Transcriptome Analysis Reveals Genes Contributed to Min Pig Villi Hair Follicle in Different Seasons
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Phenotype Observation of Hair and Follicles of Min Pigs
2.3. Data Collection
2.4. Differentially Expressed Gene (DEG) Analysis
2.5. Gene Enrichment Analysis
2.6. Construction of an mRNA Interactive Network
2.7. Identification and Validation of mRNAs by qRT-PCR
2.8. Promoter Methylation Analysis of Core Genes
3. Results
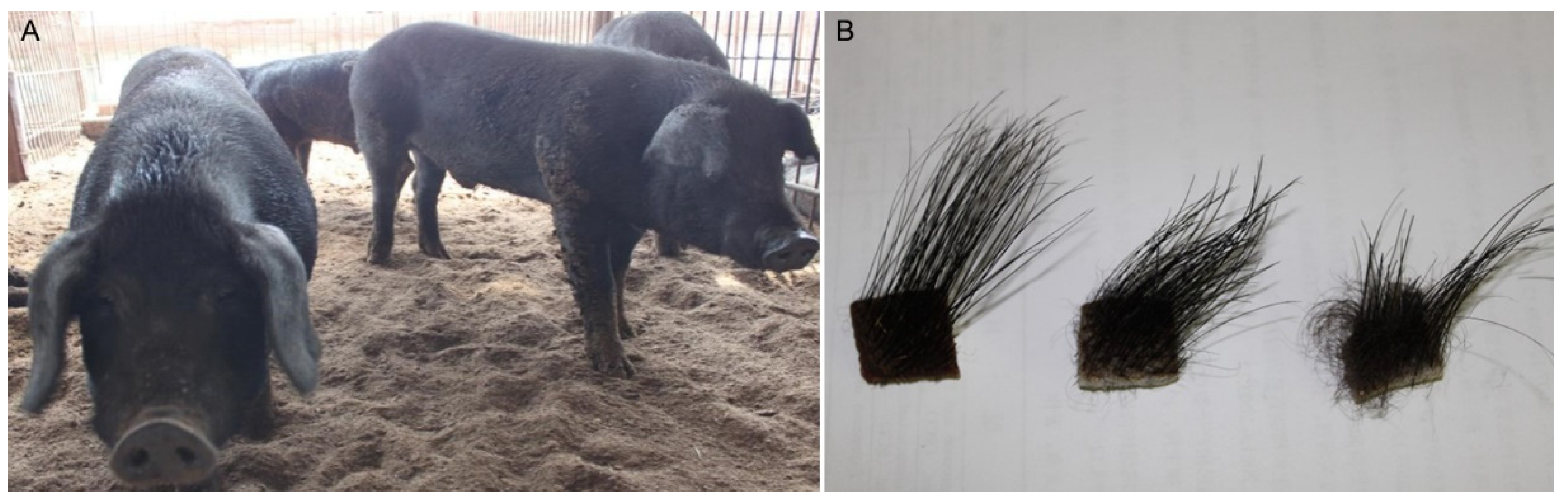
3.1. The Hair Phenotype of Min Pigs
3.1.1. Observations of Hairs of Min Pigs
3.1.2. Villus Length and Density in Min Pigs
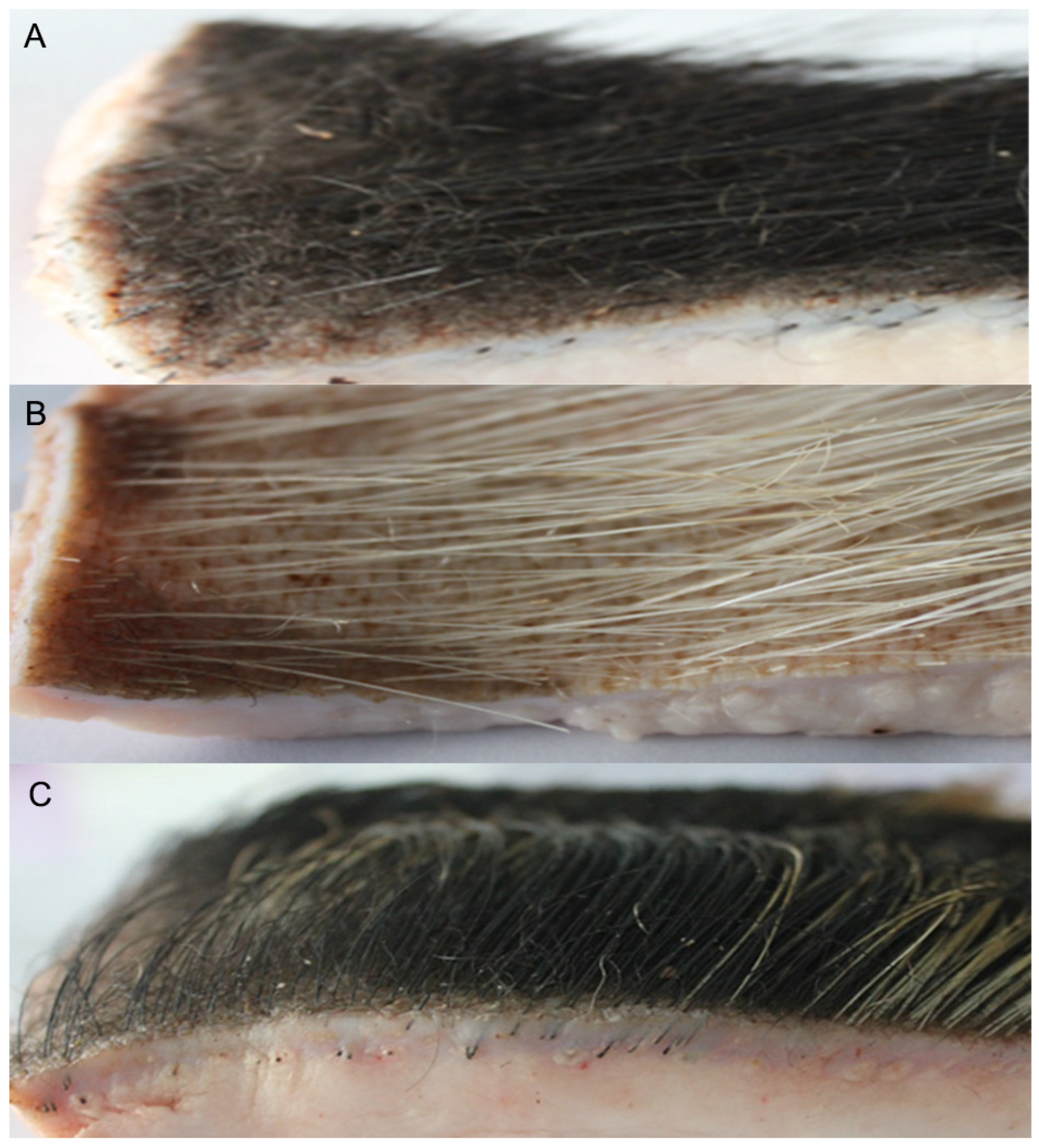
3.1.3. Tissue Analysis of Hair Follicles in Winter in Min Pigs
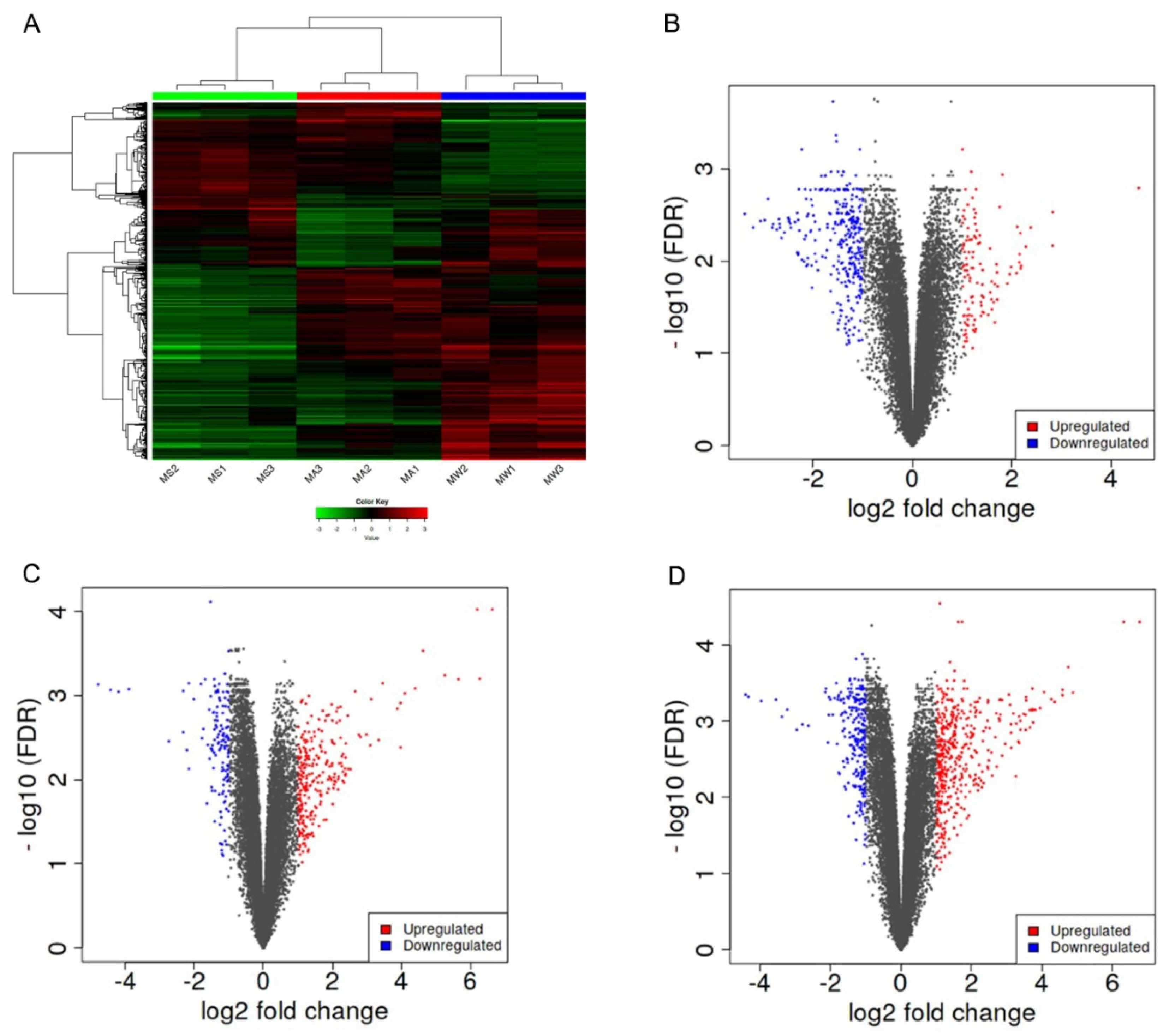
3.2. Identification of Differentially Expressed Genes (DEG) in the Skin of Pigs in Different Seasons
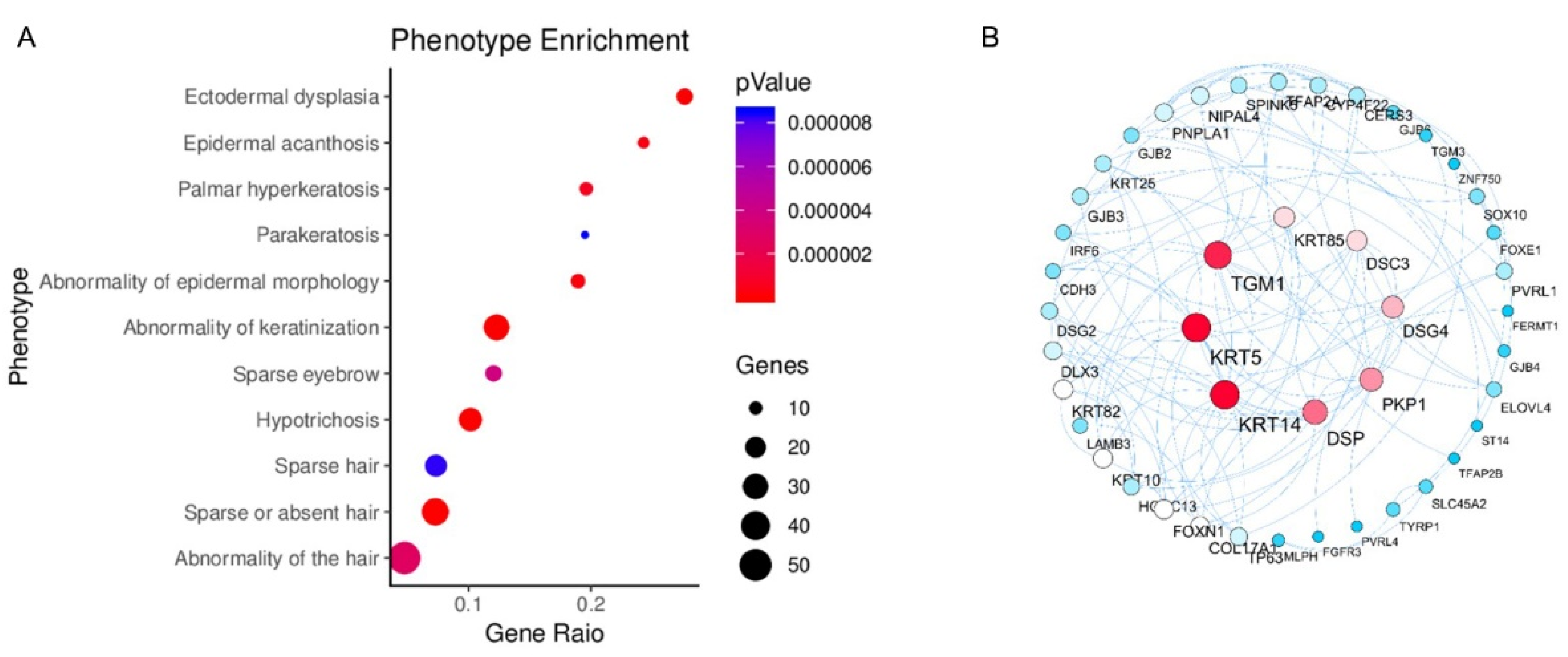
3.3. Functional Enrichment Analysis
3.4. Gene Enrichment Analysis
3.5. Identification and Validation of the Selected Genes
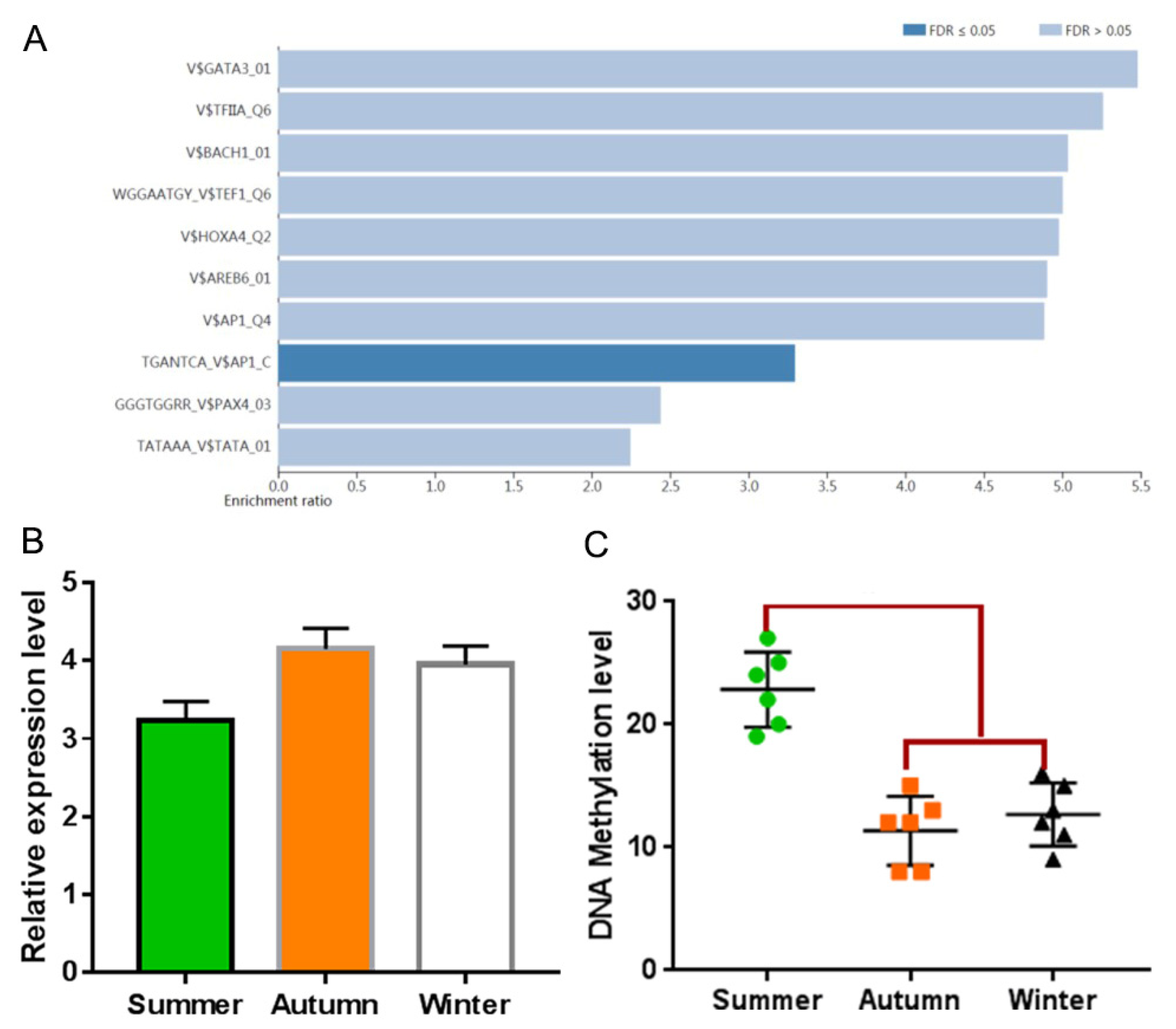
3.6. Transcription Factor Enrichment Analysis and Verification of PPI Network Genes
3.7. Methylation Analysis of Promoter of Target Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Middleton, A.D.; Kauffman, M.J.; McWhirter, D.E.; Cook, J.G.; Cook, R.C.; Nelson, A.A.; Jimenez, M.D.; Klaver, R.W. Animal migration amid shifting patterns of phenology and predation: Lessons from a Yellowstone elk herd. Ecology 2013, 94, 1245–1256. [Google Scholar] [CrossRef]
- Fokkema, W.; van der Jeugd, H.P.; Lameris, T.K.; Dokter, A.M.; Ebbinge, B.S.; de Roos, A.M.; Nolet, B.A.; Piersma, T.; Olff, H. Ontogenetic niche shifts as a driver of seasonal migration. Oecologia 2020, 193, 285–297. [Google Scholar] [CrossRef]
- Bao, P.; Luo, J.; Liu, Y.; Chu, M.; Ren, Q.; Guo, X.; Tang, B.; Ding, X.; Qiu, Q.; Pan, H.; et al. The seasonal development dynamics of the yak hair cycle transcriptome. BMC Genom. 2020, 21, 355. [Google Scholar] [CrossRef]
- Hu, S.; Li, C.; Wu, D.; Huo, H.; Bai, H.; Wu, J. The Dynamic Change of Gene-Regulated Networks in Cashmere Goat Skin with Seasonal Variation. Biochem. Genet. 2022, 60, 527–542. [Google Scholar] [CrossRef]
