Exploring a Possible Link between the Fecal Microbiota and the Production Performance of Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Management and Sample Collection
2.2. DNA Extraction and PCR Amplification
2.3. 16. S Ribosomal RNA Sequencing and Analysis
2.3.1. Quality Control and Reads Merge
2.3.2. Alpha and Beta Diversity Analysis
2.3.3. Function Prediction
2.4. Meishan Pigs Feeding Experiments
3. Results
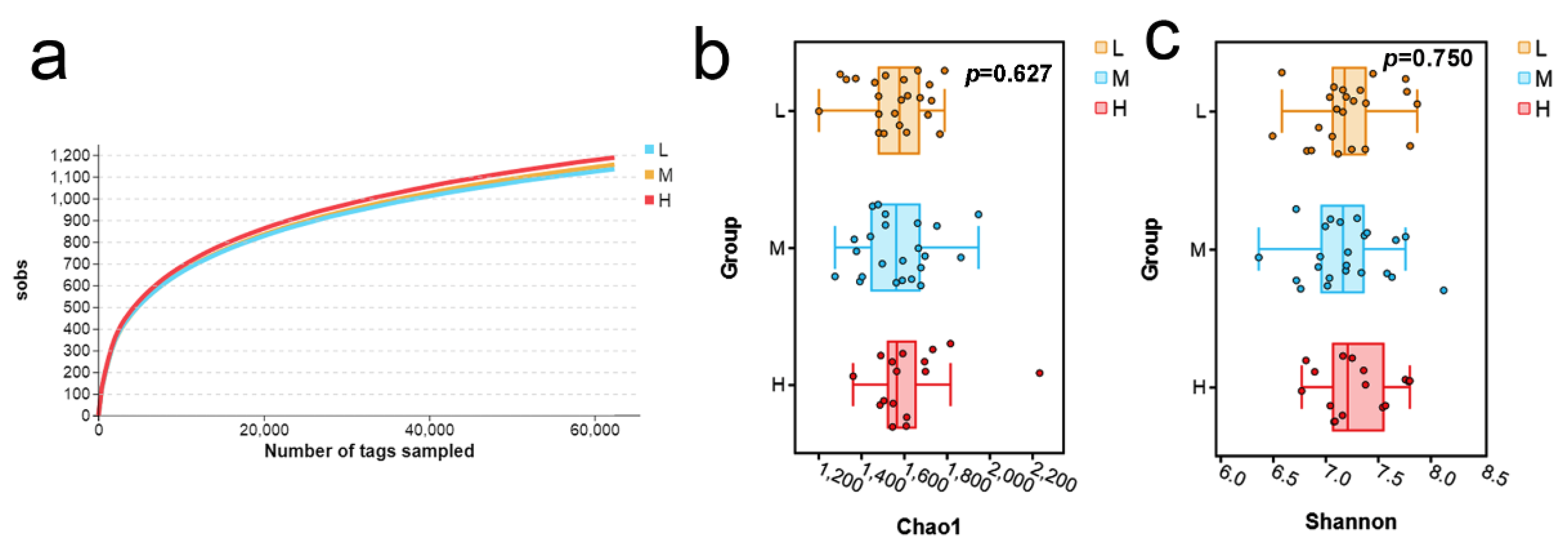
3.1. The Richness of Fecal Microbes in Pigs with Different Backfat Thickness
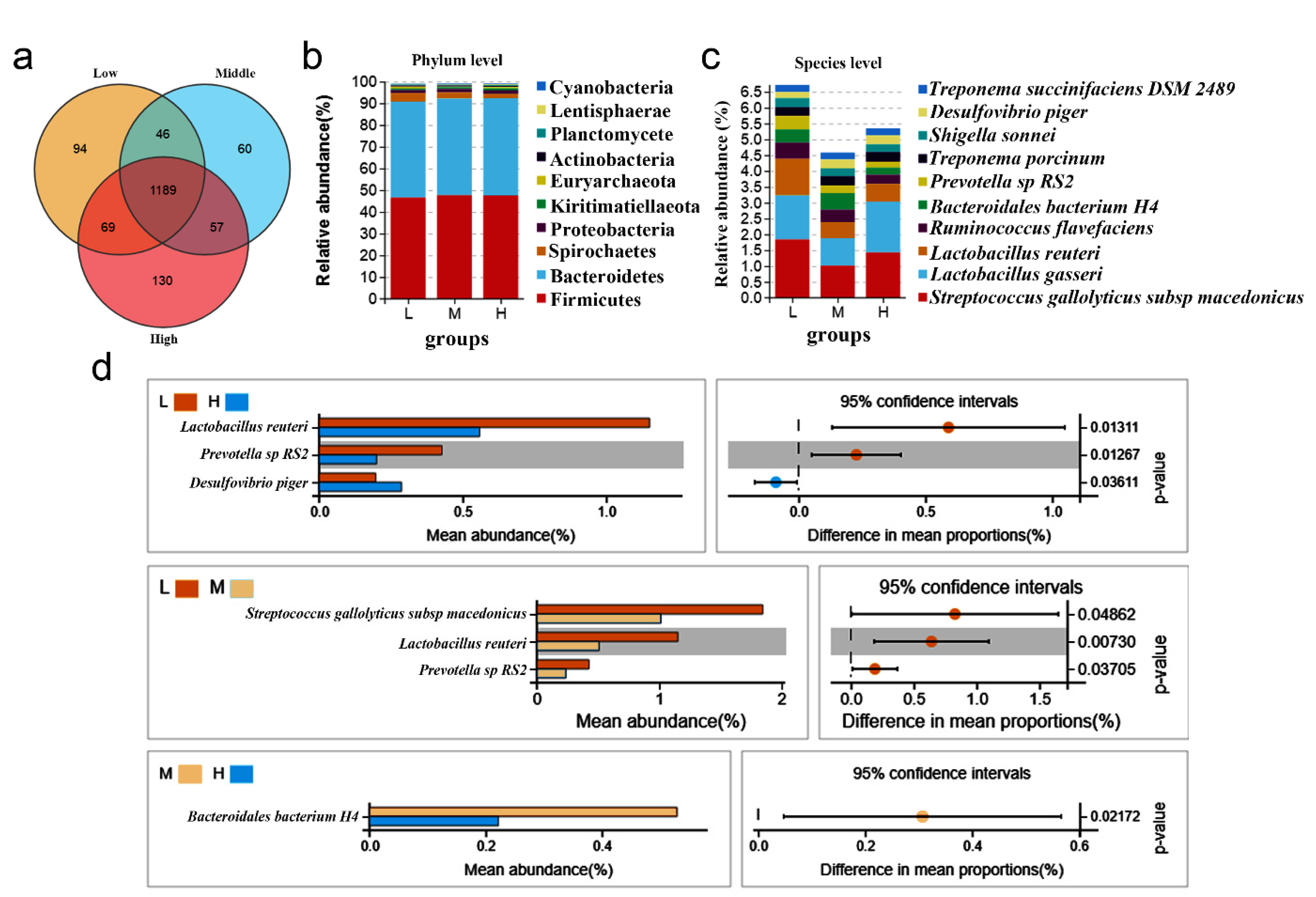
3.2. Analysis of Microbial Community and Structure Composition
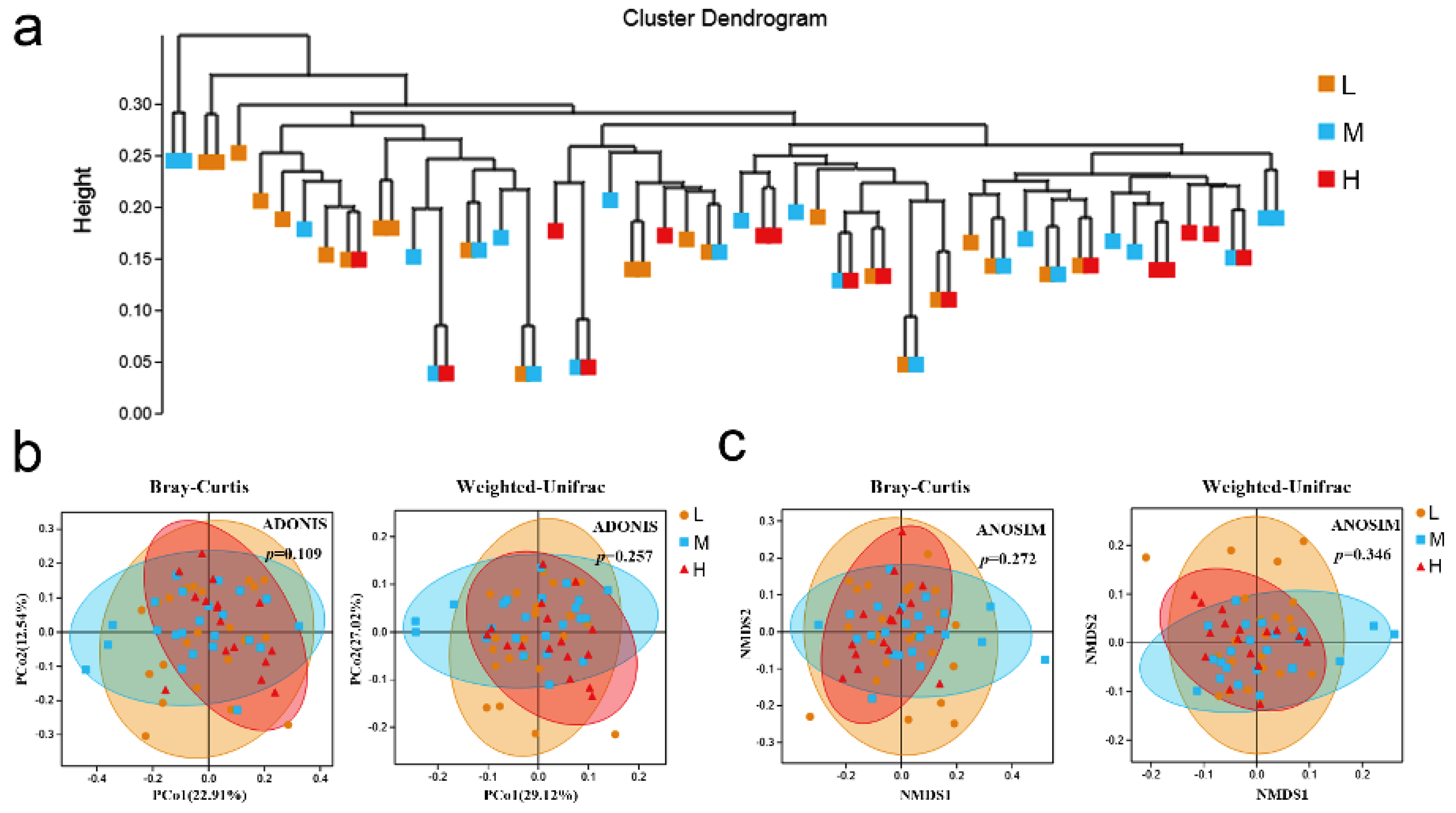
3.3. Community Structures of Different Backfat Thickness Groups
3.4. Functional Profiling of Microbial Communities in Different Backfat Thickness Groups
3.5. Feeding L. reuteri Improved Piglets Production Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amaral Filha, W.S.; Bernardi, M.L.; Wentz, I.; Bortolozzo, F.P. Reproductive performance of gilts according to growth rate and backfat thickness at mating. Anim. Reprod. Sci. 2010, 121, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, T.; Cai, A.; Wu, Y.; Wei, H.; Jiang, S.; Peng, J. Excessive backfat of sows at 109 d of gestation induces lipotoxic placental environment and is associated with declining reproductive performance. J. Anim. Sci. 2018, 96, 250–257. [Google Scholar] [CrossRef]
