First Isolation and Molecular Characterization of Pseudorabies Virus in a Hunting Dog in Sicily (Southern Italy)
Abstract
1. Introduction
2. Materials and Methods
2.1. Cases Presentation
2.2. Histopathology
2.3. Immunohistochemistry
2.4. Viral Nucleic Acids Extraction
2.5. Polymerase Chain Reaction (PCR) and Sequencing
2.6. Sequence Analysis
3. Results
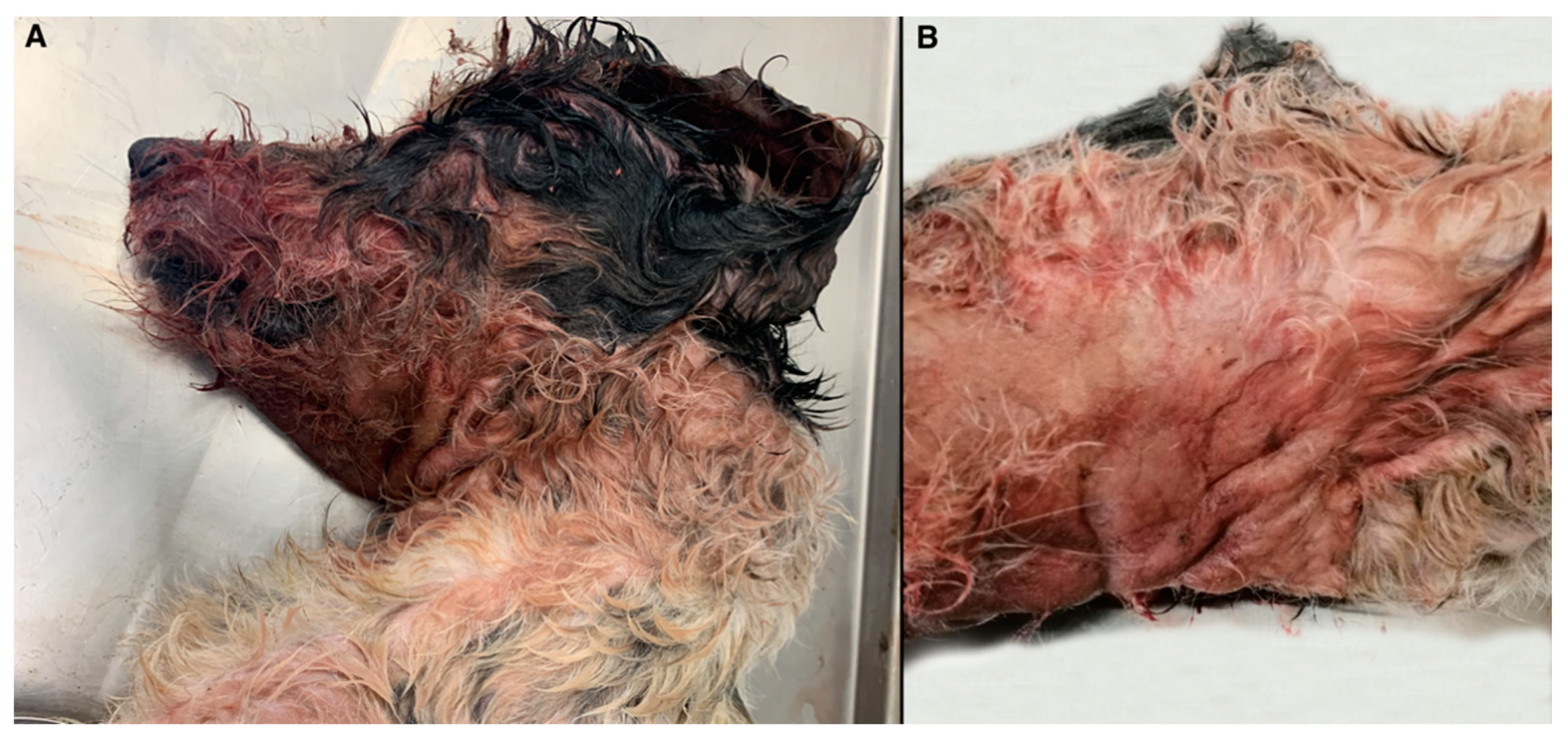
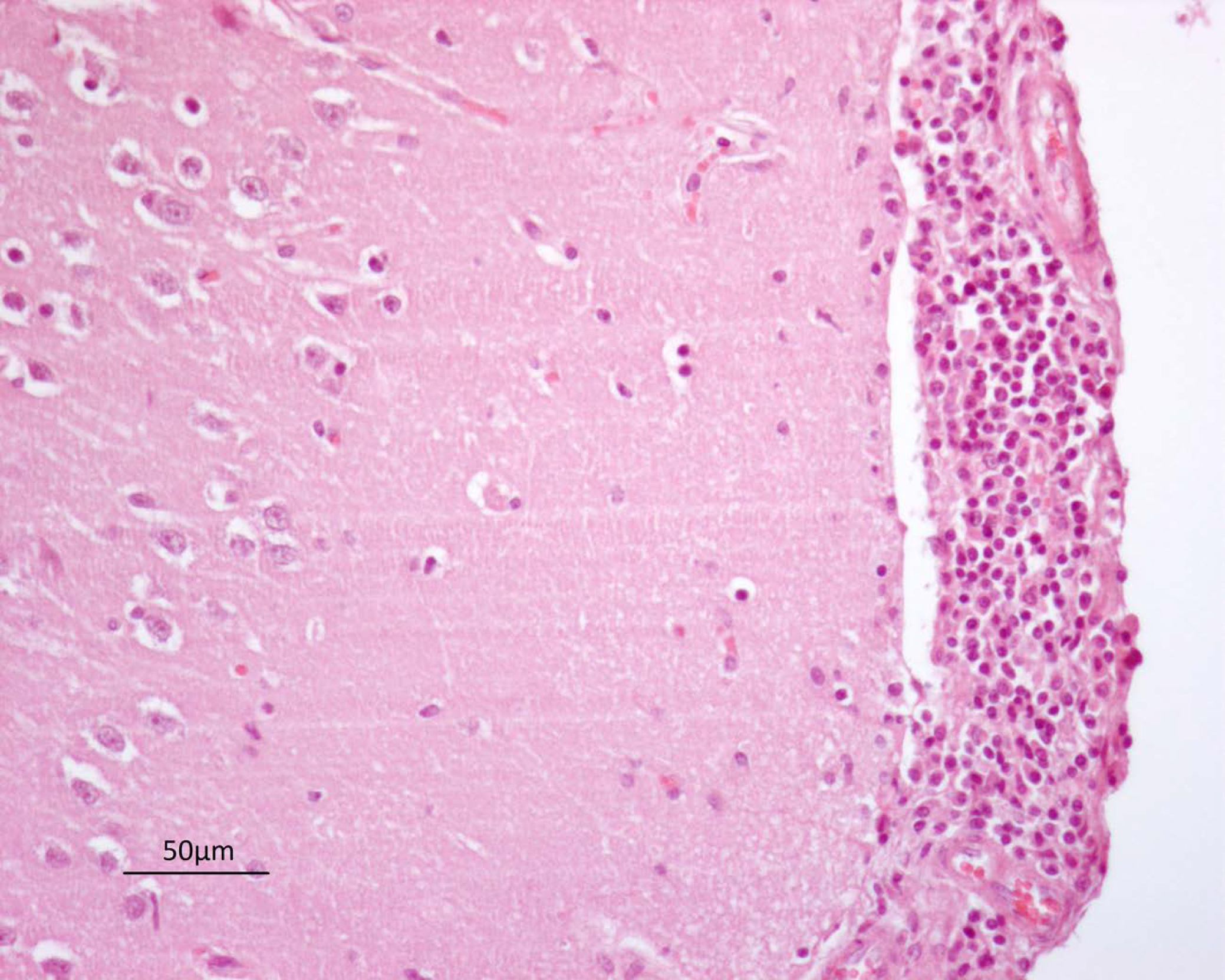
3.1. Gross and Histopathological Findings
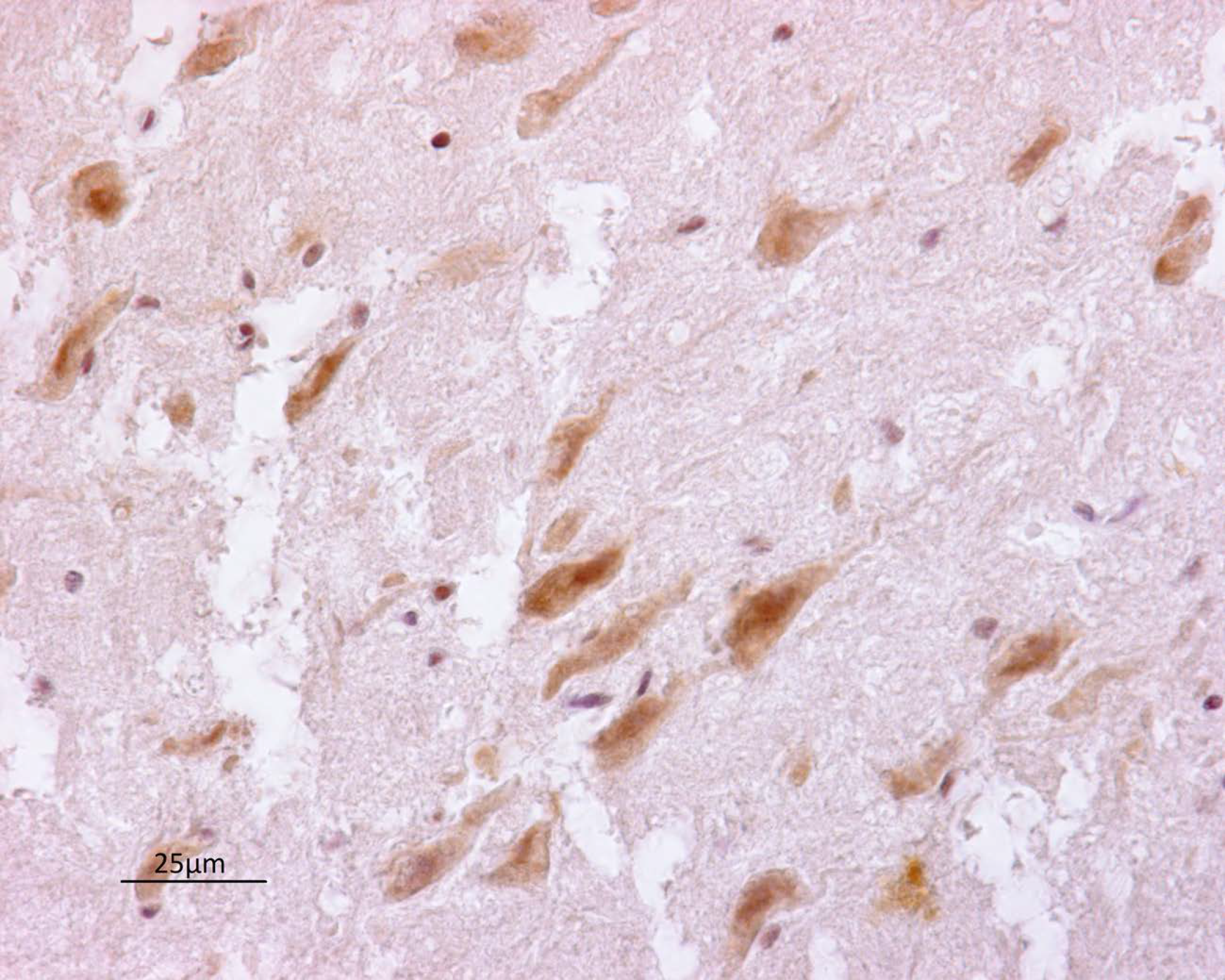
3.2. Immunohistochemistry
3.3. Molecular Identification of Pseudorabies Virus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sozzi, E.; Moreno, A.; Lelli, D.; Cinotti, S.; Alborali, G.L.; Nigrelli, A.; Luppi, A.; Bresaola, M.; Catella, A.; Cordioli, P. Genomic Characterization of Pseudorabies Virus Strains Isolated in I taly. Transbound. Emerg. Dis. 2014, 61, 334–340. [Google Scholar] [CrossRef]
- Yang, X.; Guan, H.; Li, C.; Li, Y.; Wang, S.; Zhao, X.; Zhao, Y.; Liu, Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. 2019, 87, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.-W.; Weng, S.-S.; Cheng, Q.; Cui, P.; Li, Y.-J.; Wu, H.-L.; Zhu, Y.-M.; Xu, B.; Zhang, W.-H. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerg. Infect. Dis. 2018, 24, 1087. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Hahn, E.C.; Tottewitz, F.; Kramer, M.; Klupp, B.G.; Mettenleiter, T.C.; Freuling, C. Pseudorabies virus in wild swine: A global perspective. Arch. Virol. 2011, 156, 1691. [Google Scholar] [CrossRef] [PubMed]
