2,5-Hexanedione Affects Ovarian Granulosa Cells in Swine by Regulating the CDKN1A Gene: A Transcriptome Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Porcine Ovarian Granulosa Cells (pGCs) Culture In Vitro and under 2,5-HD Treatment
2.2. Cell Morphological Observations
2.3. Cell Apoptosis Detection
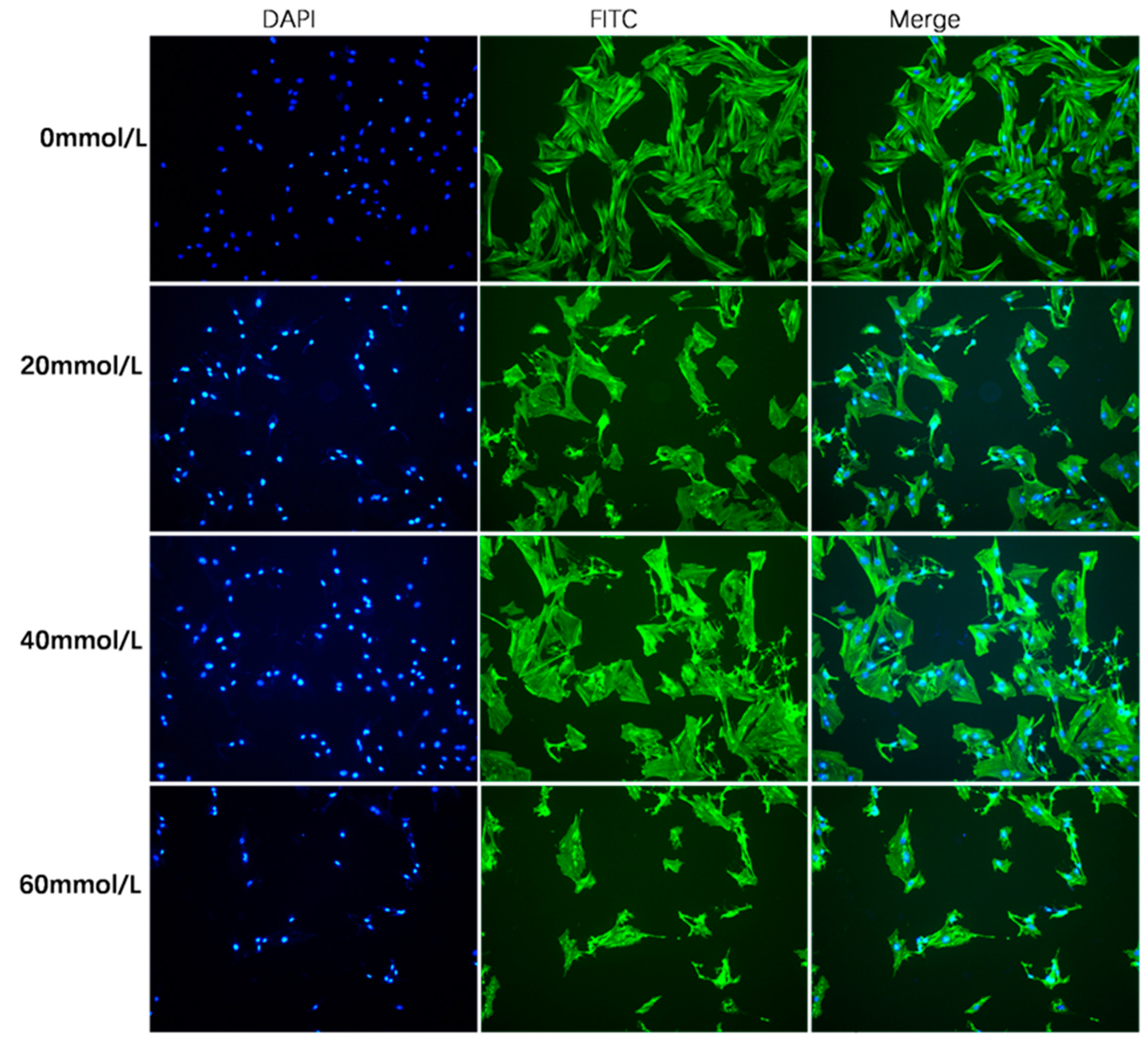
2.4. Phalloidin Staining
2.5. EDU Staining
2.6. Total RNA Extraction, Library Construction, and Sequencing Analysis
2.7. Quantitative PCR
2.8. CDKN1A Interference
2.9. Statistical Analysis
3. Results
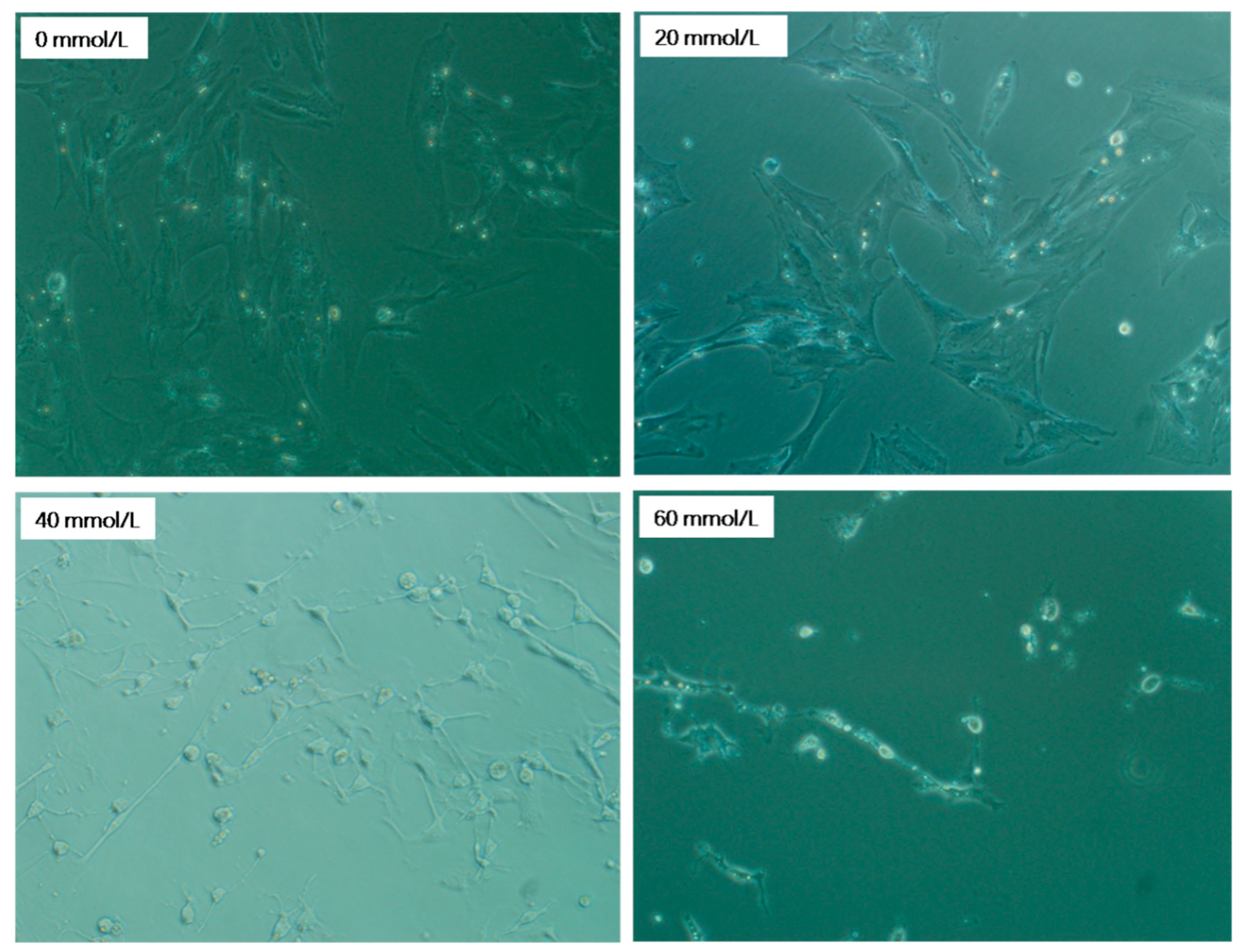
3.1. Effect of 2,5-HD on pGC Morphology
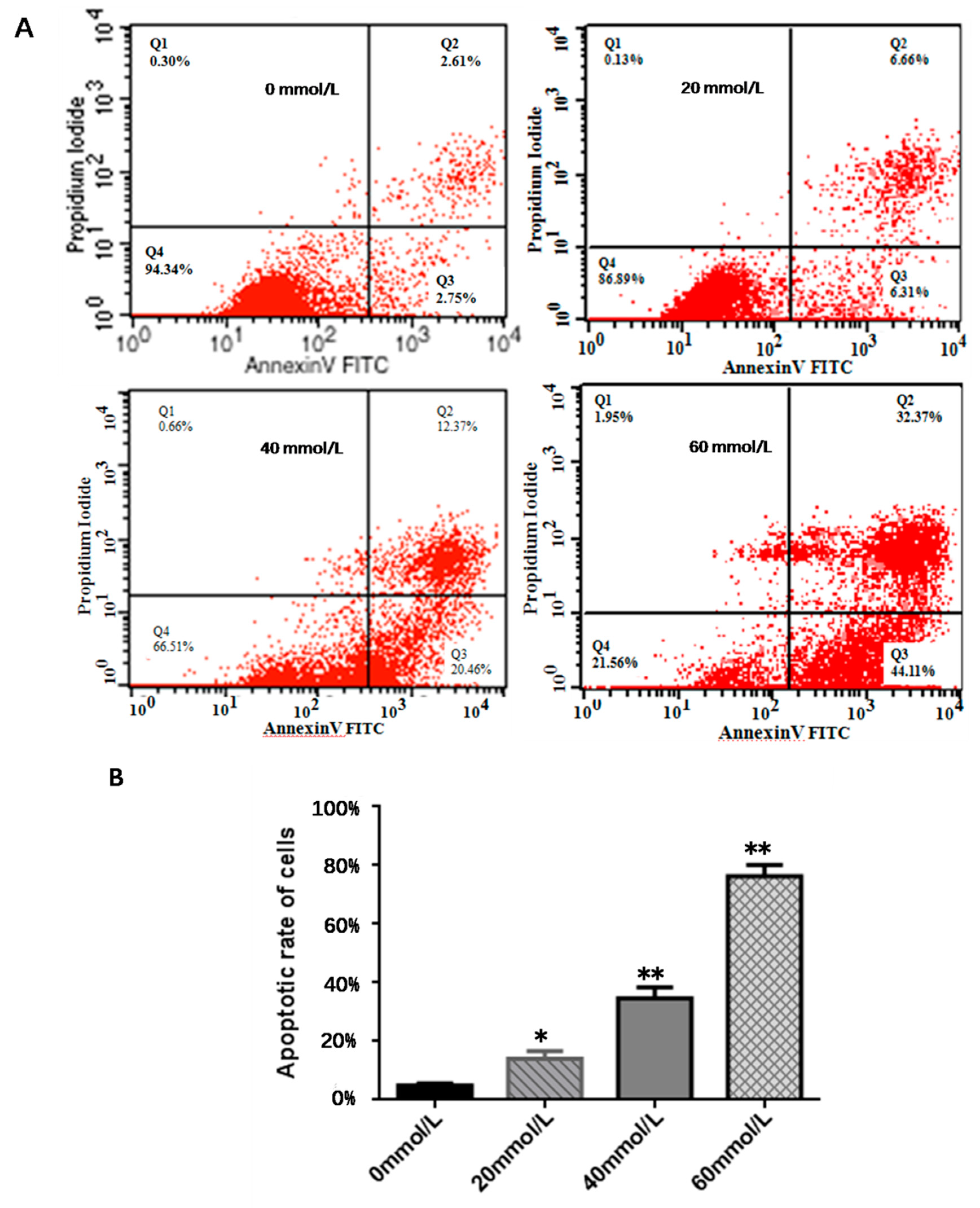
3.2. Effect of 2,5-HD on Apoptosis of pGCs
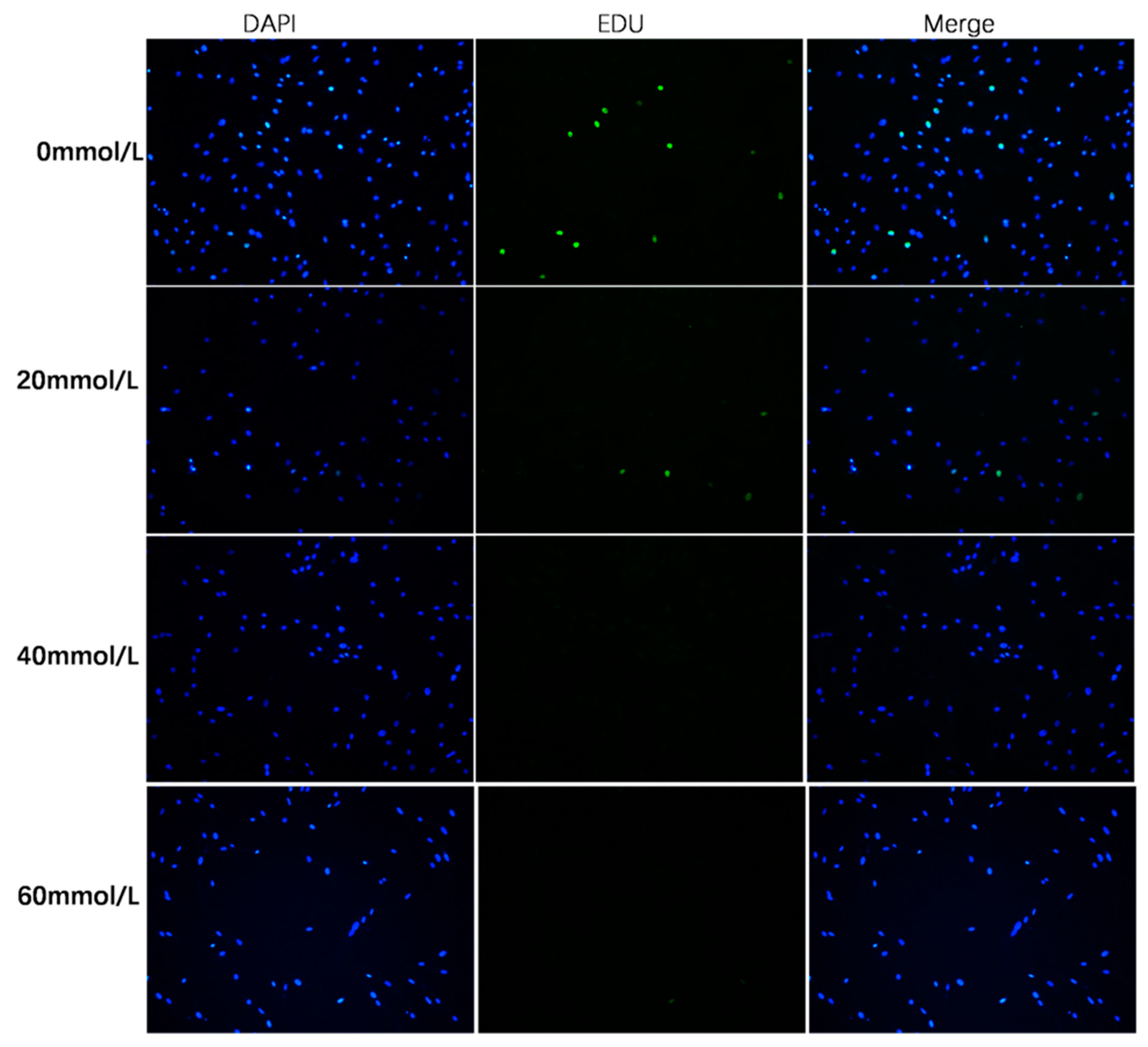
3.3. Effect of 2,5-HD on pGC Proliferation
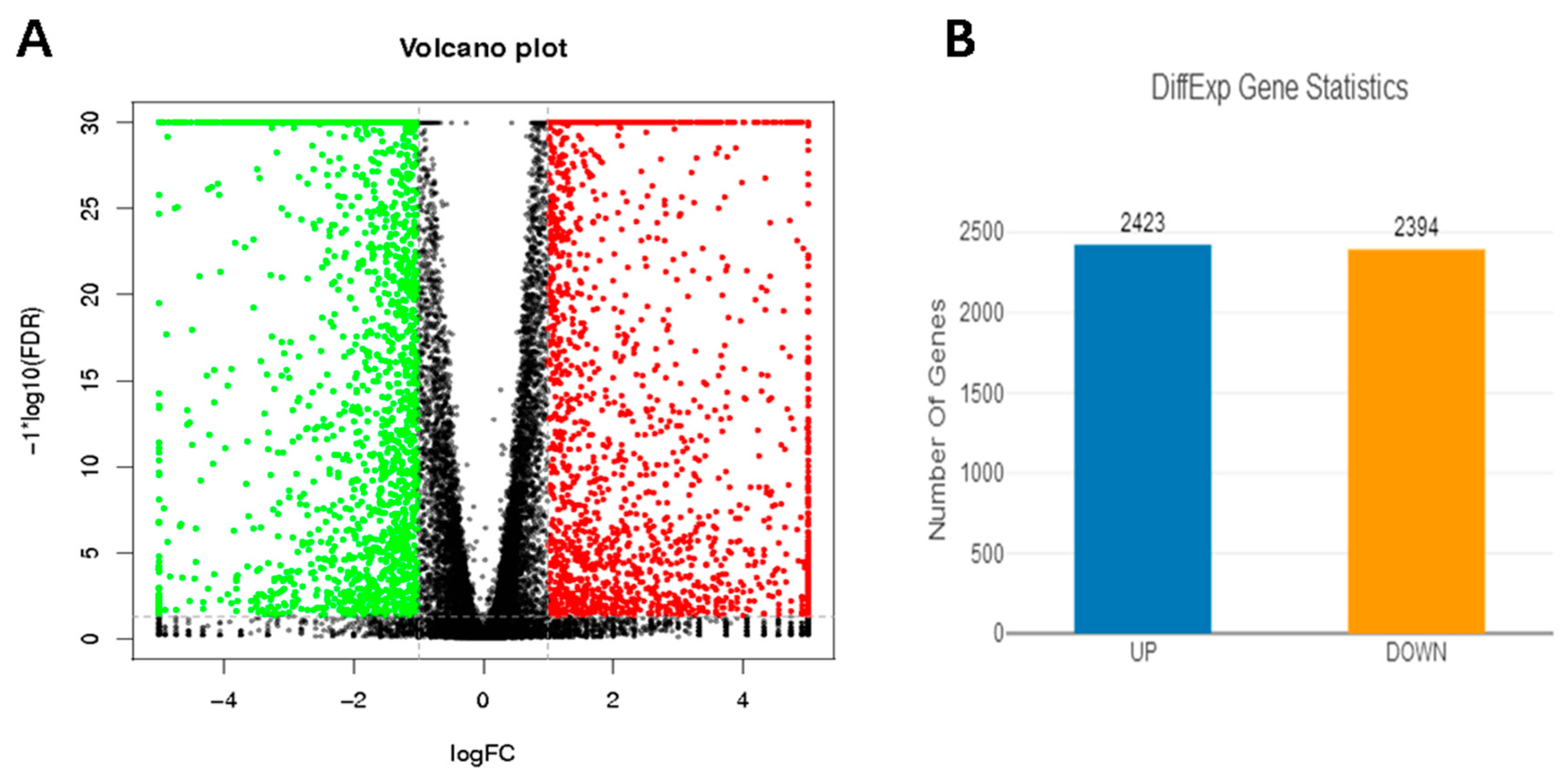
3.4. Gene Expression Profiling
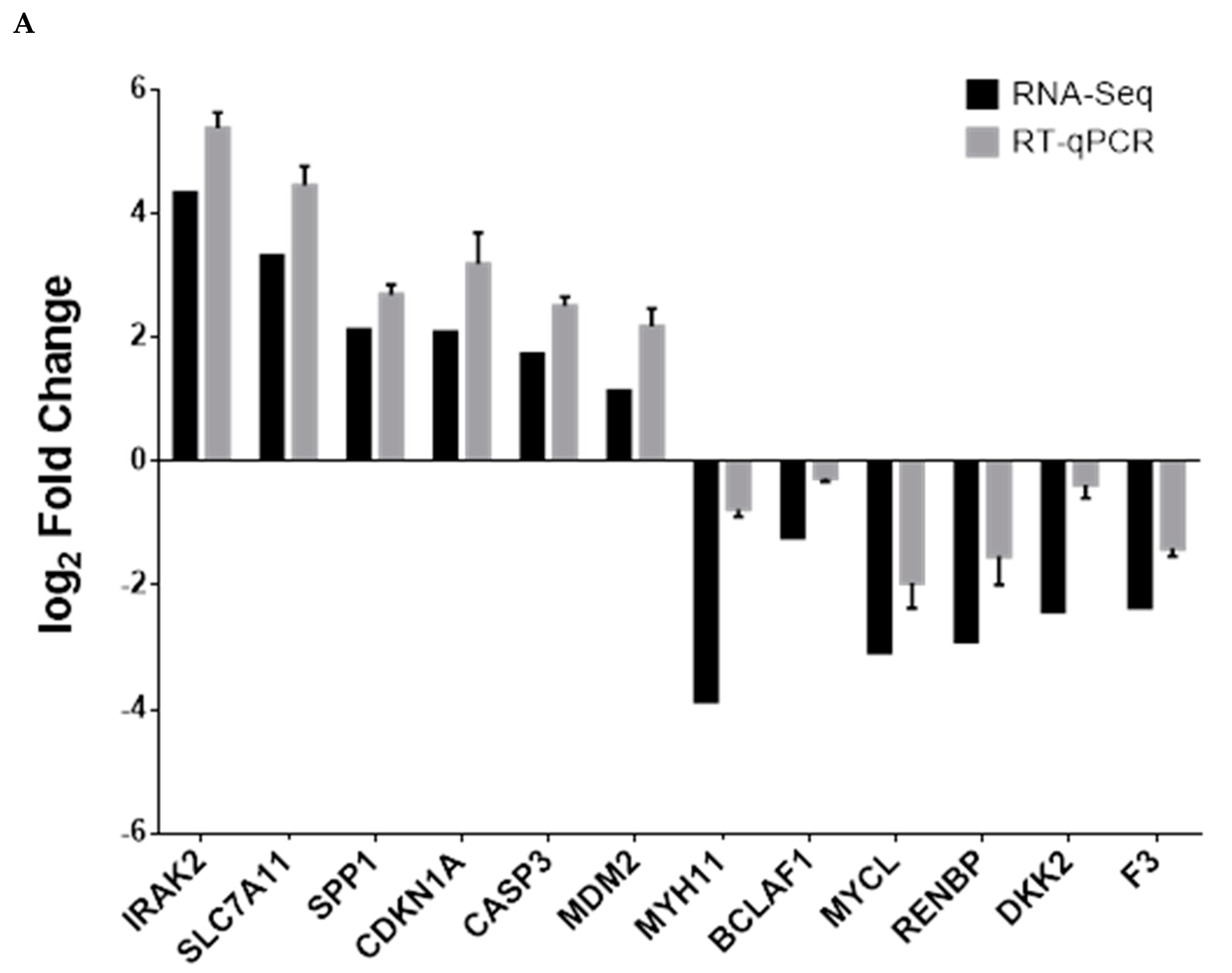
3.5. Analysis of DEGs
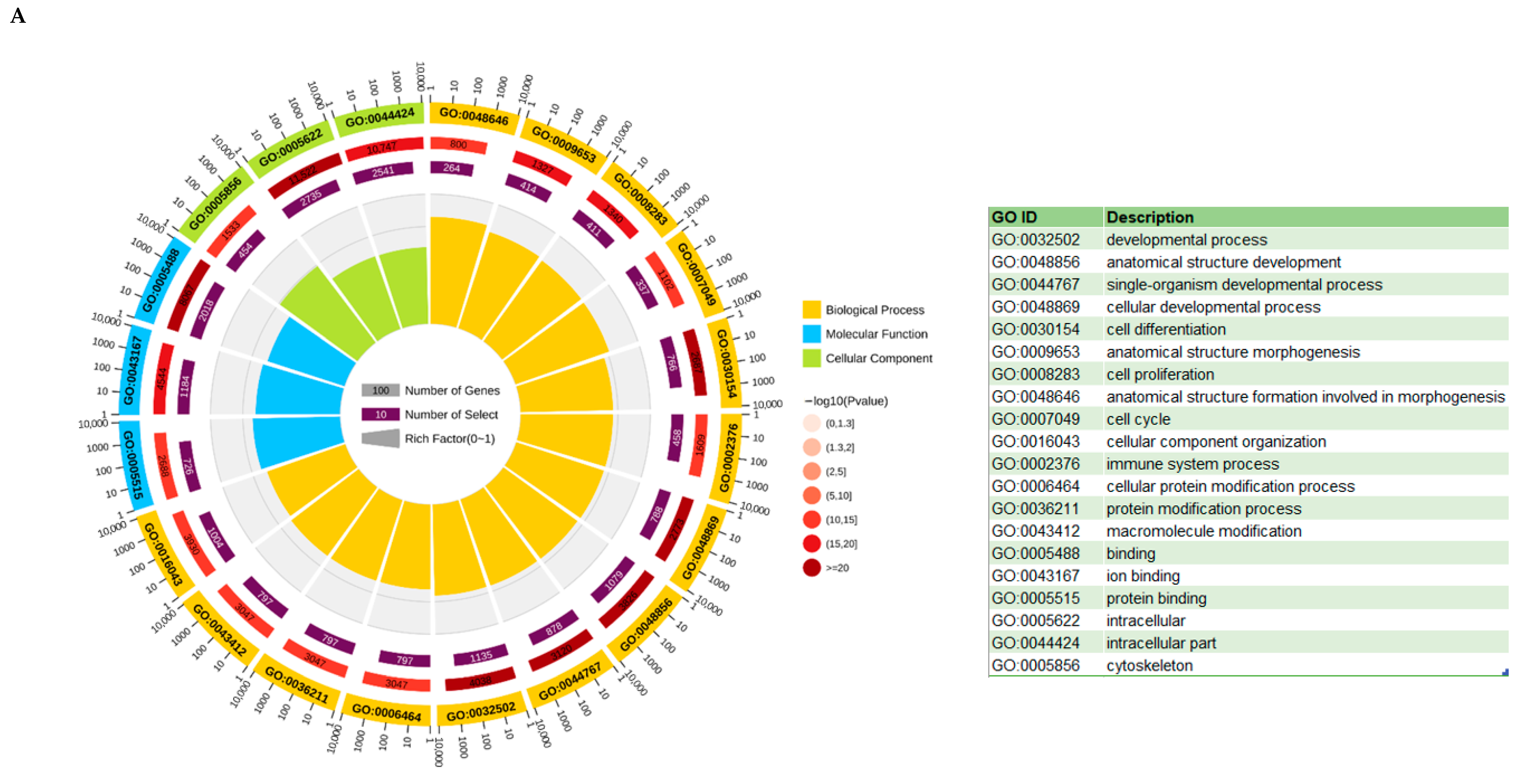
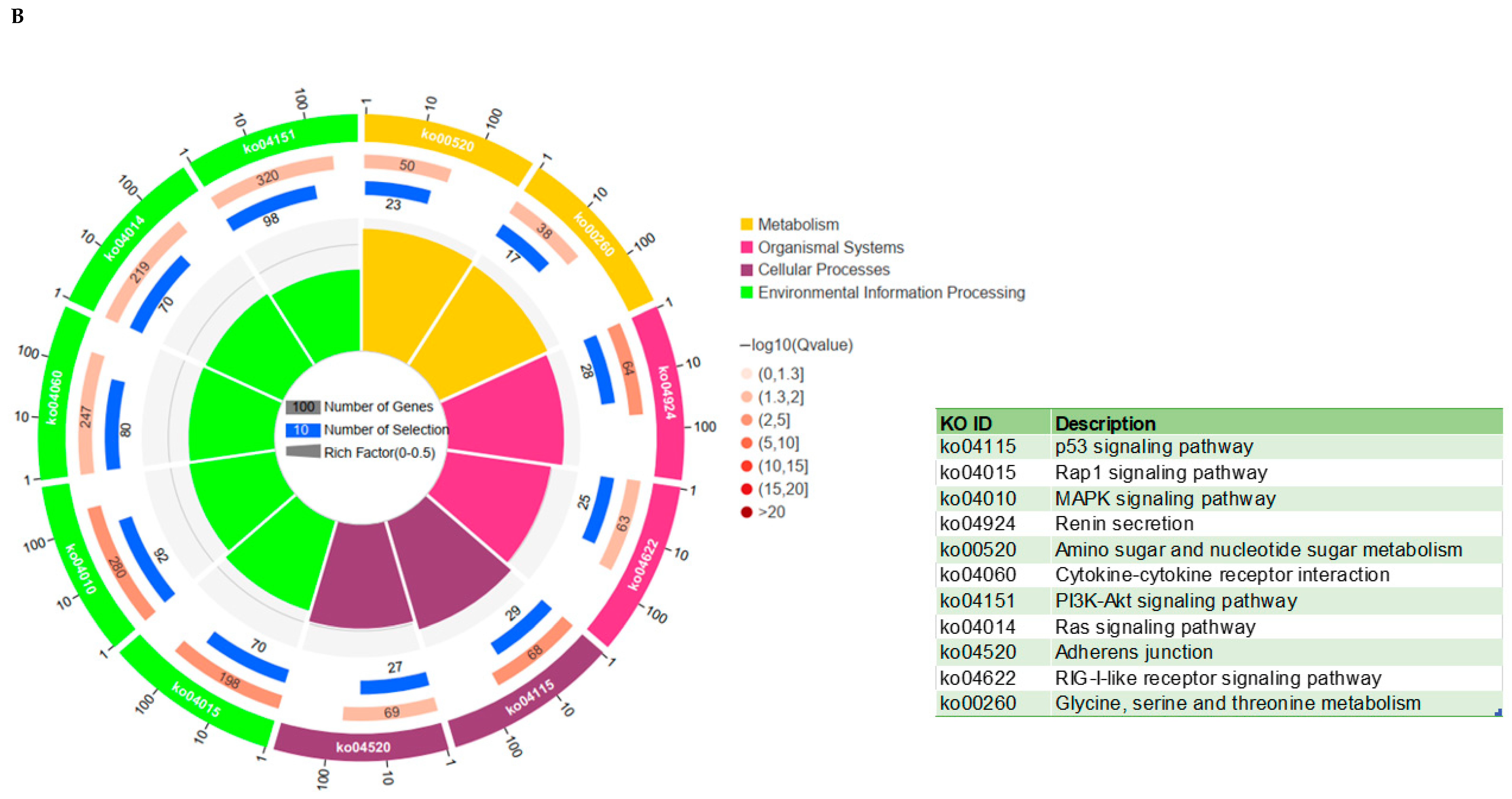
3.6. KEGG and GO Analysis
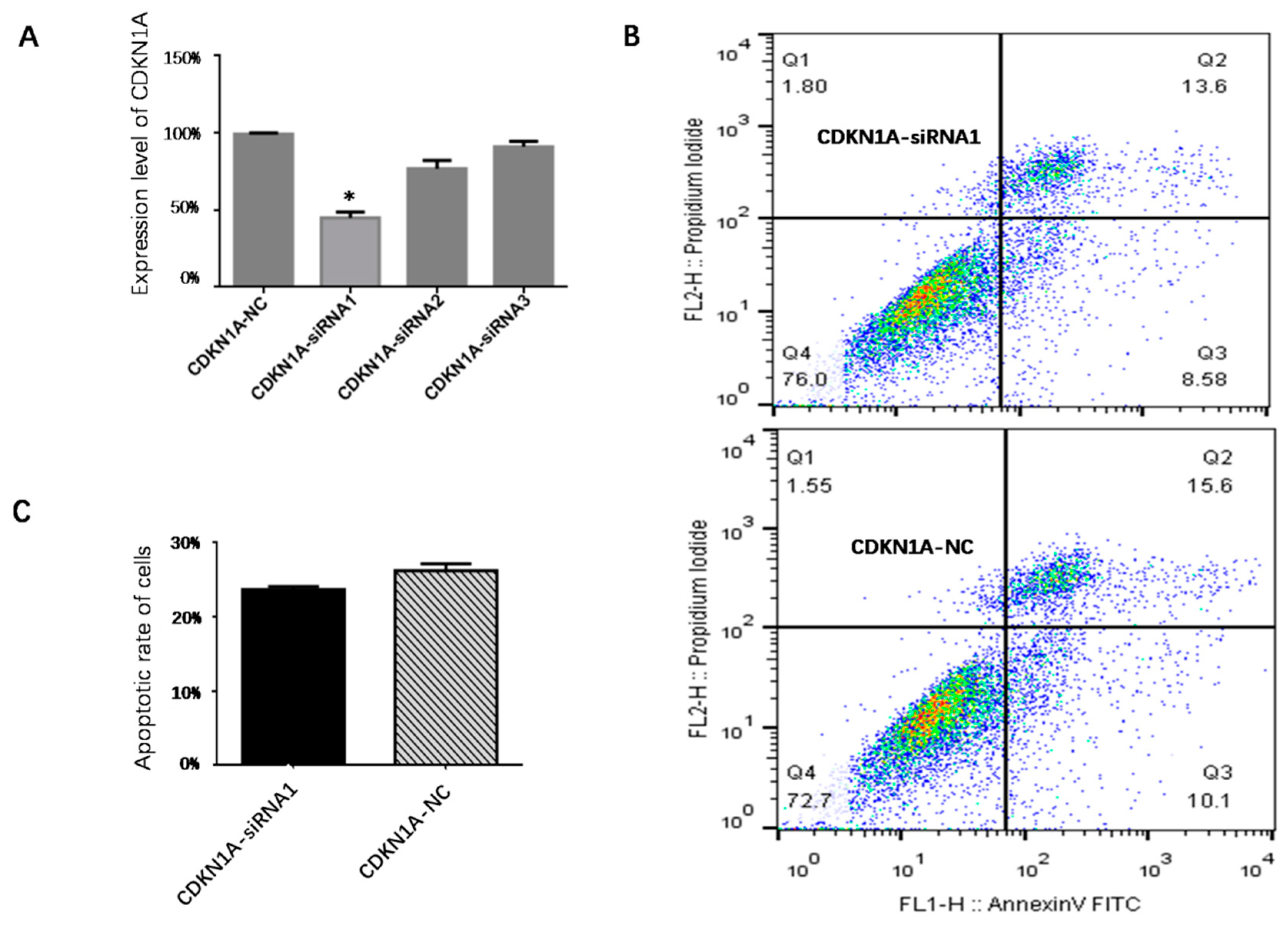
3.7. Effect of CDKN1A on Apoptosis of pGCs
3.8. Effect of CDKN1A on the Cycle of pGCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.L.; Liu, S.; Piao, F.Y.; Wang, Z.M.; Qi, Y.; Li, S.Y.; Zhang, D.M.; Shen, J.S. 2,5-hexanedione induced apoptosis in mesenchymal stem cells from rat bone marrow via mitochondria-dependent caspase-3 pathway. Ind. Health 2015, 53, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.F.; Zhang, X.Y.; Lin, Q.; Chen, Y.P.; Zhang, Z.Q.; Lin, L. Effects of n-hexane on learning and memory and the expressions of nerve growth factor mRNA and nerve growth factor receptor mRNA of brain tissue in mice exposed to N-hexane. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi Zhonghua Laodong Weisheng Zhiyebing Zazhi Chin. J. Ind. Hyg. Occup. Dis. 2019, 37, 217–220. [Google Scholar] [CrossRef]
- Pradhan, S.; Tandon, R. N-hexane neuropathy with vertigo and cold allodynia in a silk screen printer: A case study. Int. J. Occup. Med. Environ. Health 2015, 28, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Puri, V.; Gupta, A.D.; Chaudhry, N.; Saran, R.K. Reversible cerebral and brain stem dysfunction in n: Hexane neuropathy. Ann. Indian Acad. Neurol. 2015, 18, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.B.; Jortner, B.S.; Sills, R.C. Animal models of peripheral neuropathy due to environmental toxicants. ILAR J. 2014, 54, 315–323. [Google Scholar] [CrossRef]
- Tshala-Katumbay, D.; Monterroso, V.; Kayton, R.; Lasarev, M.; Sabri, M.; Spencer, P. Probing mechanisms of axonopathy. Part II: Protein targets of 2,5-hexanedione, the neurotoxic metabolite of the aliphatic solvent n-hexane. Toxicol. Sci. 2009, 107, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Han, X.Y.; Cheng, D.; Song, F.Y.; Zeng, T.; An, L.H.; Xie, K.Q. Decelerated transport and its mechanism of 2,5-hexanedione on middle-molecular-weight neurofilament in rat dorsal root ganglia cells. Neuroscience 2014, 269, 192–198. [Google Scholar] [CrossRef]