- Tian, L.; Huang, J.; Wen, A.; Yan, P. Impaired mitochondrial function results from oxidative stress in the full-term placenta of sows with excessive back-fat. Animals 2020, 10, 360. [Google Scholar] [CrossRef]
- Amdi, C.; Giblin, L.; Ryan, T.; Stickland, N.C.; Lawlor, P.G. Maternal backfat depth in gestating sows has a greater influence on offspring growth and carcass lean yield than maternal feed allocation during gestation. Anim. Int. J. Anim. Biosci. 2014, 8, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Yang, D.D.; Liu, Z.L.; Zeng, Y.Q.; Chen, W. Expression of lipid metabolism genes provides new insights into intramuscular fat deposition in Laiwu pigs. Asian-Australas J. Anim. Sci. 2020, 33, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Cui, X.; Liu, R.; Li, Q.; Zheng, M.; Zhao, G.; Ge, C.; Wen, J.; Hu, Y.; Cui, H. Identification of differentially expressed genes and pathways for abdominal fat deposition in ovariectomized and sham-operated chickens. Genes 2019, 10, 155. [Google Scholar] [CrossRef]
- Han, X.; Yang, H.; Jiang, T.; Zhang, Q.; Zeng, C.; Fan, B.; Liu, B. Investigation of four candidate genes (IGF2, JHDM1A, COPB1 and TEF1) for growth rate and backfat thickness traits on SSC2q in Large White pigs. Mol. Biol. Rep. 2014, 41, 309–315. [Google Scholar] [CrossRef]