- Mulder, W.A.M.; Pol, J.M.A.; Gruys, E.; Jacobs, L.; De Jong, M.C.M.; Peeters, B.P.H.; Kimman, T.G. Pseudorabies virus infections in pigs. Role of viral proteins in virulence, pathogenesis and transmission. Vet. Res. 1997, 28, 1–17. [Google Scholar] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Chiari, M.; Ferrari, N.; Bertoletti, M.; Avisani, D.; Cerioli, M.; Zanoni, M.; Alborali, L.G.; Lanfranchi, P.; Lelli, D.; Martin, A.M. Long-term surveillance of Aujeszky’s disease in the alpine wild boar (Sus scrofa). Ecohealth 2015, 12, 563–570. [Google Scholar] [CrossRef]
- Moreno, A.; Sozzi, E.; Grilli, G.; Gibelli, L.R.; Gelmetti, D.; Lelli, D.; Chiari, M.; Prati, P.; Alborali, G.L.; Boniotti, M.B. Detection and molecular analysis of Pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet. Microbiol. 2015, 177, 359–365. [Google Scholar] [CrossRef]
- Verin, R.; Varuzza, P.; Mazzei, M.; Poli, A. Serologic, molecular, and pathologic survey of pseudorabies virus infection in hunted wild boars (Sus scrofa) in Italy. J. Wildl. Dis. 2014, 50, 559–565. [Google Scholar] [CrossRef]
- Müller, T.; Klupp, B.G.; Freuling, C.; Hoffmann, B.; Mojcicz, M.; Capua, I.; Palfi, V.; Toma, B.; Lutz, W.; Ruiz-Fon, F. Characterization of pseudorabies virus of wild boar origin from Europe. Epidemiol. Infect. 2010, 138, 1590–1600. [Google Scholar] [CrossRef][Green Version]
- Meng, X.; Lindsay, D.S.; Sriranganathan, N. Wild boars as sources for infectious diseases in livestock and humans. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Fons, F.; Segalés, J.; Gortázar, C. A review of viral diseases of the European wild boar: Effects of population dynamics and reservoir role. Vet. J. 2008, 176, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Conraths, F.J.; Hahn, E.C. Pseudorabies virus infection (Aujeszky’s disease) in wild swine. Infect. Dis. Rev. 2000, 2, 27–34. [Google Scholar]