- Hirai, T.; Mizutani, M.; Kimura, T.; Ochiai, K.; Umemura, T.; Itakura, C. Neurotoxic Effects of 2,5-Hexanedione on Normal and Neurofilament-Deficient Quail. Toxicol. Pathol. 1999, 27, 348–353. [Google Scholar] [CrossRef]
- Zhang, Y.-s.; Liu, Q.; Liu, Q.-j.; Duan, H.-w.; He, F.-s.; Zheng, Y.-x. Effect of 2,5-hexanedione on calcium homeostasis of motor neuron. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi Zhonghua Laodong Weisheng Zhiyebing Zazhi Chin. J. Ind. Hyg. Occup. Dis. 2006, 24, 270–272. [Google Scholar]
- Boekelheide, K.; Fleming, S.L.; Allio, T.; Embree-Ku, M.E.; Hall, S.J.; Johnson, K.J.; Kwon, E.J.; Patel, S.R.; Rasoulpour, R.J.; Schoenfeld, H.A.; et al. 2,5-hexanedione-induced testicular injury. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, G.; Ma, Z.L.; Chen, Y.Y.; Fan, J.J.; Zhang, Z.L.; Lee, K.K.; Luo, H.M.; Yang, X. Exposure to 2,5-hexanedione can induce neural malformations in chick embryos. Neurotoxicology 2012, 33, 1239–1247. [Google Scholar] [CrossRef]
- Kim, M.S.; Park, H.R.; Park, M.; Kim, S.J.; Kwon, M.; Yu, B.P.; Chung, H.Y.; Kim, H.S.; Kwack, S.J.; Kang, T.S.; et al. Neurotoxic effect of 2,5-hexanedione on neural progenitor cells and hippocampal neurogenesis. Toxicology 2009, 260, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, S.; Su, D.; Chen, F.; Lei, T.; Chen, H.; Dong, W.; Jiang, Y.; Sun, X.; Sun, W. 2,5-Hexanedione increases the percentage of proliferative Sox2+ cells in rat hippocampus. Toxicol. Ind. Health 2018, 34, 589–595. [Google Scholar] [CrossRef]
- Hatzirodos, N.; Hummitzsch, K.; Irving-Rodgers, H.F.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genom. 2014, 15, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, H.L.; Pang, F.; Zhang, W.C. The effect of n-hexane on the gonad toxicity of female mice. Biomed. Environ. Sci. 2012, 25, 189–196. [Google Scholar] [CrossRef]
- Sun, Y.; Lin, Y.; Li, H.; Liu, J.; Sheng, X.; Zhang, W. 2,5-Hexanedione induces human ovarian granulosa cell apoptosis through BCL-2, BAX, and CASPASE-3 signaling pathways. Arch. Toxicol. 2012, 86, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, K.T.; Allard, E.K.; Boekelheide, K. Fate of germ cells in 2,5-hexanedione-induced testicular injury. I. Apoptosis is the mechanism of germ cell death. Toxicol. Appl. Pharmacol. 1996, 137, 141–148. [Google Scholar] [CrossRef]
- Bryant, B.H.; Yamasaki, H.; Sandrof, M.A.; Boekelheide, K. Spermatid head retention as a marker of 2,5-hexanedione-induced testicular toxicity in the rat. Toxicol. Pathol. 2008, 36, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Tao, Q.Q.; Shang, J.N.; Xu, Y.L.; Zhang, L.; Ma, Y.C.; Zhu, W.H.; Yang, M.; Ding, Y.Y.; Yin, Z.J. MiR-26a promotes apoptosis of porcine granulosa cells by targeting the 3 beta-hydroxysteroid-Delta 24-reductase gene. Asian-Australas. J. Anim. Sci. 2020, 33, 547–555. [Google Scholar] [CrossRef]