- Wen, C.; Yan, W.; Sun, C.; Ji, C.; Zhou, Q.; Zhang, D.; Zheng, J.; Yang, N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. Isme J. 2019, 13, 1422–1436. [Google Scholar] [CrossRef]
- Chauhan, V.; Kanwar, S.S. Lipopeptide(s) associated with human microbiome as potent cancer drug. Semin. Cancer Biol. 2021, 70, 128–133. [Google Scholar] [CrossRef]
- Sanchez, M.-P.; Riquet, J.; Iannuccelli, N.; Gogué, J.; Billon, Y.; Demeure, O.; Caritez, J.-C.; Burgaud, G.; Fève, K.; Bonnet, M.; et al. Effects of quantitative trait loci on chromosomes 1, 2, 4, and 7 on growth, carcass, and meat quality traits in backcross Meishan × Large White pigs1. J. Anim. Sci. 2006, 84, 526–537. [Google Scholar] [CrossRef]
- Niu, Q.; Zhang, G.; Zhang, L.; Ma, Y.; Shi, Q.; Fu, W. Purification and characterization of a thermophilic 1,3-1,4-β-glucanase from Bacillus methylotrophicus S2 isolated from booklice. J. Biosci. Bioeng. 2016, 121, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Meade, K.G.; O’Farrelly, C. β-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 2018, 9, 3072. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.K.; Cadwell, K. Regulation of interferon signaling in response to gut microbes by autophagy. Gut Microbes 2020, 11, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Li, J.; Xing, T.; Jiang, Y.; Zhang, L.; Gao, F. Dietary corn-resistant starch suppresses broiler abdominal fat deposition associated with the reduced cecal Firmicutes. Poult. Sci. 2020, 99, 5827–5837. [Google Scholar] [CrossRef]