- Capua, I.; Fico, R.; Banks, M.; Tamba, M.; Calzetta, G. Isolation and characterisation of an Aujeszky’s disease virus naturally infecting a wild boar (Sus scrofa). Vet. Microbiol. 1997, 55, 141–146. [Google Scholar] [CrossRef]
- Müller, T.F.; Teuffert, J.; Zellmer, R.; Conraths, F.J. Experimental infection of European wild boars and domestic pigs with pseudorabies viruses with differing virulence. Am. J. Vet. Res. 2001, 62, 252–258. [Google Scholar] [CrossRef]
- Steinrigl, A.; Revilla-Fernandez, S.; Kolodziejek, J.; Wodak, E.; Bago, Z.; Nowotny, N.; Schmoll, F.; Köfer, J. Detection and molecular characterization of Suid herpesvirus type 1 in Austrian wild boar and hunting dogs. Vet. Microbiol. 2012, 157, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.G.; Di Concilio, D.; D’Alessio, N.; Veneziano, V.; Galiero, G.; Fusco, G. Canine parvovirus and pseudorabies virus coinfection as a cause of death in a wolf (Canis lupus). Vet. Med. Sci. 2020, 6, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Cano-Terriza, D.; Martínez, R.; Moreno, A.; Pérez-Marín, J.E.; Jiménez-Ruiz, S.; Paniagua, J.; Borge, C.; García-Bocanegra, I. Survey of Aujeszky’s disease virus in hunting dogs from Spain. Ecohealth 2019, 16, 351–355. [Google Scholar] [CrossRef]
- Serena, M.S.; Metz, G.E.; Lozada, M.I.; Aspitia, C.G.; Nicolino, E.H.; Pidone, C.L.; Fossaroli, M.; Balsalobre, A.; Quiroga, M.A.; Echeverria, M.G. First isolation and molecular characterization of Suid herpesvirus type 1 from a domestic dog in Argentina. Open Vet. J. 2018, 8, 131–139. [Google Scholar] [CrossRef]