- Yang, M.; Wu, X.D.; Zhang, W.; Ye, P.F.; Wang, Y.L.; Zhu, W.H.; Tao, Q.Q.; Xu, Y.L.; Shang, J.N.; Zhao, D.D.; et al. Transcriptional analysis of deoxynivalenol-induced apoptosis of sow ovarian granulosa cell. Reprod. Domest. Anim. 2020, 55, 217–228. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, L.; Kong, C.; Liu, J.; Luo, L.; Huang, H. Apoptosis of rat ovarian granulosa cells by 2,5-hexanedione in vitro and its relevant gene expression. J. Appl. Toxicol. 2013, 33, 661–669. [Google Scholar] [CrossRef]
- Zang, W.Q.; Wang, Y.Y.; Wang, T.; Du, Y.W.; Chen, X.N.; Li, M.; Zhao, G.Q. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Mol. Cancer 2015, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.J.; Zhang, M.Y.; Wu, H.; Ding, X.; Li, D.Y.; Dong, X.; Hu, X.X.; Su, S.; Shang, W.D.; Wu, J.X.; et al. SAIL: A new conserved anti-fibrotic lncRNA in the heart. Basic Res. Cardiol. 2021, 116, 15. [Google Scholar] [CrossRef]
- Jiang, Q.; Liu, C.; Li, C.P.; Xu, S.S.; Yao, M.D.; Ge, H.M.; Sun, Y.N.; Li, X.M.; Zhang, S.J.; Shan, K.; et al. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Investig. 2020, 130, 3833–3847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, L.; Wu, T.; Feng, Y.; Ding, Y.; Ye, P.; Yin, Z. Transcriptomic Analysis of Ovaries from Pigs with High And Low Litter Size. PLoS ONE 2015, 10, e0139514. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.J.; Sui, B.D.; Fan, W.X.; Lei, L.; Zhou, L.; Yang, L.; Diao, Y.J.; Zhang, Y.; Li, Z.; Liu, J.Y.; et al. Exosomes derived from osteogenic tumor activate osteoclast differentiation and concurrently inhibit osteogenesis by transferring COL1A1-targeting miRNA-92a-1-5p. J. Extracell. Vesicles 2021, 10, e12056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Nan, A.R.; Chen, L.J.; Li, X.; Jia, Y.Y.; Qiu, M.Y.; Dai, X.; Zhou, H.Y.; Zhu, J.L.; Zhang, H.; et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 2020, 19, 101. [Google Scholar] [CrossRef]
- Li, Z.Z.; Zhang, C.; Wang, Z.T.; Shen, J.; Xiang, P.P.; Chen, X.; Nan, J.L.; Lin, Y.N. Lipofectamine 2000/siRNA complexes cause endoplasmic reticulum unfolded protein response in human endothelial cells. J. Cell. Physiol. 2019, 234, 21166–21181. [Google Scholar] [CrossRef]
- Zuo, A.J.; Li, J.; Zhao, X.Y.; Li, T.T.; Lei, S.Y.; Chen, J.Y.; Xu, D.; Song, C.X.; Li, N.; Ruan, S.Y.; et al. Globular CTRP9 protects cardiomyocytes from palmitic acid-induced oxidative stress by enhancing autophagic flux. Chem.-Biol. Interact. 2020, 329, 109094. [Google Scholar] [CrossRef]