- Xu, J.; Fu, Y.; Hu, Y.; Yin, L.; Tang, Z.; Yin, D.; Zhu, M.; Yu, M.; Li, X.; Zhou, Y.; et al. Whole genome variants across 57 pig breeds enable comprehensive identification of genetic signatures that underlie breed features. J. Anim. Sci. Biotechnol. 2020, 11, 115. [Google Scholar] [CrossRef]
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. CB 2009, 19, R132–R142. [Google Scholar] [CrossRef]
- Reya, T.; Clevers, H. Wnt signalling in stem cells and cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef]
- Jung, H.S.; Francis-West, P.H.; Widelitz, R.B.; Jiang, T.X.; Ting-Berreth, S.; Tickle, C.; Wolpert, L.; Chuong, C.M. Local inhibitory action of BMPs and their relationships with activators in feather formation: Implications for periodic patterning. Dev. Biol. 1998, 196, 11–23. [Google Scholar] [CrossRef]
- Yuhki, M.; Yamada, M.; Kawano, M.; Iwasato, T.; Itohara, S.; Yoshida, H.; Ogawa, M.; Mishina, Y. BMPR1A signaling is necessary for hair follicle cycling and hair shaft differentiation in mice. Development 2004, 131, 1825–1833. [Google Scholar]
- Millar, S.E. Molecular mechanisms regulating hair follicle development. J. Investig. Dermatol. 2002, 118, 216–225. [Google Scholar] [CrossRef]
- Andrés-Manzano, M.J.; Andrés, V.; Dorado, B. Oil Red O and Hematoxylin and Eosin Staining for Quantification of Atherosclerosis Burden in Mouse Aorta and Aortic Root. Methods Mol. Biol. 2015, 1339, 85–99. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Tian, M.; He, X.; Wang, W.; Liu, D.; Meng, Q. Differential microRNA expression profiling and target gene prediction in the muscle tissues of clenbuterol-fed Chinese miniature swine. Acta Agric. Scand. Sect. A—Anim. Sci. 2017, 67, 9–14. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, D.; Wang, W.; He, X.; Peng, F.; Wang, L.; Guo, Z.; Fu, B.; Wu, S.; Li, Z.; et al. A comparative study of the effects of long-term cold exposure, and cold resistance in Min Pigs and Large White Pigs. Acta Agric. Scand. Sect. A—Anim. Sci. 2017, 67, 34–39. [Google Scholar] [CrossRef]