- Suárez-Zamorano, N.; Fabbiano, S.; Chevalier, C.; Stojanović, O.; Colin, D.J.; Stevanović, A.; Veyrat-Durebex, C.; Tarallo, V.; Rigo, D.; Germain, S.; et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015, 21, 1497–1501. [Google Scholar] [CrossRef]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Sung, H.W.; Chen, C.N.; Liang, H.F.; Hong, M.H. A natural compound (reuterin) produced by Lactobacillus reuteri for biological-tissue fixation. Biomaterials 2003, 24, 1335–1347. [Google Scholar] [CrossRef]
- Taranto, M.P.; Vera, J.L.; Hugenholtz, J.; De Valdez, G.F.; Sesma, F. Lactobacillus reuteri CRL1098 produces cobalamin. J. Bacteriol. 2003, 185, 5643–5647. [Google Scholar] [CrossRef]
- White, B.R.; Lan, Y.H.; McKeith, F.K.; Novakofski, J.; Wheeler, M.B.; McLaren, D.G. Growth and body composition of Meishan and Yorkshire barrows and gilts. J. Anim. Sci. 1995, 73, 738–749. [Google Scholar] [CrossRef]
- Legault, C. Selection of breeds, strains and individual pigs for prolificacy. J. Reprod. Fertil. Suppl. 1985, 33, 151–166. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W. Ggplot2: An Implementation of the Grammar of Graphics. Rpackage Version 0.7. Available online: https://www.semanticscholar.org/paper/ggplot%3A-An-implementation-of-the-Grammar-of-in-R-Wickham/7f3e2207d2ef8fc0cee74069879c8adf35303a91 (accessed on 26 February 2022).
