Abstract
Latin American countries produce more than a quarter of the world’s beef and are a major global supplier of livestock protein. Tick-borne diseases (TBDs) are a major constraint to the livestock industry worldwide, including in Latin America. The aim of this study was to detect and characterise tick-borne pathogens in cattle from Santa Cruz, Bolivia, where no detailed epidemiological data are available. Blood samples were collected from 104 cattle. Apicomplexan parasites were detected by nested PCR amplification of the 18S ribosomal RNA gene (rDNA), and Anaplasmataceae was screened by the PCR amplification of 16S rDNA, followed by characterisation based on the heat shock protein and citrate synthase gene sequences. Babesia infection was observed in nine cattle (one Babesia bovis and eight Babesia bigemina), while Anaplasmataceae infection was detected in thirty-two cattle. A sequencing analysis confirmed the presence of Anaplasma marginale and Anaplasma platys-like. These results provide the first molecular evidence for the four above-mentioned tick-borne pathogens in cattle in Bolivia. This information improves our understanding of the epidemiology of TBDs and will help in formulating appropriate and improved pathogen control strategies.
1. Introduction
Latin American countries produce a substantial portion of the world’s beef supply [1]. Beef production in Latin America has increased by 29.8% over nearly two decades (between 2000 and 2018), and the livestock sector accounts for 46% of the agricultural gross domestic product in Latin America [1,2]. Diseases transmitted by ticks, so-called tick-borne diseases (TBDs), are a major issue in the livestock industry, causing considerable economic losses worldwide, including in Latin America. For instance, in Brazil, TBDs cause an annual economic loss of around 3.24 billion USD [3]. However, in many Latin American countries, the significance of TBDs has not been evaluated, in part owing to a lack of relevant epidemiological data.
In Latin American countries, the most severe and prevalent TBDs are babesiosis and anaplasmosis [4,5,6,7,8]. Bovine babesiosis is a globally distributed tick-borne hemoprotozoan disease caused by pathogenic species, such as Babesia bovis, Babesia bigemina, and Babesia divergens. The geographical distribution of the disease is defined by the prevalence of vector tick species [9]. The disease is clinically manifested by anaemia, fever, haemoglobinuria, and marked splenomegaly, sometimes resulting in death [10]. In Latin America, epidemiological studies have confirmed the presence of two pathogenic species, B. bovis and B. bigemina, which are transmitted by Rhipicephalus microplus [7]. The rate of tick transmission is generally higher for B. bigemina than for B. bovis under natural conditions, and B. bovis is more pathogenic than B. bigemina [11]. In Colombia, a molecular study of cattle (n = 1432) revealed that the Babesia-positive rate is 31.6% (24.2% for B. bigemina and 14.4% for B. bovis) [7].
Bovine anaplasmosis is a major tick-borne bacterial disease in cattle [12]. The causative agents in the genus Anaplasma include Anaplasma marginale, Anaplasma centrale, Anaplasma phagocytophilum, and Anaplasma bovis [12,13]. Anaplasma marginale infects erythrocytes and is highly pathogenic in cattle with a wide distribution in tropical and subtropical regions [13,14]. In cattle aged > 2 years, A. marginale causes persistent fever, lethargy, icterus, weight loss, abortion, decreased milk yield, and death in more than 50% of untreated animals [14,15]. Anaplasma centrale, which is less pathogenic than A. marginale, causes mild symptoms in cattle [13]. Anaplasma phagocytophilum is an obligate intracellular bacterium that infects granulocytes and is distributed worldwide [13,15]. It is a zoonotic pathogen that causes tick-borne fever in ruminants and induces high fever, respiratory symptoms, leucopoenia, abortion, and sudden decreases in milk yield [13,15]. Anaplasma bovis, a monocytotropic species, has been detected in ruminants in many countries. Asymptomatic infection has been documented; however, it can cause fever, anaemia, weight loss, and occasional abortion and death [13]. Anaplasma platys usually infects dogs and is generally transmitted by brown dog ticks (Rhipicephalus sanguineus). However, in recent studies, strains genetically related to A. platys (A. platys-like) were detected in ruminants (sheep, goats, deer, camels, and cattle) [16,17,18,19]. For instance, a survey of beef cattle (n = 400) in the Brazilian Pantanal detected A. platys-like in 4.75% of the tested animals showing no anaemia or other clinical signs [20].
Cattle are of substantial economic importance for the livestock industry in Bolivia. The expansion of cattle ranching was expected since Bolivia began exporting beef to China in 2019 [21]. In Bolivia, only a few serological studies have investigated the prevalence of TBDs more than two decades ago [22,23]. Since then, detailed epidemiological reports including genetic data for pathogens circulating in the area are lacking. Therefore, this study aimed to detect and characterise tick-borne pathogens in different cattle breeds in Santa Cruz, Bolivia, using molecular methods.