- Barkinge, J.L.; Gudi, R.; Sarah, H.; Chu, F.; Borthakur, A.; Prabhakar, B.S.; Prasad, K.V. The p53-induced Siva-1 plays a significant role in cisplatin-mediated apoptosis. J. Carcinog. 2009, 8, 2. [Google Scholar] [CrossRef]
- Yang, L.; Lv, Q.; Liu, J.; Qi, S.; Fu, D. miR-431 regulates granulosa cell function through the IRS2/PI3K/AKT signaling pathway. J. Reprod. Dev. 2020, 66, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.S.; Sui, H.S.; Han, Z.B.; Li, W.; Luo, M.J.; Tan, J.H. Apoptosis in granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004, 14, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Ozer, A.; Erdost, H.; Zik, B. Histological investigations on the effects of feeding a diet containing red hot pepper on the reproductive organs of the chicken. Phytother. Res. 2005, 19, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Mori, H.; Dakeishi, M.; Onozaki, I.; Murata, K. Effects of mixed organic solvents on neuromotor functions among workers in Buddhist altar manufacturing factories. J. Occup. Health 2005, 47, 143–148. [Google Scholar] [CrossRef]
- Sallmen, M.; Neto, M.; Mayan, O.N. Reduced fertility among shoe manufacturing workers. Occup. Environ. Med. 2008, 65, 518–524. [Google Scholar] [CrossRef]
- Ou, Y.J.; Zhang, W.C.; Wang, J.L. Experimental study of toxic effect on female gonad induced by N-hexane in rats. Strait J. Prev. Med. 2009, 15, 4–6. [Google Scholar]
- Zilz, T.R.; Griffiths, H.R.; Coleman, M.D. Apoptotic and necrotic effects of hexanedione derivatives on the human neuroblastoma line SK-N-SH. Toxicology 2007, 231, 210–214. [Google Scholar] [CrossRef]
- Qi, B.; Xu, S.; Liang, Y.; Wang, J.; Zhang, Z.; Li, J.; Zhou, J. Proapoptotic effects of 2,5hexanedione on pheochromocytoma cells via oxidative injury. Mol. Med. Rep. 2019, 20, 3249–3255. [Google Scholar] [CrossRef]
- Zuo, E.; Zhang, C.; Mao, J.; Gao, C.; Hu, S.; Shi, X.; Piao, F. 2,5-Hexanedione mediates neuronal apoptosis through suppression of NGF via PI3K/Akt signaling in the rat sciatic nerve. Biosci. Rep. 2019, 39, BSR20181122. [Google Scholar] [CrossRef]
- Hartikainen, J.M.; Tengstrom, M.; Kosma, V.M.; Kinnula, V.L.; Mannermaa, A.; Soini, Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012, 72, 5537–5546. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Li, D.; Terasaka, T.; Nicholas, D.A.; Knight, V.S.; Yang, J.J.; Lawson, M.A. SRXN1 Is Necessary for Resolution of GnRH-Induced Oxidative Stress and Induction of Gonadotropin Gene Expression. Endocrinology 2019, 160, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zeng, Y.; Zhou, J.M.; Nie, S.L.; Peng, Q.; Gong, J.; Huo, J.R. MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol. Med. Rep. 2016, 13, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Moulik, M.; Vatta, M.; Witt, S.H.; Arola, A.M.; Murphy, R.T.; McKenna, W.J.; Boriek, A.M.; Oka, K.; Labeit, S.; Bowles, N.E.; et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J. Am. Coll. Cardiol. 2009, 54, 325–333. [Google Scholar] [CrossRef]