- Tamaki, T.; Horinouchi, S.; Fukaya, M.; Okumura, H.; Kawamura, Y.; Beppu, T. Nucleotide sequence of the membrane-bound aldehyde dehydrogenase gene from Acetobacter polyoxogenes. J. Biochem. 1989, 106, 541–544. [Google Scholar] [CrossRef]
- Desposito, D.; Chollet, C.; Taveau, C.; Descamps, V.; Alhenc-Gelas, F.; Roussel, R.; Bouby, N.; Waeckel, L. Improvement of skin wound healing in diabetic mice by kinin B2 receptor blockade. Clin. Sci. 2016, 130, 45–56. [Google Scholar] [CrossRef]
- Kandyba, E.; Kobielak, K. Wnt7b is an important intrinsic regulator of hair follicle stem cell homeostasis and hair follicle cycling. Stem Cells 2014, 32, 886–901. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, K.; Yang, K.; Ye, J.X.; Xing, Y.Z.; Guo, H.Y.; Deng, F.; Lian, X.H.; Yang, T. Adenovirus-mediated Wnt10b overexpression induces hair follicle regeneration. J. Investig. Dermatol. 2013, 133, 42–48. [Google Scholar] [CrossRef]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. WNT signals are required for the initiation of hair follicle development. Dev. Cell 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Wang, B.; Fallon, J.F.; Beachy, P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 2000, 100, 423–434. [Google Scholar] [CrossRef]
- Nanba, D.; Nakanishi, Y.; Hieda, Y. Role of Sonic hedgehog signaling in epithelial and mesenchymal development of hair follicles in an organ culture of embryonic mouse skin. Dev. Growth Differ. 2003, 45, 231–239. [Google Scholar] [CrossRef]
- Wang, X.J.; Liefer, K.M.; Tsai, S.; O’Malley, B.W.; Roop, D.R. Development of gene-switch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc. Natl. Acad. Sci. USA 1999, 96, 8483–8488. [Google Scholar] [CrossRef]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef]
- Kobielak, K.; Pasolli, H.A.; Alonso, L.; Polak, L.; Fuchs, E. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J. Cell Biol. 2003, 163, 609–623. [Google Scholar] [CrossRef]
- Botchkarev, V.A.; Botchkareva, N.V.; Sharov, A.A.; Funa, K.; Huber, O.; Gilchrest, B.A. Modulation of BMP signaling by noggin is required for induction of the secondary (nontylotrich) hair follicles. J. Investig. Dermatol. 2002, 118, 3–10. [Google Scholar] [CrossRef]