2. Materials and Methods
2.1. Blood Sampling and DNA Extraction
Blood samples were collected from pastured cattle (age of over 18 months, regardless of sex) at three farms managed by the Autonomous University Gabriel Rene Moreno (University Farm 1 at El Plado, University Farm 2 at Todos Santos, and University Farm 3 at Yabare) and three private farms located in San Juan, Santa Cruz, between December 2019 and March 2020 (Figure 1). A total of 104 individuals from the following 8 breeds were included: Nelore (n = 41), Holstein (n = 10), Gyr (n = 10), Gyrolando (n = 8), Mestizo (n = 2), Senepol (n = 10), Neloblanca (n = 1), and Criollo (n = 22). The sampled animals were randomly selected from each farm. Approximately 2–3 mL of cattle blood was collected from the jugular vein into EDTA tubes. DNA was extracted from 500 µL of blood using DNAzol (MRC, Cincinnati, OH, USA). All animal handling procedures were conducted in accordance with the guidelines established by the Animal Experiment Committee of the Graduate School of Veterinary Medicine, Hokkaido University (Sapporo, Japan). This study was approved by the Institutional Committee for the Care and Use of Experimental Animals at Autonomous University Gabriel Rene Moreno (CICUAE, 2015, No. 008/19).
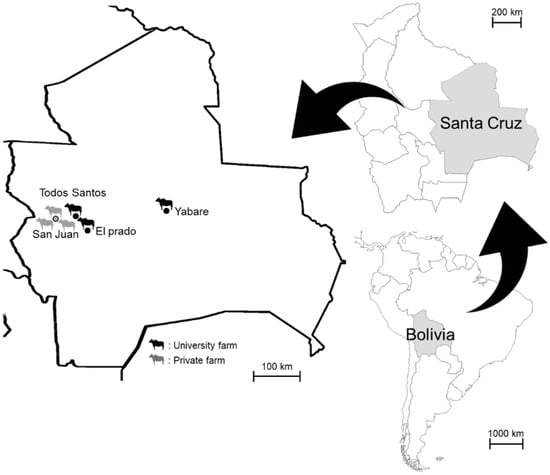
Figure 1.
Geographical location of the study farms in Santa Cruz, Bolivia.
2.2. Detection and Characterisation of Babesia spp.
Babesia, Theileria, and Hepatozoon were screened by nested BTH PCR using the primer sets BTH 1st F/BTH 1st R and BTH 2nd F/BTH 2nd R for the primary and secondary rounds, respectively (Table 1). This PCR amplified nearly the full length of the 18S ribosomal RNA gene (rDNA) (1400–1600 bp) [24]. The PCR was carried out in a 20.0 μL reaction mixture containing 10.0 μL of 2× Gflex PCR Buffer (Mg2+, dNTP plus), 400 nM of Tks Gflex™ DNA Polymerase (Takara Bio, Shiga, Japan), 400 nM of each primer, 1.0 μL of DNA template (or 10-fold diluted first PCR product), and sterilized water. The reaction was performed at 94 °C for 1 min, followed by 45 cycles at 98 °C for 10 s, 55 or 60 °C for 15 s, and 68 °C for 90 s, and a final step at 68 °C for 5 min. The PCR products were separated by electrophoresis on a 1.5% agarose gel stained with Gel-Red (Biotium, Hayward, CA, USA) and visualised under UV light (Supplementary Figure S1). Each assay included Theileria parva DNA detected in our previous study [25] and sterilised water as positive and negative controls, respectively.

Table 1.
Primers used in this study.
2.3. Detection and Characterisation of Anaplasmataceae
Anaplasmataceae was screened by EHR PCR using the primers EHR16SD and EHR16SR (Table 1). This PCR amplified approximately 345 bp of the V1 hypervariable region of the 16S rDNA of Anaplasmataceae [26].
To further characterise Anaplasmataceae detected by EHR PCR, additional semi-nested PCRs targeting the heat shock protein (groEL) and citrate synthase (gltA) genes were employed [16]. A partial gltA gene sequence (approximately 630 bp) of A. platys and related strains was amplified in either of the two semi-nested PCRs. Initially, PCR was conducted using the primer sets Pglt-F/Pglt-R1 and Pglt-F/Pglt-R2 for the primary and secondary rounds of semi-nested PCR, respectively (Table 1). Alternatively, when the reaction was negative, another PCR was conducted using the primer sets Pglt-L-F1/Pglt-L-R and Pglt-L-F2/Pglt-L-R for the primary and secondary rounds, respectively (Table 1). Approximately 373 bp of the groEL gene sequence was amplified with either of the following primer combinations: Pgro-F-F/Pgro-F-R1 for the primary and Pgro-F-F/Pgro-F-R2 for the secondary round, Pgro-L-F1/Pgro-L-R for the primary and Pgro-L-F2/Pgro-L-R for the secondary round, or Pgro-F1/Pgro-R for the primary and Pgro-F2/Pgro-R for the secondary round (Table 1).
The PCR was conducted as described above with the annealing temperatures listed in Table 1. Tick DNA samples positive for Anaplasma in our previous study [27,28] and sterilised water were included in each PCR run as positive and negative controls, respectively. The PCR products were separated by electrophoresis on a 1.0% agarose gel stained with Gel-Red and visualised under UV light (Supplementary Figure S1).
2.4. Sanger Sequencing
All amplicons of the second groEL and second gltA PCRs for Anaplasmataceae and the second BHT PCR were purified using ExoSAP-IT (USB Corporation, Cleveland, OH, USA). The purified products were sequenced using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems).