- McKnight, A.J.; Patterson, C.C.; Pettigrew, K.A.; Savage, D.A.; Kilner, J.; Murphy, M.; Sadlier, D.; Maxwell, A.P.; the Warren 3/U.K. Genetics of Kidneys in Diabetes (GoKinD) Study Group. A GREM1 gene variant associates with diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21, 773–781. [Google Scholar] [CrossRef]
- Mueller, C.; Baudler, S.; Welzel, H.; Bohm, M.; Nickenig, G. Identification of a novel redox-sensitive gene, Id3, which mediates angiotensin II-induced cell growth. Circulation 2002, 105, 2423–2428. [Google Scholar] [CrossRef]
- Soria, G.; Ben-Baruch, A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008, 267, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Hu, W.; Feng, Z. The P53 pathway: What questions remain to be explored? Cell Death Differ. 2006, 13, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.L.; Wüstenberg, R.; Rübsam, A.; Schmitz-Salue, C.; Warnecke, G.; Bücker, E.M.; Pettkus, N.; Speidel, D.; Rohde, V.; Schulz-Schaeffer, W.; et al. Chloroquine activates the p53 pathway and induces apoptosis in human glioma cells. Neuro-Oncol. 2010, 12, 389–400. [Google Scholar] [CrossRef]
- Qi, B.; Yi, J.; Tang, G.; Miao, J.; Guo, J. The experimental study of n-hexane on lipid peroxidation and DNA damage of hepatic cell in rats. J. Xian Jiaotong Univ. 2007, 28, 145–148. [Google Scholar]
- Abolaji, A.O.; Adedara, I.A.; Soladogun, A.; Salau, V.; Oguaka, M.; Farombi, E.O. Exposure to 2,5-hexanedione is accompanied by ovarian and uterine oxidative stress and disruption of endocrine balance in rats. Drug Chem. Toxicol. 2014, 300, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2017, 25, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Liu, H.; You, B.; Gu, M.; Shi, S.; Shan, Y.; Li, L.; Chen, J.; You, Y. Overexpression of IGFBP3 is associated with poor prognosis and tumor metastasis in nasopharyngeal carcinoma. Tumor Biol. 2016, 37, 15043–15052. [Google Scholar] [CrossRef] [PubMed]
- Joseph, E.K.; Levine, J.D. Caspase signalling in neuropathic and inflammatory pain in the rat. Eur. J. Neurosci. 2015, 20, 2896–2902. [Google Scholar] [CrossRef]
- Qi, D.S.; Tao, J.H.; Zhang, L.Q.; Li, M.; Wang, M.; Qu, R.; Zhang, S.C.; Liu, P.; Liu, F.; Miu, J.C.; et al. Neuroprotection of Cilostazol against ischemia/reperfusion-induced cognitive deficits through inhibiting JNK3/caspase-3 by enhancing Aktl. Brain Res. 2016, 1653, 67–74. [Google Scholar] [CrossRef]
- Hamzehzadeh, L.; Atkin, S.L.; Majeed, M.; Butler, A.E.; Sahebkar, A. The versatile role of curcumin in cancer prevention and treatment: A focus on PI3K/AKT pathway. J. Cell. Physiol. 2018, 233, 6530–6537. [Google Scholar] [CrossRef]