- McMahon, J.A.; Takada, S.; Zimmerman, L.B.; Fan, C.M.; Harland, R.M.; McMahon, A.P. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998, 12, 1438–1452. [Google Scholar] [CrossRef]
- Sharov, A.A.; Fessing, M.; Atoyan, R.; Sharova, T.Y.; Haskell-Luevano, C.; Weiner, L.; Funa, K.; Brissette, J.L.; Gilchrest, B.A.; Botchkarev, V.A. Bone morphogenetic protein (BMP) signaling controls hair pigmentation by means of cross-talk with the melanocortin receptor-1 pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 93–98. [Google Scholar] [CrossRef]
- Sharov, A.A.; Sharova, T.Y.; Mardaryev, A.N.; Tommasi di Vignano, A.; Atoyan, R.; Weiner, L.; Yang, S.; Brissette, J.L.; Dotto, G.P.; Botchkarev, V.A. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. USA 2006, 103, 18166–18171. [Google Scholar] [CrossRef]
- Kurzen, H.; Moll, I.; Moll, R.; Schafer, S.; Simics, E.; Amagai, M.; Wheelock, M.J.; Franke, W.W. Compositionally different desmosomes in the various compartments of the human hair follicle. Differentiation 1998, 63, 295–304. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.A.; Wessagowit, V. Human hair abnormalities resulting from inherited desmosome gene mutations. Keio J. Med. 2005, 54, 72–79. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brooke, M.A.; Nitoiu, D.; Kelsell, D.P. Cell-cell connectivity: Desmosomes and disease. J. Pathol. 2012, 226, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Loussouarn, G.; Garcel, A.L.; Lozano, I.; Collaudin, C.; Porter, C.; Panhard, S.; Saint-Leger, D.; de La Mettrie, R. Worldwide diversity of hair curliness: A new method of assessment. Int. J. Derm. 2007, 46 (Suppl. 1), 2–6. [Google Scholar] [CrossRef]
- Tanaka, A.; Lai-Cheong, J.E.; Cafe, M.E.; Gontijo, B.; Salomao, P.R.; Pereira, L.; McGrath, J.A. Novel truncating mutations in PKP1 and DSP cause similar skin phenotypes in two Brazilian families. Br. J. Derm. 2009, 160, 692–697. [Google Scholar] [CrossRef]
- Xue, K.; Zheng, Y.; Cui, Y. A novel heterozygous missense mutation of DSP in a Chinese Han pedigree with palmoplantar keratoderma. J. Cosmet. Derm. 2019, 18, 371–376. [Google Scholar] [CrossRef]