2.5. Data Analysis
Raw sequence data were edited by merging the forward and reverse sequences, followed by the removal of primer annealing sites using ATGC version 9.1 (GENETYX Corporation, Tokyo, Japan). Phylogenetic trees were constructed based on the partial sequences of groEL and gltA for Anaplasma and 18S rDNA for Babesia. The nucleotide sequences were aligned with representative sequences of known Anaplasma and Babesia species available in GenBank as implemented in MEGA7 [29]. Phylogenetic trees were constructed using the maximum likelihood (ML) method with 1000 bootstrap replicates. The best evolutionary models for the sequence data were determined based on the Akaike information criterion using MEGA7 [29]. The sequences obtained in this study were submitted to the DNA Data Bank of Japan (DDBJ) (18S rDNA sequences of B. bigemina, LC645216-LC645223; 18S rDNA sequence of B. bovis, LC645224; gltA gene sequences of A. platys-like, LC645225-LC645237; gltA gene sequence of A. marginale: LC645238; groEL gene sequence of A. platys-like, LC645239-LC645260).
3. Results
3.1. Detection and Characterisation of Babesia
BTH PCR was positive in nine cattle: five at University Farm 1, three at University Farm 2, and one at University Farm 3. The cattle breeds that were positive for infection were Nelore (n = 2), Holstein (n = 5), Gyrolando (n = 1), and Criollo (n = 1). All amplicons were successfully sequenced using the Sanger method. The species detected were B. bigemina (n = 8) and B. bovis (n = 1) (Figure 2).
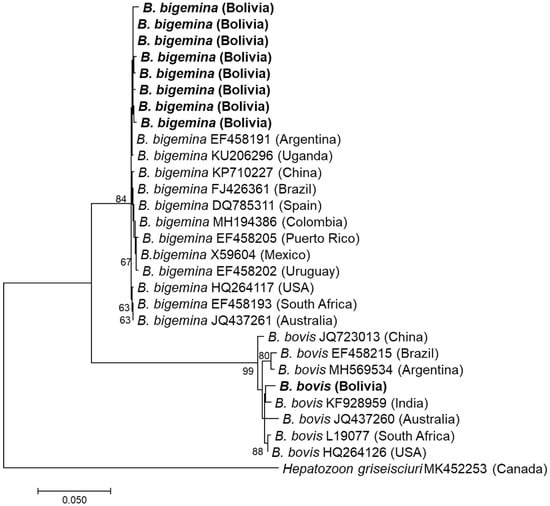
Figure 2.
Phylogenetic tree based on 18S rDNA sequences of Babesia species. The phylogenetic tree was constructed using MEGA7 based on the maximum likelihood method using the Tamura 3-parameter model. Only bootstrap values greater than 60% are indicated. The sequences obtained in this study are shown in bold. The geographic origin (country) of each sequence/strain is provided in parentheses.
Eight 18S rDNA sequences of B. bigemina showed one and seven mismatched bases and showed the highest sequence identity (99.5–99.9%) with 18S rDNA of B. bigemina vaccine strain ‘S1A’ reported from Argentina (EF458191). One sequence of B. bovis had the highest identity (1387/1403, 98.9%) with 18S rDNA of a B. bovis strain from the blood of cattle in India (KF928959).
3.2. Detection and Characterisation of Anaplasmataceae
By EHR PCR detection, Anaplasmataceae infection was positive in 32 cattle: 8 from University Farm 1, 15 from University Farm 2, 7 from University Farm 3, 1 from Private Farm 2, and 1 from Private Farm 3. The infected cattle breeds were Nelore (n = 4), Holstein (n = 8), Gyr (n = 6), Gyrolando (n = 7), and Criollo (n = 7).
All samples positive by EHR PCR were subjected to semi-nested PCRs targeting the gltA and groEL genes. Although PCR amplicons were obtained from all 32 samples, gltA and groEL were only successfully sequenced in 14 and 22 samples, respectively (Supplementary Table S1). In the remaining samples, sequencing failed due to mixed signals. Finally, a sequencing analysis of the purified amplicons identified four and three different gltA and groEL sequences, respectively. Three gltA sequences obtained from thirteen samples showed the highest sequence identity (628–630/630, 99.7–100.0%) with the A. platys-like strain reported from R. microplus in China (MH716426), while one gltA sequence had the highest identity (611/633, 96.5%) with A. marginale reported from R. microplus in China (KX987364). The three groEL sequences obtained had 1–5 nucleotide mismatches with each other and showed the highest identity (370–372/373, 99.2–99.7%) with an A. platys-like strain reported from R. microplus in China (MH716435). Phylogenetic trees based on gltA and groEL are shown in Figure 3 and Figure 4, respectively. The gltA sequences of A. platys were divided into two clades in both trees. The clade including A. platys-like detected in this study was composed of A. platys-like detected from cattle ticks (R. microplus), cattle, camels, and water buffalo, while the other clade consisted of A. platys reported from dogs and brown dog ticks (R. sanguineus) (Figure 3 and Figure 4).
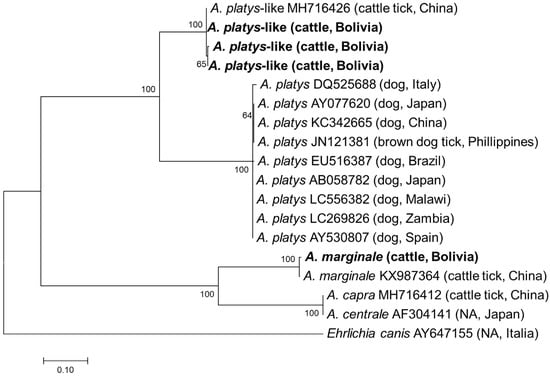
Figure 3.
Phylogenetic tree based on gltA sequences in Anaplasma species. The phylogenetic tree was constructed using MEGA7 based on the maximum likelihood method using the Kimura 2-parameter model. Only bootstrap values greater than 60% are indicated. The sequences obtained in this study are shown in bold. The host animal and geographic origin (country) of the sequence/strain are provided in parentheses. NA stands for not available.