- Korkolopoulou, P.; Levidou, G.; Trigka, E.A.; Prekete, N.; Karlou, M.; Thymara, I.; Sakellariou, S.; Fragkou, P.; Isaiadis, D.; Pavlopoulos, P. A comprehensive immunohistochemical and molecular approach to the PI3K/AKT/mTOR (phosphoinositide 3-kinase/v-akt murine thymoma viral oncogene/mammalian target of rapamycin) pathway in bladder urothelial carcinoma. BJU Int. 2013, 110, E1237–E1248. [Google Scholar] [CrossRef]
- Wang, Z.; Qiu, Z.; Gao, C.; Sun, Y.; Dong, W.; Zhang, Y.; Chen, R.; Qi, Y.; Li, S.; Guo, Y.; et al. 2,5-hexanedione downregulates nerve growth factor and induces neuron apoptosis in the spinal cord of rats via inhibition of the PI3K/Akt signaling pathway. PLoS ONE 2017, 12, e0179388. [Google Scholar] [CrossRef]
- Mousses, S.; Ozcelik, H.; Lee, P.D.; Malkin, D.; Bull, S.B.; Andrulis, I.L. Two variants of the CIP1/WAF1 gene occur together and are associated with human cancer. Hum. Mol. Genet. 1995, 4, 1089–1092. [Google Scholar] [CrossRef]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-Interacting Protein Cipl Is a Potent Inhibitor of Gl Cyclin-Dependent Kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Parker, S.B.; Eichele, G.; Zhang, P.; Rawls, A.; Sands, A.T.; Bradley, A.; Olson, E.N.; Harper, J.W.; Elledge, S.J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 1995, 267, 1024–1027. [Google Scholar] [CrossRef]
- Dai, X.L.; Guo, X.; Liu, J.J.; Cheng, A.Q.; Peng, X.D.; Zha, L.; Wang, Z.W. Circular RNA circGRAMD1B inhibits gastric cancer progression by sponging miR-130a-3p and regulating PTEN and p21 expression. Aging 2019, 11, 9689–9708. [Google Scholar] [CrossRef]
- Lei, K.; Li, W.; Huang, C.; Li, Y.; Alfason, L.; Zhao, H.; Miyagishi, M.; Wu, S.; Kasim, V. Neurogenic differentiation factor 1 promotes colorectal cancer cell proliferation and tumorigenesis by suppressing the p53/p21 axis. Cancer Sci. 2020, 111, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, S.; Vigneault, F.; Lessard, M.; Leclerc, S.; Drouin, R.; Guerin, S.L. Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nucleic Acids Res. 2006, 34, 6472–6487. [Google Scholar] [CrossRef]
- Waldman, T.; Kinzler, K.W.; Vogelstein, B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995, 55, 5187–5190. [Google Scholar] [PubMed]
- Kralj, M.; Husnjak, K.; Korbler, T.; Pavelic, J. Endogenous p21(WAF1/CIP1) status predicts the response of human tumor cells to wild-type p53 and p21(WAF1/CIP1) overexpression. Cancer Gene Ther. 2003, 10, 457–467. [Google Scholar] [CrossRef]
- Torgovnick, A.; Heger, J.M.; Liaki, V.; Isensee, J.; Schmitt, A.; Knittel, G.; Riabinska, A.; Beleggia, F.; Laurien, L.; Leeser, U.; et al. The Cdkn1a(SUPER) Mouse as a Tool to Study p53-Mediated Tumor Suppression. Cell Rep. 2018, 25, 1027–1039.e6. [Google Scholar] [CrossRef]
- Afshari, C.A.; Nichols, M.A.; Xiong, Y.; Mudryj, M. A role for a p21-E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1996, 7, 979–988. [Google Scholar] [CrossRef]