- Cramer, S.D.; Campbell, G.A.; Njaa, B.L.; Morgan, S.E.; Smith, S.K.; McLin, W.R., IV; Brodersen, B.W.; Wise, A.G.; Scherba, G.; Langohr, I.M. Pseudorabies virus infection in Oklahoma hunting dogs. J. Vet. diagnostic Investig. 2011, 23, 915–923. [Google Scholar] [CrossRef]
- Minamiguchi, K.; Kojima, S.; Sakumoto, K.; Kirisawa, R. Isolation and molecular characterization of a variant of Chinese gC-genotype II pseudorabies virus from a hunting dog infected by biting a wild boar in Japan and its pathogenicity in a mouse model. Virus Genes 2019, 55, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Chiapponi, C.; Sozzi, E.; Morelli, A.; Silenzi, V.; Gobbi, M.; Lavazza, A.; Paniccià, M. Detection of a gE-deleted Pseudorabies virus strain in an Italian red fox. Vet. Microbiol. 2020, 244, 108666. [Google Scholar] [CrossRef]
- Kong, H.; Zhang, K.; Liu, Y.; Shang, Y.; Wu, B.; Liu, X. Attenuated live vaccine (Bartha-K16) caused pseudorabies (Aujeszky’s disease) in sheep. Vet. Res. Commun. 2013, 37, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Shao, Y.; Tan, C.; Shen, Y.; Zhang, X.; Xiao, J.; Wu, Y.; He, L.; Shao, G.; Han, M. Commercial vaccine against pseudorabies virus: A hidden health risk for dogs. Vet. Microbiol. 2019, 233, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Monroe, W.E. Clinical signs associated with pseudorabies in dogs. J. Am. Vet. Med. Assoc. 1989, 195, 599–602. [Google Scholar]