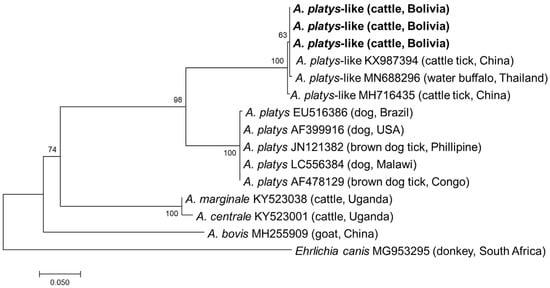
Figure 4.
Phylogenetic tree based on the groEL sequences of Anaplasma species. The phylogenetic tree was constructed using MEGA7 based on the maximum likelihood method, using the Tamura 3-parameter model. Only bootstrap values greater than 60% are indicated. The sequences obtained in this study are shown in bold. The geographic origin (country) of each sequence/strain is provided in parentheses.
3.3. Co-Infection
Seven of nine cattle infected with B. bigemina were also infected with A. platys-like (Supplementary Table S2). These included four animals at University Farm 1 and three animals at University Farm 2. One B. bigemina-infected animal at University Farm 1 was co-infected with A. marginale (Supplementary Table S1). The cattle breeds that were positive for co-infection were Nelore (n = 2), Holstein (n = 5), and Gyrolando (n = 1) (Table 2, Supplementary Table S2).

Table 2.
Infection rate of Babesia and Anaplasma.
4. Discussion
Babesiosis and anaplasmosis are among the most important TBDs in cattle. Although there is increasing evidence for their high prevalence worldwide, few studies have focused on resource-limited countries, such as Bolivia. The lack of epidemiological data for the occurrence of TBDs not only leads to misdiagnosis and treatment delays but also hinders the design of proper tick control measures.
Bovine babesiosis is a worldwide TBD caused by B. bovis and B. bigemina in Latin America [7]. This study is the first molecular survey of TBDs in cattle in Bolivia and confirmed the presence of both B. bovis and B. bigemina. Most Babesia detected in this study were B. bigemina, which is pathogenic and thus poses a significant challenge to livestock health. These results are in contrast to those of previous serological studies showing that the seroprevalence rate of B. bovis is higher than that of B. bigemina in two different surveyed locations, Bolivian Chaco and Santa Cruz [22,23]. However, molecular studies in other Latin American countries, including Argentina, Brazil, and Colombia, have also reported a higher molecular prevalence of B. bigemina than B. bovis [8,30,31]. The presence of two pathogenic Babesia species indicates poor tick management procedures in the surveyed farms. Although diminazene aceturate and imidocarb dipropionate have been used to treat animal babesiosis, several studies have suggested the possible development of diminazene aceturate resistance in Babesia parasites [32,33]. Therefore, proper tick control programs as well as the appropriate use of chemotherapeutic agents are necessary.
Anaplasma infection in cattle is mainly caused by A. marginale, A. centrale, A. phagocytophilum, and A. bovis. Other Anaplasma species of unknown pathogenicity, such as A. platys-like and Anaplasma capra, have also been detected in cattle [16,19,34,35]. In the present study, A. marginale, a pathogenic species, was detected. In addition, A. platys-like infection in cattle in Bolivia was first reported in this study. Although A. platys is primarily infectious and pathogenic to dogs, there is also evidence that A. platys-like infects ruminants, including cattle in Algeria, Brazil, and Egypt, goats in Tunisia, camels in Egypt, water buffalo in Thailand, sheep in Tunisia, and red deer in China [18,19,20,26,36,37,38,39]. The sequences of A. platys-like detected in this study clustered with those detected in R. microplus in China, cattle in Egypt, and water buffalo in Thailand, and formed a distinct cluster from those reported in dogs and brown dog ticks (Figure 3 and Figure 4). These results support the geographically widespread distribution of A. platys-like infection in ruminants. Anaplasma platys-like strains were found to infect ruminant neutrophils instead of platelets [20]. Although the pathogenicity of these A. platys-like strains is unknown, a study from Brazil reported that the cattle infected with A. platys-like did not show any clinical signs, including anaemia [20]. Nonetheless, further studies are needed to investigate their interaction with host ticks and other microorganisms, including pathogenic Anaplasma.
This study employed cattle reared on farms with different tick control measures. In private farms, external pour-on antiparasitics (ACIENDEL PLUS, Biogénesis Bagó, Casa Matriz, Argentina) and injection (Dectomax® Injectable solution, Zoetis, NJ, USA) were used for tick prevention twice a year (March and September). In contrast, in the university farms, acaricide (ECTOSULES 6% SPILLED, Laboratorio Microsules, Uruguay) was used for tick prevention twice a year (February and August). In recent years, acaricide resistance in ticks has been a remarkable problem worldwide, and resistance to almost all chemicals has been observed in R. microplus [40,41]. The difference in tick control measures may explain the higher infection rate of TBDs in university farms than in private farms. Additionally, the cattle breeds differed among farms. Of note, University Farm 1, where the Holsteins were kept, had the highest infection rate (Table 2). Several studies have shown that genetic factors are associated with resistance to tick infestations and babesiosis, with Bos taurus indicus (e.g., Nelore) being more resistant than Bos taurus taurus (e.g., Holstein) [42,43]. In fact, B. taurus indicus is less likely to be infected with tick-borne pathogens because it shows tick resistance [44]. In Brazil, Angus cattle show a significantly higher rate of Babesia infection than that of Nelore cattle [42,44]. Another possible explanation for the high infection rate in Holstein cattle is poor adaptation to the climate in Bolivia. Holstein is a cold-hardy European breed that may be less resistant to the tropical climate of South America. In fact, although the health status of cattle was not evaluated in the present study, Holstein cattle showed a higher frequency of clinical signs, such as loss of appetite and decrease in body weight, than that of Nelore cattle (data not shown). In Bolivia, a variety of hybrids are bred locally, and these may be highly adaptable to the Bolivian environment.