- Linke, S.P.; Harris, M.P.; Neugebauer, S.E.; Clarkin, K.C.; Shepard, H.M.; Maneval, D.C.; Wahl, G.M. p53-mediated accumulation of hypophosphorylated pRb after the G1 restriction point fails to halt cell cycle progression. Oncogene 1997, 15, 337–345. [Google Scholar] [CrossRef]




| Production Number | Sample | Concentration-ng/uL | Volume/uL | Total/μg | RIN Value | Library Type | Conclusion |
|---|---|---|---|---|---|---|---|
| R18081118 | HD0-1 | 498 | 41 | 20.42 | 9.3 | RNA-seq | A |
| R18081119 | HD0-2 | 465 | 41 | 19.07 | 10.0 | RNA-seq | A |
| R18081120 | HD0-3 | 412 | 42 | 17.30 | 9.4 | RNA-seq | A |
| R18081121 | HD40-1 | 308 | 40 | 12.32 | 9.5 | RNA-seq | A |
| R18081122 | HD40-2 | 282 | 38 | 10.72 | 9.2 | RNA-seq | A |
| R18081123 | HD40-3 | 225 | 40 | 9.00 | 9.7 | RNA-seq | A |
| Gene Symbol | Primer Sequence (5′-3′) | Up/Down |
|---|---|---|
| MYH11 | GCGTCCATGCCAGATAACAC | Down |
| CGCCCGACTTTGATACGTG | ||
| MYCL | GGACCCCTGCATGAAACACT | Down |
| CTGCCTCCTCTTCCTTTTCG | ||
| RENBP | GAGTGGGCCATGAAGCTCT | Down |
| CGAAACTGGCGAAACGTGTA | ||
| DKK2 | TTTGCTGTGCACGTCACTTC | Down |
| TTCTTGCGCTGCTTGGTACA | ||
| F3 | ACGCCCTACCTGGACACAAA | Down |
| TGCCGTTCACCCTGACTAAG | ||
| BCLAF1 | GATTCGGAAGGGGATGACAC | Down |
| CCTCCTCAGTATTCCGGTGA | ||
| MDM2 | ACGACAAAGAAAGCGCCACA | Up |
| ACTCACACCAGCGTCGAGAT | ||
| CASP3 | CGGACAGTGGGACTGAAGAT | Up |
| CGCCAGGAATAGTAACCAGG | ||
| CDKN1A | GACCATGTGGACCTGTTGCT | Up |
| GGCGTTTGGAGTGGTAGAAA | ||
| SPP1 | AGAGACCCTGCCAAGCAAGT | Up |
| ATGAGACTCGTCGGATCGGT | ||
| SLC7A11 | TATCTCTGGCATTTGGACGC | Up |
| GCACTCCAGCTGACACTCA | ||
| IRAK2 | GCTCAGGTCCAGGATTGATTG | Up |
| GCCCAGCAGAGGTAAGATGTT | ||
| GAPDH | ATTCCACCCACGGCAAGTT | GAPDH-F |
| TTTGATGTTGGCGGGATCT | GAPDH-R |
| Gene | Target Sequence |
|---|---|
| si-ssc-CDKN1A_001 | CCAGCATGACAGATTTCTA |
| si-ssc-CDKN1A_002 | CCAAACGCCGGCTGATCTT |
| si-ssc-CDKN1A_003 | GCCGGCTGATCTTCTCCAA |
| Sample | Before Filter | After Filter | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clean | Q20 (%) | Q30 (%) | N (%) | GC (%) | HQ Clean | Q20 (%) | Q30 (%) | N (%) | GC (%) | |
| Data (bp) | Data (bp) | |||||||||
| HD0-1 | 8,546,605,500 | 8,344,655,111 (97.64%) | 8,047,846,082 (94.16%) | 1,288,743 (0.02%) | 4,550,166,659 (53.24%) | 8,224,210,048 | 8,107,811,882 (98.58%) | 7,867,744,147 (95.67%) | 857,487 (0.01%) | 4,374,980,230 (53.20%) |
| HD0-2 | 10,701,057,900 | 10,470,640,137 (97.85%) | 10,123,251,944 (94.60%) | 1,615,274 (0.02%) | 5,683,390,145 (53.11%) | 10,322,343,616 | 10,187,549,091 (98.69%) | 9,903,589,358 (95.94%) | 1,073,648 (0.01%) | 5,478,669,969 (53.08%) |
| HD0-3 | 9,492,620,100 | 9,284,411,677 (97.81%) | 8,972,299,225 (94.52%) | 1,432,089 (0.02%) | 5,053,549,739 (53.24%) | 9,147,813,875 | 9,026,488,425 (98.67%) | 8,771,971,839 (95.89%) | 949,030 (0.01%) | 4,866,908,860 (53.20%) |