- Nekrasova, O.; Green, K.J. Desmosome assembly and dynamics. Trends Cell Biol. 2013, 23, 537–546. [Google Scholar] [CrossRef]
- Lee, J.Y.W.; McGrath, J.A. Mutations in genes encoding desmosomal proteins: Spectrum of cutaneous and extracutaneous abnormalities. Br. J. Derm. 2020, 184, 596–605. [Google Scholar] [CrossRef]
- Kljuic, A.; Bazzi, H.; Sundberg, J.P.; Martinez-Mir, A.; O’Shaughnessy, R.; Mahoney, M.G.; Levy, M.; Montagutelli, X.; Ahmad, W.; Aita, V.M.; et al. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: Evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 2003, 113, 249–260. [Google Scholar] [CrossRef]
- Favre, B.; Begre, N.; Borradori, L. A recessive mutation in the DSP gene linked to cardiomyopathy, skin fragility and hair defects impairs the binding of desmoplakin to epidermal keratins and the muscle-specific intermediate filament desmin. Br. J. Derm. 2018, 179, 797–799. [Google Scholar] [CrossRef]
- Alibardi, L.; Bernd, N. Immunolocalization of junctional proteins in human hairs indicates that the membrane complex stabilizes the inner root sheath while desmosomes contact the companion layer through specific keratins. Acta Histochem. 2013, 115, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Norgett, E.E.; Lucke, T.W.; Bowers, B.; Munro, C.S.; Leigh, I.M.; Kelsell, D.P. Early death from cardiomyopathy in a family with autosomal dominant striate palmoplantar keratoderma and woolly hair associated with a novel insertion mutation in desmoplakin. J. Investig. Dermatol. 2006, 126, 1651–1654. [Google Scholar] [CrossRef] [PubMed]
- Maruthappu, T.; Posafalvi, A.; Castelletti, S.; Delaney, P.J.; Syrris, P.; O’Toole, E.A.; Green, K.J.; Elliott, P.M.; Lambiase, P.D.; Tinker, A.; et al. Loss-of-function desmoplakin I and II mutations underlie dominant arrhythmogenic cardiomyopathy with a hair and skin phenotype. Br. J. Derm. 2019, 180, 1114–1122. [Google Scholar] [CrossRef]
- Al-Owain, M.; Wakil, S.; Shareef, F.; Al-Fatani, A.; Hamadah, E.; Haider, M.; Al-Hindi, H.; Awaji, A.; Khalifa, O.; Baz, B.; et al. Novel homozygous mutation in DSP causing skin fragility-woolly hair syndrome: Report of a large family and review of the desmoplakin-related phenotypes. Clin. Genet. 2011, 80, 50–58. [Google Scholar] [CrossRef]