In this study, most cattle infected with Babesia were infected with A. platys-like. Co-infection with Babesia and Anaplasma is a common finding in cattle: A. marginale and B. bigemina, and Anaplasma sp. and B. bigemina have been simultaneously detected in the same individuals in Egypt and Ethiopia, respectively [45,46]. The effect of mixed infections on clinical outcomes needs to be evaluated in the future.
In the past two serological studies conducted in Bolivia, three tick-borne pathogens, namely B. bovis, B. bigemina, and A. marginale, were detected from cattle, and relatively high seroprevalence was observed: B. bovis (66.1% and 64.2%), B. bigemina (32.1% and 46.3%), and A. marginale (20.5% and 38.8%) [22,23]. Though this study also detected the same pathogens, overall infection rates were lower than those observed in the previous serological studies. PCR-based assays detect pathogens’ DNA, and the results depend on the presence of the target DNA in the tested blood. In contrast, serological assays detect the antibodies resulting from immune response to the infection and thus do not show infection status at the time of sampling. Although this difference hinders a direct comparison between studies, the results correctively indicate that these pathogens have been endemic for at least decades in Bolivia. There is a need to conduct a longitudinal survey to monitor the prevalence of TBDs in Bolivia.
One of the biggest limitations of this study is that blood samples were collected from a limited number of cattle. The variation in sample size among cattle breeds makes it difficult to conduct a statistical analysis of the effect of breed on TBDs. Additionally, the occurrence of TBDs is attributed to interactions between arthropod vectors and hosts, and these interactions are influenced by climatic factors, such as temperature, relative humidity, and precipitation [47]. Moreover, the samples were only collected from geographically limited areas, which may have masked other TBDs prevalent in Bolivia. Thus, this cross-sectional survey may have underestimated the TBD status in cattle. Further studies employing larger sample sizes from geographically diverse locations are needed to evaluate the overall significance of TBDs in the cattle industry in Bolivia. It is also of great importance to conduct a survey on ticks, including R. microplus, which has been observed on the body surface of the cattle examined in this study (data not shown), since the presence of Babesia and Anaplasma has been detected from this tick species in other countries [11,12,14].
5. Conclusions
This is the first report to provide molecular evidence for four tick-borne pathogens, namely B. bovis, B. bigemina, A. marginale, and A. platys-like, in cattle in Bolivia. The results of this study provide information about the prevalence of TBDs as well as updated molecular data for the studied areas. This information is important for understanding the epidemiology of TBDs and interactions among pathogens and is expected to help in formulating appropriate and improved control strategies for pathogens in the area, consequently reducing losses in the cattle industry. Since A. platys-like infection in cattle is insufficiently understood, further efforts are needed to clarify their pathogenicity in animals and their interaction with other pathogens in ticks. Longitudinal sampling of cattle from wider geographic origins is needed to obtain a comprehensive view of the prevalence and epidemiological consequences of TBDs in Bolivia.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/vetsci8090188/s1, Table S1: Results of PCR and Sanger sequencing of Anaplasmataceae-positive samples. Table S2: List of samples co-infected with Babesia and Anaplasma. Figure S1: Agarose gel electrophoresis of (a) BTH PCR products, (b) EHR PCR products, (c) gltA PCR (primer set 1) products, and (d) groEL PCR (primer set 1) products.
Author Contributions
Conceptualisation, S.O.; Data curation, Y.O., H.S., and N.N.; Formal analysis, K.M., Y.O., and H.S.; Investigation, S.O., J.A.C.P., L.V.A.J., and H.P.G.C.; Methodology, K.M., F.K., and R.N.; Project administration, J.A.C.P. and R.N.; Funding acquisition, R.N.; Resources, J.A.C.P., L.V.A.J., H.P.G.C., and F.K.; Software, K.M.; Supervision, R.N.; Visualization, S.O.; Writing—original draft, S.O.; Writing—review and editing, J.A.C.P., L.V.A.J., H.P.G.C., K.M., Y.O., H.S., F.K., N.N., and R.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI (16H06431, 19H03118, 20K21358, and 20KK0151).
Institutional Review Board Statement
This study was approved by the Institutional Committee for the Care and Use of Experimental Animals at Autonomous University Gabriel Rene Moreno (CICUAE, 2015, No. 008/19).
Informed Consent Statement
Informed consent was sought from the owners of animals. Blood samples were only collected when farmers agreed to have their cattle sampled.
Data Availability Statement
Babesia and Anaplasma sequences obtained in this study were submitted to the DNA Data Bank of Japan (DDBJ) under the following accession numbers (18S rDNA sequences of B. bigemina, LC645216-LC645223; 18S rDNA sequence of B. bovis, LC645224; gltA gene sequences of A. platys-like, LC645225-LC645237; gltA gene sequence of A. marginale: LC645238; groEL gene sequence of A. platys-like, LC645239-LC645260).