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, H.; Oksanen, M. The Vegan Package. Vegan: Community Ecology Package. 2010. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 26 February 2022).
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Tummaruk, P.; Sumransap, P.; Jiebna, N. Fat and whey supplementation influence milk composition, backfat loss, and reproductive performance in lactating sows. Trop. Anim. Health Prod. 2014, 46, 753–758. [Google Scholar] [CrossRef]
- Cheng, C.; Wu, X.; Zhang, X.; Zhang, X.; Peng, J. Obesity of sows at late pregnancy aggravates metabolic disorder of perinatal sows and affects performance and intestinal health of piglets. Animals 2019, 10, 49. [Google Scholar] [CrossRef]
- De Rensis, F.; Gherpelli, M.; Superchi, P.; Kirkwood, R.N. Relationships between backfat depth and plasma leptin during lactation and sow reproductive performance after weaning. Anim. Reprod. Sci. 2005, 90, 95–100. [Google Scholar] [CrossRef]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X.; et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, X.M.; Xia, R.W.; Huo, Y.J.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Association between the MUC4 g.243A > G polymorphism and immune and production traits in Large White pigs. Turk. J. Vet. Anim. Sci. 2015, 39, 141–146. [Google Scholar] [CrossRef]
- Muñoz-Garach, A.; Diaz-Perdigones, C.; Tinahones, F.J. Gut microbiota and type 2 diabetes mellitus. Endocrinol. Y Nutr. Organo De La Soc. Esp. De Endocrinol. Y Nutr. 2016, 63, 560–568. [Google Scholar] [CrossRef]
- John, G.K.; Mullin, G.E. The Gut Microbiome and Obesity. Curr. Oncol. Rep. 2016, 18, 45. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Torres-Fuentes, C.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. The microbiota–gut–brain axis in obesity. Lancet Gastroenterol. Hepatol. 2017, 2, 747–756. [Google Scholar] [CrossRef]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 2017, 19, 95–105. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Fang, S.; Huang, X.; Zhao, Y.; Ke, S.; Yang, H.; Li, Z.; Gao, J.; Chen, C.; Huang, L. Evaluating the Contribution of Gut Microbiota to the Variation of Porcine Fatness with the Cecum and Fecal Samples. Front. Microbiol. 2016, 7, 2108. [Google Scholar] [CrossRef]
- Jiang, J.; Feng, N.; Zhang, C.; Liu, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W.J.F.M.L. Lactobacillus reuteri A9 and Lactobacillus mucosae A13 isolated from Chinese superlongevity people modulate lipid metabolism in a hypercholesterolemia rat model. FEMS Microbiol. Lett. 2019, 366, fnz254. [Google Scholar] [CrossRef]
- Sun, Y.; Tang, Y.; Hou, X.; Wang, H.; Huang, L.; Wen, J.; Niu, H.; Zeng, W.; Bai, Y.J.F.i.V.S. Novel Lactobacillus reuteri HI120 affects lipid metabolism in C57BL/6 obese mice. Front. Vet. Sci. 2020, 7, 560241. [Google Scholar] [CrossRef]
- Zhang, C.; Fang, R.; Lu, X.; Zhang, Y.; Yang, M.; Su, Y.; Jiang, Y.; Man, C. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 2022, 13, 6688–6701. [Google Scholar] [CrossRef]
- Haley, C.S.; Lee, G.J. Genetic basis of prolificacy in Meishan pigs. J. Reprod. Fertility. Suppl. 1993, 48, 247–259. [Google Scholar]