- Tang, X.N.; Zhu, Y.Q.; Marcelo, C.L.; Ritchie, H.H. Expression of mineralized tissue associated proteins: Dentin sialoprotein and phosphophoryn in rodent hair follicles. J. Dermatol. Sci. 2011, 64, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Samuelov, L.; Sprecher, E. Inherited desmosomal disorders. Cell Tissue Res. 2015, 360, 457–475. [Google Scholar] [CrossRef]
- Olivry, T.; Linder, K.E.; Wang, P.; Bizikova, P.; Bernstein, J.A.; Dunston, S.M.; Paps, J.S.; Casal, M.L. Deficient plakophilin-1 expression due to a mutation in PKP1 causes ectodermal dysplasia-skin fragility syndrome in Chesapeake Bay retriever dogs. PLoS ONE 2012, 7, e32072. [Google Scholar] [CrossRef]
- McGrath, J.A.; McMillan, J.R.; Shemanko, C.S.; Runswick, S.K.; Leigh, I.M.; Lane, E.B.; Garrod, D.R.; Eady, R.A. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat. Genet. 1997, 17, 240–244. [Google Scholar] [CrossRef]
- Boyce, A.E.; McGrath, J.A.; Techanukul, T.; Murrell, D.F.; Chow, C.W.; McGregor, L.; Warren, L.J. Ectodermal dysplasia-skin fragility syndrome due to a new homozygous internal deletion mutation in the PKP1 gene. Australas. J. Dermatol. 2012, 53, 61–65. [Google Scholar] [CrossRef]
- Waseem, A.; Dogan, B.; Tidman, N.; Alam, Y.; Purkis, P.; Jackson, S.; Lalli, A.; Machesney, M.; Leigh, I.M. Keratin 15 expression in stratified epithelia: Downregulation in activated keratinocytes. J. Investig. Dermatol. 1999, 112, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Yay, A.; Goktepe, O.; Bahadir, A.; Ozdamar, S.; Oktem, I.S.; Coruh, A.; Baran, M. Assessment of markers expressed in human hair follicles according to different skin regions. Adv. Clin. Exp. Med. 2018, 27, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wildermoth, J.E.; Wallace, O.A.; Gordon, S.W.; Maqbool, N.J.; Maclean, P.H.; Nixon, A.J.; Pearson, A.J. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp. Derm. 2011, 20, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Ramot, Y.; Zlotogorski, A. Keratins: The hair shaft’s backbone revealed. Exp. Derm. 2015, 24, 416–417. [Google Scholar] [CrossRef] [PubMed]