Acknowledgments
We would like to thank all collaborators who supported sample collection in Bolivia. We would like to show our greatest appreciation to TOBITATE! Young Ambassador Program, MEXT, Japan.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Williams, G.W.; Anderson, D.P. The Latin American Livestock Industry: Growth and Challenges. Choices 2020, 34, 1–11. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Livestock Production in Latin America and the Caribbean. 2017. Available online: http://www.fao.org/americas/priorities/produccion-pecuaria/en/ (accessed on 2 September 2021).
- Grisi, L.; Leite, R.C.; Martins, J.R.D.S.; De Barros, A.T.M.; Andreotti, R.; Cançado, P.H.D.; De León, A.A.P.; Pereira, J.B.; Villela, H.S. Reassessment of the potential economic impact of cattle parasites in Brazil. Bras. Parasitol. Vet. 2014, 23, 150–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, L.L.; Canever, M.F.; Cardozo, L.L.; Cardozo, C.P.; Herkenhoff, M.E.; Neto, A.T.; Vogel, C.I.G.; Miletti, L.C. Prevalence of Anaplasma marginale, Babesia bovis, and Babesia bigemina in cattle in the Campos de Lages region, Santa Catarina state, Brazil, estimated by multiplex-PCR. Parasite Epidemiol. Control 2019, 6, e00114. [Google Scholar] [CrossRef] [PubMed]
- Parodi, P.; Corbellini, L.G.; Leotti, V.B.; Rivero, R.; Miraballes, C.; Correa, F.R.; Venzal, J.M.; Fernández, M.T.A. Validation of a multiplex PCR assay to detect Babesia spp. and Anaplasma marginale in cattle in Uruguay in the absence of a gold standard test. J. Vet. Diagn. Investig. 2021, 33, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Obregón, D.; Cruz, A.C.; Armas, Y.; Silva, J.B.; Fonseca, A.H.; André, M.R.; Alfonso, P.; Oliveira, M.C.S.; Machado, R.Z.; González, B.C. High co-infection rates of Babesia bovis, Babesia bigemina, and Anaplasma marginale in water buffalo in Western Cuba. Parasitol. Res. 2019, 118, 955–967. [Google Scholar] [CrossRef]
- Dueñez, J.J.; Chávez, O.T.; Rocha, A.H.; Castaño, A.T.; Jaramillo, A.M.M. Molecular surveillance and phylogenetic traits of Babesia bigemina and Babesia bovis in cattle (Bos taurus) and water buffaloes (Bubalus bubalis) from Colombia. Parasites Vectors 2018, 11, 510. [Google Scholar] [CrossRef]
- Paoletta, M.S.; Arias, L.L.; Fournière, S.; Guillemi, E.C.; Luciani, C.; Sarmiento, N.F.; Mosqueda, J.; Farber, M.D.; Wilkowsky, S.E. Epidemiology of Babesia, Anaplasma and Trypanosoma species using a new expanded reverse line blot hybridization assay. Ticks Tick Borne Dis. 2018, 9, 155–163. [Google Scholar] [CrossRef]
- Hakimi, H.; Yamagishi, J.; Kegawa, Y.; Kaneko, O.; Kawazu, S.; Asada, S. Establishment of transient and stable transfection systems for Babesia ovata. Parasites Vectors 2016, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Terkawi, M.A.; Thekisoe, O.M.M.; Katsande, C.; Latif, A.A.; Mans, B.J.; Matthee, O.; Mkize, N.; Mabogoane, N.; Marais, F.; Yokoyama, N.; et al. Serological survey of Babesia bovis and Babesia bigemina in cattle in South Africa. Vet. Parasitol. 2011, 182, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Carter, P.D. Babesiosis. In MSD Veterinary Manual; Merck & Co. Inc.: Kenilworth, NJ, USA, 2015; Available online: https://www.msdvetmanual.com/circulatory-system/blood-parasites/babesiosis (accessed on 2 September 2021).