- Yoon, H.A.; Eo, S.K.; Aleyas, A.G.; Park, S.O.; Lee, J.H.; Chae, J.S.; Cho, J.G.; Song, H.J. Molecular survey of latent pseudorabies virus infection in nervous tissues of slaughtered pigs by nested and real-time PCR. J. Microbiol. 2005, 43, 430–436. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Lari, A.; Lorenzi, D.; Nigrelli, D.; Brocchi, E.; Faccini, S.; Poli, A. Pseudorabies virus in European wild boar from central Italy. J. Wildl. Dis. 2006, 42, 319–324. [Google Scholar] [CrossRef]
- Deblanc, C.; Oger, A.; Simon, G.; Le Potier, M.-F. Genetic Diversity among Pseudorabies Viruses Isolated from Dogs in France from 2006 to 2018. Pathogens 2019, 8, 266. [Google Scholar] [CrossRef]
- Abbate, J.M.; Arfuso, F.; Iaria, C.; Arestia, G.; Lanteri, G. Prevalence of Bovine Tuberculosis in Slaughtered Cattle in Sicily, Southern Italy. Animals 2020, 10, 1473. [Google Scholar] [CrossRef]
- Ciarello, F.P.; Capucchio, M.T.; Ippolito, D.; Colombino, E.; Gibelli, L.R.M.; Fiasconaro, M.; Moreno Martin, A.M.; Di Marco Lo Presti, V. First report of a severe outbreak of Aujeszky’s disease in cattle in Sicily (Italy). Pathogens 2020, 9, 954. [Google Scholar] [CrossRef] [PubMed]
- Di Marco Lo Presti, V.; Moreno, A.; Castelli, A.; Ippolito, D.; Aliberti, A.; Amato, B.; Vitale, M.; Fiasconaro, M.; Pruiti Ciarello, F. Retrieving Historical Cases of Aujeszky’s Disease in Sicily (Italy): Report of a Natural Outbreak Affecting Sheep, Goats, Dogs, Cats and Foxes and Considerations on Critical Issues and Perspectives in Light of the Recent EU Regulation 429/2016. Pathogens 2021, 10, 1301. [Google Scholar] [CrossRef]
- Thompson, K. Bones and joints. Jubb Kennedy Palmers Pathol. Domest. Anim. 2007, 1, 1–184. [Google Scholar]