- Amico, S.; Ged, C.; Taieb, A.; Morice-Picard, F. Compound heterozygosity for novel KRT85 variants associated with pure hair and nail ectodermal dysplasia. J. Eur. Acad. Derm. Venereol. 2019, 33, e458–e459. [Google Scholar] [CrossRef] [PubMed]
- Karim, N.; Phinney, B.S.; Salemi, M.; Wu, P.W.; Naeem, M.; Rice, R.H. Human stratum corneum proteomics reveals cross-linking of a broad spectrum of proteins in cornified envelopes. Exp. Derm. 2019, 28, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Nomura, T.; Sugiyama, T.; Miyauchi, T.; Suzuki, S.; Fujita, Y.; Shimizu, H. Compound heterozygous missense mutations p.Leu207Pro and p.Tyr544Cys in TGM1 cause a severe form of lamellar ichthyosis. J. Derm. 2018, 45, 1463–1467. [Google Scholar] [CrossRef]
- Rorke, E.A.; Adhikary, G.; Young, C.A.; Rice, R.H.; Elias, P.M.; Crumrine, D.; Meyer, J.; Blumenberg, M.; Eckert, R.L. Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function. Cell Death Dis. 2015, 6, e1647. [Google Scholar] [CrossRef]
- Rishikaysh, P.; Dev, K.; Diaz, D.; Qureshi, W.M.; Filip, S.; Mokry, J. Signaling involved in hair follicle morphogenesis and development. Int. J. Mol. Sci. 2014, 15, 1647–1670. [Google Scholar] [CrossRef]
- Lee, J.; Bscke, R.; Tang, P.C.; Hartman, B.H.; Heller, S.; Koehler, K.R. Hair Follicle Development in Mouse Pluripotent Stem Cell-Derived Skin Organoids. Cell Rep. 2018, 22, 242–254. [Google Scholar] [CrossRef]



| mRNA | Forward Primer | Reverse Primer |
|---|---|---|
| DSP | CGGGTACAATGACCCCGAAA | TTCCGGCCACGGACATCATC |
| KRT15 | GATCGAGAGCCTGAACGAGG | GAGGCTTCTCTGGTGCCAAT |
| KRT4 | ATCAGCTGACTCTTCACCGC | GCCTTCTCCCCAAGGAACAAA |
| KRT85 | CATGAATGTGTTGTCCCGCA | GGTAGGTGGCGATCTCGATG |
| TGM1 | TACAAGGAGTACCAGCCCCA | ACTGCCAGCCATGCTTCTTA |
| DSG4 | GTGCGCACAATGTCCAGTTT | GCAGTCACACTCGGAAGACA |
| DSC3 | GAGGGAGTTCCCACCTGTTG | AGCCCATCTTCTCTTGGCAC |
| PKP1 | TGGCCTACGAATGCTTCCAG | GCGGTCCCGTAGTTGTTGTA |
| transcription factor AP-1 | ACTTTCCTCCTTCACGGTCC | ACTGGATTATCAGGCGCTCG |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
