAGCCAGCAATTCA | ||
| miR-708-5p | F | TCGGCAGGAAGGAGCTTACAAT | 60 |
| R | CTCAACTGGTGTCGTGGA | ||
| miR-708-5p | RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAGCTA | |
| Cdk4 | F | CGAGCGTAAGGCTGATGGAT | 60 |
| R | CCAGGCCGCTTAGAAACTGA | ||
| Ccna | F | GAGCTCCCAAGCTCTACTGC | 60 |
| R | TTTTCATGGGCAGTCCTGGT | ||
| Ccnd | F | GAAGGAGGTAAGGGAAGCACTC | 60 |
| R | TAGCGCTCCTCGATGGTCAA | ||
| Pcna | F | GCCGAGACCTTAGCCACATT | 60 |
| R | GTAGGAGACAGTGGAGTGGC | ||
| Myhc | F | AGCCAAGTGCTAGACCGAAA | 60 |
| R | CGTCGTGAGCCCAAAACTTC | ||
| Myog | F | CAGCCCAGCGAGGGAATTTA | 60 |
| R | GGTCAGGGCACTCATGTCTC | ||
| Myod | F | AAGACGACTCTCACGGCTTG | 60 |
| R | GCAGGTCTGGTGAGTCGAAA | ||
| Pik3r3 | F | TTGCAGACGGGGAAGTGAAG | 60 |
| R | GACGTTGAGGGAGTCGTTGT | ||
| Pik3ca | F | CTCAGCTCTCACCCTCCTCT | 60 |
| R | GGTCTCTCTTTCCGCTCACA | ||
| Irs1 | F | AATGAGGGCAACTCCCCAAG | 60 |
| R | ATTGACGATCCTCTGGCTGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Lu, H.; Xu, D.; Yu, Z.; Ai, N.; Wang, K.; Li, X.; He, J.; Jiang, J.; Ma, H.; et al. miR-708-5p Regulates Myoblast Proliferation and Differentiation. Vet. Sci. 2022, 9, 641. https://doi.org/10.3390/vetsci9110641
Xu X, Lu H, Xu D, Yu Z, Ai N, Wang K, Li X, He J, Jiang J, Ma H, et al. miR-708-5p Regulates Myoblast Proliferation and Differentiation. Veterinary Sciences. 2022; 9(11):641. https://doi.org/10.3390/vetsci9110641
Chicago/Turabian StyleXu, Xueli, Hui Lu, Dong Xu, Zonggang Yu, Nini Ai, Kaiming Wang, Xintong Li, Jun He, Jun Jiang, Haiming Ma, and et al. 2022. "miR-708-5p Regulates Myoblast Proliferation and Differentiation" Veterinary Sciences 9, no. 11: 641. https://doi.org/10.3390/vetsci9110641
APA StyleXu, X., Lu, H., Xu, D., Yu, Z., Ai, N., Wang, K., Li, X., He, J., Jiang, J., Ma, H., & Zhang, Y. (2022). miR-708-5p Regulates Myoblast Proliferation and Differentiation. Veterinary Sciences, 9(11), 641. https://doi.org/10.3390/vetsci9110641