- Zhang, L.; Zhong, C.; Wang, J.; Lu, Z.; Liu, L.; Yang, W.; Lyu, Y. Pathogenesis of natural and experimental Pseudorabies virus infections in dogs. Virol. J. 2015, 12, 1–10. [Google Scholar] [CrossRef]
- Kotnik, T.; Suhadolc, S.; Juntes, P.; Gombač, M.; Toplak, I.; Hostnik, P.; Malovrh, T.; Barlič-Maganja, D.; Grom, J. Case report of a pseudorabies (Aujeszky’s disease) in a bitch. Slov. Vet. Res 2006, 43, 143–145. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbate, J.M.; Giannetto, A.; Iaria, C.; Riolo, K.; Marruchella, G.; Hattab, J.; Calabrò, P.; Lanteri, G. First Isolation and Molecular Characterization of Pseudorabies Virus in a Hunting Dog in Sicily (Southern Italy). Vet. Sci. 2021, 8, 296. https://doi.org/10.3390/vetsci8120296
Abbate JM, Giannetto A, Iaria C, Riolo K, Marruchella G, Hattab J, Calabrò P, Lanteri G. First Isolation and Molecular Characterization of Pseudorabies Virus in a Hunting Dog in Sicily (Southern Italy). Veterinary Sciences. 2021; 8(12):296. https://doi.org/10.3390/vetsci8120296
Chicago/Turabian StyleAbbate, Jessica Maria, Alessia Giannetto, Carmelo Iaria, Kristian Riolo, Giuseppe Marruchella, Jasmine Hattab, Placido Calabrò, and Giovanni Lanteri. 2021. "First Isolation and Molecular Characterization of Pseudorabies Virus in a Hunting Dog in Sicily (Southern Italy)" Veterinary Sciences 8, no. 12: 296. https://doi.org/10.3390/vetsci8120296
APA StyleAbbate, J. M., Giannetto, A., Iaria, C., Riolo, K., Marruchella, G., Hattab, J., Calabrò, P., & Lanteri, G. (2021). First Isolation and Molecular Characterization of Pseudorabies Virus in a Hunting Dog in Sicily (Southern Italy). Veterinary Sciences, 8(12), 296. https://doi.org/10.3390/vetsci8120296