- Aubry, P.; Geale, D.W. A review of Bovine anaplasmosis. Transbound. Emerg. Dis. 2011, 58, 1–30. [Google Scholar] [CrossRef]
- Belkahia, H.; Said, M.B.; Alberti, A.; Abdi, K.; Issaoui, Z.; Hattab, D.; Gharbi, M.; Messadi, L. First molecular survey and novel genetic variants’ identification of Anaplasma marginale, A. centrale and A. bovis in cattle from Tunisia. Infect. Genet. Evol. 2015, 34, 361–371. [Google Scholar] [CrossRef]
- Kocan, K.M.; Fuente, J.; Guglielmone, A.A.; Meléndez, R.D. Antigens and Alternatives for Control of Anaplasma marginale Infection in Cattle. Clin. Microbiol. Rev. 2003, 16, 698–712. [Google Scholar] [CrossRef] [Green Version]
- M’Ghirbi, Y.; Bèji, M.; Oporto, B.; Khrouf, F.; Hurtado, A.; Bouattour, A. Anaplasma marginale and A. phagocytophilum in cattle in Tunisia. Parasites Vectors 2016, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.P.; Zhang, B.; Wang, Y.H.; Xu, G.; Wang, X.; Ni, X.; Zhou, E.M. Molecular identification and characterization of Anaplasma capra and Anaplasma platys-like in Rhipicephalus microplus in Ankang, Northwest China. BMC Infect. Dis. 2019, 19, 434. [Google Scholar] [CrossRef] [PubMed]
- Zobba, R.; Anfossi, A.G.; Parpaglia, M.L.P.; Dore, G.M.; Chessa, B.; Spezzigu, A.; Rocca, S.; Visco, S.; Pittau, M.; Alberti, A. Molecular investigation and phylogeny of Anaplasma spp. in mediterranean ruminants reveal the presence of neutrophil-tropic strains closely related to A. platys. Appl. Environ. Microbiol. 2014, 80, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, W.M.A.; Ali, A.O.; Mahmoud, H.Y.A.H.; Omar, M.A.; Chatanga, E.; Salim, B.; Naguib, D.; Anders, J.L.; Nonaka, N.; Moustafa, M.A.M.; et al. Exploring Prokaryotic and Eukaryotic Microbiomes Helps in Detecting Tick-Borne Infectious Agents in the Blood of Camels. Pathogens 2021, 10, 351. [Google Scholar] [CrossRef]
- Said, M.B.; Belkahia, H.; El Mabrouk, N.; Saidani, M.; Alberti, A.; Zobba, R.; Cherif, A.; Mahjoub, T.; Bouattour, A.; Messadi, L. Anaplasma platys-like strains in ruminants from Tunisia. Infect. Genet. Evol. 2017, 49, 226–233. [Google Scholar] [CrossRef]
- André, M.R.; Calchi, A.C.; Herrera, H.M.; Zanatto, D.C.S.; Horta, B.C.L.S.; Tasso, J.B.; Ramos, I.S.A.; Mello, V.V.C.; Machado, R.Z. The co-infection with Ehrlichia minasensis, Anaplasma marginale and Anaplasma platys is not associated with anemia in beef cattle in the Brazilian Pantanal. Vet. Parasitol. Reg. Stud. Rep. 2020, 21, 100437. [Google Scholar] [CrossRef]
- Ray, R.; Albright, Z.C.; Wang, K. China-Latin America Economic Bulletin, 2021; Global Development Policy Center: Boston, MA, USA, 2021; Available online: https://www.bu.edu/gdp/2021/02/22/china-latin-america-economic-bulletin-2021/ (accessed on 2 September 2021).
- Mas, J.J.C.; Widdowson, M.A.; Cuéllar, A.M.; Ribera, H.; Walker, A.R. Risk of babesiosis and anaplasmosis in different ecological zones of Santa Cruz Department, Bolivia. Vet. Parasitol. 2000, 93, 29–38. [Google Scholar] [CrossRef]
- Carrique, J.J.; Morales, G.J.; Edelsten, M. Endemic Instability for Babesiosis and Anaplasmosis in Cattle in the Bolivian Chaco. Vet. J. 2000, 160, 162–164. [Google Scholar] [CrossRef]
- Masatani, T.; Hayashi, K.; Andoh, M.; Tateno, M.; Endo, Y.; Asada, M.; Kusakisako, K.; Tanaka, T.; Gokuden, M.; Hozumi, N.; et al. Detection and molecular characterization of Babesia, Theileria, and Hepatozoon species in hard ticks collected from Kagoshima, the southern region in Japan. Ticks Tick Borne Dis. 2017, 8, 581–587. [Google Scholar] [CrossRef]
- Chatanga, E.; Hayashida, K.; Muleya, W.; Kusakisako, K.; Moustafa, M.A.M.; Salim, B.; Katakura, K.; Sugimoto, C.; Nonaka, N.; Nakao, R. Genetic diversity and sequence polymorphism of two genes encoding Theileria parva antigens recognized by CD8+ T cells among vaccinated and unvaccinated cattle in Malawi. Pathogens 2020, 9, 334. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Roux, V.; Camicas, J.L.; Baradji, I.; Brouqui, P.; Raoult, D. Detection of ehrlichiae in African ticks by polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 707–708. [Google Scholar] [CrossRef]
- Qiu, Y.; Kaneko, C.; Kajihara, M.; Ngonda, S.; Simulundu, E.; Muleya, W.; Thu, M.J.; Hang’ombe, M.B.; Katakura, K.; Takada, A.; et al. Tick-borne haemoparasites and Anaplasmataceae in domestic dogs in Zambia. Ticks Tick Borne Dis. 2018, 9, 988–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adenyo, C.; Ohya, K.; Qiu, Y.; Takashima, Y.; Ogawa, H.; Matsumoto, T.; Thu, M.J.; Sato, K.; Kawabata, H.; Katayama, Y.; et al. Bacterial and protozoan pathogens/symbionts in ticks infecting wild grasscutters (Thryonomys swinderianus) in Ghana. Acta Trop. 2020, 205, 105388. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Dueñez, J.J.; Chávez, O.T.; Jaramillo, A.M.M. Parasitological and molecular surveys reveal high rates of infection with vector-borne pathogens and clinical anemia signs associated with infection in cattle from two important livestock areas in Colombia. Ticks Tick Borne Dis. 2017, 8, 290–299. [Google Scholar] [CrossRef]