| HD40-1 | 8,278,856,700 | 809,754,7348 (97.81%) | 7,826,959,983 (94.54%) | 1,251,797 (0.02%) | 4,324,792,272 (52.24%) | 7,978,267,014 | 7,872,909,409 (98.68%) | 7,652,604,412 (95.92%) | 830,497 (0.01%) | 4,163,722,726 (52.19%) |
| HD40-2 | 11,571,998,400 | 11,316,248,473 (97.79%) | 10,935,522,921 (94.50%) | 1,738,051 (0.02%) | 6,103,038,102 (52.74%) | 11,151,334,819 | 11,003,287,936 (98.67%) | 10,693,484,565 (95.89%) | 1,160,264 (0.01%) | 5,875,747,248 (52.69%) |
| HD40-3 | 8,262,103,500 | 8,082,046,730 (97.82%) | 7,811,943,104 (94.55%) | 1,245,894 (0.02%) | 4,320,317,791 (52.29%) | 7,967,279,552 | 7,862,124,495 (98.68%) | 7,641,947,568 (95.92%) | 830,065 (0.01%) | 4,162,633,359 (52.25%) |
| Gene ID | Gene Name | Control | 2,5-HD | Log2 Fold Change | Up/Down |
|---|---|---|---|---|---|
| ENSSSCG00000007554 | ZFAND2A | 411.333 | 8812.667 | 4.457916894 | Up |
| ENSSSCG00000023298 | SRXN1 | 1613.667 | 24,429.000 | 4.006772443 | Up |
| ENSSSCG00000008988 | CCNG2 | 411.667 | 4900.333 | 3.610374422 | Up |
| ENSSSCG00000001488 | GCLC | 1608.333 | 17,005.667 | 3.483567018 | Up |
| ENSSSCG00000022649 | SLC7A11 | 602.000 | 5672.333 | 3.33201828 | Up |
| ENSSSCG00000010461 | ANKRD1 | 15,720.333 | 299.333 | 4.606115928 | Down |
| ENSSSCG00000033657 | GREM1 | 6506.000 | 358.000 | 4.085838065 | Down |
| ENSSSCG00000039514 | ID3 | 11,693.000 | 722.333 | 3.919259823 | Down |
| ENSSSCG00000017723 | CCL2 | 16,245.333 | 1019.000 | 3.912958487 | Down |
| ENSSSCG00000000146 | MYH11 | 24,112.870 | 1581.530 | 3.875688189 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Kong, C.; Yang, M.; Liu, Y.; Han, Z.; Xu, L.; Zheng, X.; Ding, Y.; Yin, Z.; Zhang, X. 2,5-Hexanedione Affects Ovarian Granulosa Cells in Swine by Regulating the CDKN1A Gene: A Transcriptome Analysis. Vet. Sci. 2023, 10, 201. https://doi.org/10.3390/vetsci10030201
Chen Y, Kong C, Yang M, Liu Y, Han Z, Xu L, Zheng X, Ding Y, Yin Z, Zhang X. 2,5-Hexanedione Affects Ovarian Granulosa Cells in Swine by Regulating the CDKN1A Gene: A Transcriptome Analysis. Veterinary Sciences. 2023; 10(3):201. https://doi.org/10.3390/vetsci10030201
Chicago/Turabian StyleChen, Yige, Chengcheng Kong, Min Yang, Yangguang Liu, Zheng Han, Liming Xu, Xianrui Zheng, Yueyun Ding, Zongjun Yin, and Xiaodong Zhang. 2023. "2,5-Hexanedione Affects Ovarian Granulosa Cells in Swine by Regulating the CDKN1A Gene: A Transcriptome Analysis" Veterinary Sciences 10, no. 3: 201. https://doi.org/10.3390/vetsci10030201
APA StyleChen, Y., Kong, C., Yang, M., Liu, Y., Han, Z., Xu, L., Zheng, X., Ding, Y., Yin, Z., & Zhang, X. (2023). 2,5-Hexanedione Affects Ovarian Granulosa Cells in Swine by Regulating the CDKN1A Gene: A Transcriptome Analysis. Veterinary Sciences, 10(3), 201. https://doi.org/10.3390/vetsci10030201