| Gene | Forward Primer | Reverse Primer | |
|---|---|---|---|
| DSP | M | GTAGTTGGATAAAATTAAAGTCGAT | TCAAATAATCCATCCTCTCGAA |
| U | GGTAGTTGGATAAAATTAAAGTTG | TCAAATAATCCATCCTCTCAAAA | |
| Probe | TTTCAACAAATTCTCATACTCCTCCTCCAA | ||
| KRT15 | M | GGTTGGAGTAGGAGATCGTTA | CTAACTCCTCGACGTTAATACGAA |
| U | GGTTGGAGTAGGAGATTGTTA | CTCCTCAACATTAATACAAACTTTACC | |
| Probe | CCACCTCCCAAAAAAACTTCTCTAATACCA | ||
| KRT4 | M | TAAAGGATGTTTATAGTAAGCGTG | ATCAACTCCTAATATTCACGCAA |
| U | AAAGGATGTTTATAGTAAGTGTG | ATCAACTCCTAATATTCACACAA | |
| Probe | AACCAACTCCTCCTTAACCTTCTTCAAAAC | ||
| TGM1 | M | GAGGTTTAGAAGTTTTCGGA | CAATTAAACTCTAACAAACGACC |
| U | GGAGGTTTAGAAGTTTTTGGAA | CAATTAAACTCTAACAAACAACC | |
| Probe | CCATAAAACTTAAAATTCACCCTCAAACAA | ||
| PKP1 | M | GGGATTAGGGTTGGTAGGAC | TCCGATAAAACACAATCCGA |
| U | GGGATTAGGGTTGGTAGGATG | CCAATAAAACACAATCCAAAA | |
| Probe | ACCAACCCCTCCTAACTCCTAAAACAAACC | ||
| Hair Length (mm) | Yorkshire | Berkshire | Min Pig | |
| Bristles | Bristles | Bristles | Villus | |
| 41.51 ± 4.7 | 49.48 ± 2.44 | 52.74 ± 2.05 | 30.55 ± 4.72 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, M.; He, X.; Wang, W.; Feng, Y.; Zhang, D.; Li, Z.; Liu, D. Transcriptome Analysis Reveals Genes Contributed to Min Pig Villi Hair Follicle in Different Seasons. Vet. Sci. 2022, 9, 639. https://doi.org/10.3390/vetsci9110639
Tian M, He X, Wang W, Feng Y, Zhang D, Li Z, Liu D. Transcriptome Analysis Reveals Genes Contributed to Min Pig Villi Hair Follicle in Different Seasons. Veterinary Sciences. 2022; 9(11):639. https://doi.org/10.3390/vetsci9110639
Chicago/Turabian StyleTian, Ming, Xinmiao He, Wentao Wang, Yanzhong Feng, Dongjie Zhang, Zhongqiu Li, and Di Liu. 2022. "Transcriptome Analysis Reveals Genes Contributed to Min Pig Villi Hair Follicle in Different Seasons" Veterinary Sciences 9, no. 11: 639. https://doi.org/10.3390/vetsci9110639
APA StyleTian, M., He, X., Wang, W., Feng, Y., Zhang, D., Li, Z., & Liu, D. (2022). Transcriptome Analysis Reveals Genes Contributed to Min Pig Villi Hair Follicle in Different Seasons. Veterinary Sciences, 9(11), 639. https://doi.org/10.3390/vetsci9110639