- Amorim, L.S.; Wenceslau, A.A.; Carvalho, F.S.; Carneiro, P.L.S.; Albuquerque, G. Bovine babesiosis and anaplasmosis complex: Diagnosis and evaluation of the risk factors from Bahia, Brazil. Rev. Bras. Parasitol. Veterinária 2014, 23, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.J.; Yamasaki, M.; Nakamura, K.; Sasaki, N.; Murakami, M.; Kumara, B.; Rajapakshage, W.; Ohta, H.; Maede, Y.; Takiguchi, M. Development and characterization of a strain of Babesia gibsoni resistant to diminazene aceturate in vitro. J. Vet. Med. Sci. 2010, 72, 765–771. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, M.; Watanabe, N.; Idaka, N.; Yamamori, T.; Otsuguro, K.; Uchida, N.; Iguchi, A.; Ohta, H.; Takiguchi, M. Intracellular diminazene aceturate content and adenosine incorporation in diminazene aceturate-resistant Babesia gibsoni isolate in vitro. Exp. Parasitol. 2017, 183, 92–98. [Google Scholar] [CrossRef]
- Seo, M.G.; Kwon, O.D.; Kwak, D. Genotypic analysis of piroplasms and associated pathogens from ticks infesting cattle in Korea. Microorganisms 2020, 8, 728. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.G.; Ouh, I.O.; Lee, H.; Geraldino, P.J.L.; Rhee, M.H.; Kwon, O.D.; Kwak, D. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet. Microbiol. 2018, 226, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Tumwebaze, M.A.; Lee, S.-H.; Moumouni, P.F.A.; Mohammed-Geba, K.; Sheir, S.K.; Galal-Khallaf, A.; El Latif, H.M.A.; Morsi, D.S.; Bishr, N.M.; Galon, E.M.; et al. Parasitology International First detection of Anaplasma ovis in sheep and Anaplasma platys-like variants from cattle in Menoufia governorate, Egypt. Parasitol. Int. 2020, 78, 102150. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.H.L.; Tiawsirisup, S.; Kaewthamasorn, M. Molecular detection and genetic characterization of Anaplasma marginale and Anaplasma platys-like (Rickettsiales: Anaplasmataceae) in water buffalo from eight provinces of Thailand. BMC Vet. Res. 2020, 16, 380. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, M.; Davoust, B.; Seghir, M.; Fenollar, F.; Raoult, D.; Mediannikov, O. Comparative Immunology, Microbiology and Infectious Diseases Development of a new PCR-based assay to detect Anaplasmataceae and the first report of Anaplasma phagocytophilum and Anaplasma platys in cattle from Algeria. Comp. Immunol. Microbiol. Infect. Dis. 2015, 39, 39–45. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Liu, Z.; Liu, J.; Yang, J.; Li, Q.; Li, Y.; Luo, J.; Yin, H. Molecular Survey of Anaplasma and Ehrlichia of Red Deer and Sika Deer in Gansu, China in 2013. Transbound. Emerg. Dis. 2016, 63, e228–e236. [Google Scholar] [CrossRef] [PubMed]
- Janer, E.C.; Rifran, L.; Piaggio, J.; Gil, A.; Miller, R.J.; Schumaker, T.T.S. In vitro tests to establish LC 50 and discriminating concentrations for fipronil against Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) and their standardization. Vet. Parasitol. 2009, 162, 120–128. [Google Scholar] [CrossRef]
- Bandara, K.M.U.J.; Karunaratne, S.H.P.P. Mechanisms of acaricide resistance in the cattle tick Rhipicephalus (Boophilus) microplus in Sri Lanka. Pestic. Biochem. Physiol. 2017, 139, 68–72. [Google Scholar] [CrossRef]
- Piper, E.K.; Jackson, L.A.; Ohmann, H.B.; Gondro, C.; Tabor, A.E.L.; Jonsson, N.N. Tick-susceptible Bos taurus cattle display an increased cellular response at the site of larval Rhipicephalus (Boophilus) microplus attachment, compared with tick-resistant Bos indicus cattle. Int. J. Parasitol. 2010, 40, 431–441. [Google Scholar] [CrossRef]
- Giglioti, R.; Oliveira, H.N.; Santana, C.H.; Ibelli, A.M.G.; Néo, T.A.; Bilhassi, T.B.; Rabelo, M.D.; Machado, R.Z.; Brito, L.G.; Oliveira, M.C.S. Babesia bovis and Babesia bigemina infection levels estimated by qPCR in Angus cattle from an endemic area of São Paulo state, Brazil. Ticks Tick Borne Dis. 2016, 7, 657–662. [Google Scholar] [CrossRef] [Green Version]
- Guglielmone, A.A. Epidemiology of babesiosis and anaplasmosis in South and Central America. Vet. Parasitol. 1995, 57, 109–119. [Google Scholar] [CrossRef]
- Hosary, A.L.; Vectors, P.; Hosary, A.A.L.; Răileanu, C.; Tauchmann, O.; Fischer, S.; Nijhof, A.M.; Silaghi, C. Epidemiology and genotyping of Anaplasma marginale and co-infection with piroplasms and other Anaplasmataceae in cattle and buffaloes from Egypt. Parasit. Vectors 2020, 13, 495. [Google Scholar] [CrossRef]
- Hailemariam, Z.; Krücken, J.; Baumann, M.; Ahmed, J.S.; Clausen, P.H.; Nijhof, A.M. Molecular detection of tick-borne pathogens in cattle from Southwestern Ethiopia. PLoS ONE 2017, 12, e0188248. [Google Scholar] [CrossRef] [Green Version]
- Ogden, N.H.; Lindsay, L.R. Effects of Climate and Climate Change on Vectors and Vector-Borne Diseases: Ticks Are Different. Trends Parasitol. 2016, 32, 646–656. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).