- Rothschild, M.; Jacobson, C.; Vaske, D.; Tuggle, C.; Wang, L.; Short, T.; Eckardt, G.; Sasaki, S.; Vincent, A.; McLaren, D. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc. Natl. Acad. Sci. USA 1996, 93, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Hoa, V.B.; Seo, H.W.; Seong, P.N.; Cho, S.H.; Kang, S.M.; Kim, Y.S.; Moon, S.S.; Choi, Y.M.; Kim, J.H.; Seol, K.H. Back-fat thickness as a primary index reflecting the yield and overall acceptance of pork meat. Anim. Sci. J. 2021, 92, e13515. [Google Scholar] [CrossRef] [PubMed]
- Montagne, L.; Boudry, G.; Favier, C.; Le Huërou-Luron, I.; Lalles, J.-P.; Seve, B. Main intestinal markers associated with the changes in gut architecture and function in piglets after weaning. Br. J. Nutr. 2007, 97, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, J.; Zhang, X.; Mo, S.; Tan, X.; Wang, L.; Li, J.; Li, Y.; Ding, X.; Liu, X. The production of short chain fatty acid and colonic development in weaning piglets. J. Anim. Physiol. Anim. Nutr. 2019, 103, 1530–1537. [Google Scholar] [CrossRef]
- Wang, M.; Wu, H.; Lu, L.; Jiang, L.; Yu, Q. Lactobacillus reuteri promotes intestinal development and regulates mucosal immune function in newborn piglets. Front. Vet. Sci. 2020, 7, 42. [Google Scholar] [CrossRef]
- Dawood, M.A.; Magouz, F.I.; Salem, M.F.; Elbialy, Z.I.; Abdel-Daim, H.A. Synergetic effects of Lactobacillus plantarum and β-glucan on digestive enzyme activity, intestinal morphology, growth, fatty acid, and glucose-related gene expression of genetically improved farmed tilapia. Probiotics Antimicrob. Proteins 2020, 12, 389–399. [Google Scholar] [CrossRef]
- Shen, Y.L.; Zhang, L.Q.; Yang, Y.; Yin, B.C.; Ye, B.C.; Zhou, Y. Advances in the role and mechanism of lactic acid bacteria in treating obesity. Food Bioeng. 2022, 1, 101–115. [Google Scholar] [CrossRef]
- Ni, C.; Li, X.; Wang, L.; Li, X.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Lactic acid bacteria strains relieve hyperuricaemia by suppressing xanthine oxidase activity via a short-chain fatty acid-dependent mechanism. Food Funct. 2021, 12, 7054–7067. [Google Scholar] [CrossRef]
- Lew, L.-C.; Hor, Y.-Y.; Jaafar, M.-H.; Lau, A.-S.-Y.; Lee, B.-K.; Chuah, L.-O.; Yap, K.-P.; Azlan, A.; Azzam, G.; Choi, S.-B. Lactobacillus strains alleviated hyperlipidemia and liver steatosis in aging rats via activation of AMPK. Int. J. Mol. Sci. 2020, 21, 5872. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Wang, F.; Wang, H.; Wu, S.; Bao, W. Exploring a Possible Link between the Fecal Microbiota and the Production Performance of Pigs. Vet. Sci. 2022, 9, 527. https://doi.org/10.3390/vetsci9100527
Cao Y, Wang F, Wang H, Wu S, Bao W. Exploring a Possible Link between the Fecal Microbiota and the Production Performance of Pigs. Veterinary Sciences. 2022; 9(10):527. https://doi.org/10.3390/vetsci9100527
Chicago/Turabian StyleCao, Yanan, Fei Wang, Haifei Wang, Shenglong Wu, and Wenbin Bao. 2022. "Exploring a Possible Link between the Fecal Microbiota and the Production Performance of Pigs" Veterinary Sciences 9, no. 10: 527. https://doi.org/10.3390/vetsci9100527
APA StyleCao, Y., Wang, F., Wang, H., Wu, S., & Bao, W. (2022). Exploring a Possible Link between the Fecal Microbiota and the Production Performance of Pigs. Veterinary Sciences, 9(10), 527. https://doi.org/10.3390/vetsci9100527







