Comparative Genome Analysis of a Pathogenic Erysipelothrix rhusiopathiae Isolate WH13013 from Pig Reveals Potential Genes Involve in Bacterial Adaptions and Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Bioinformatical Analysis and Comparative Genomics
3. Result and Discussion
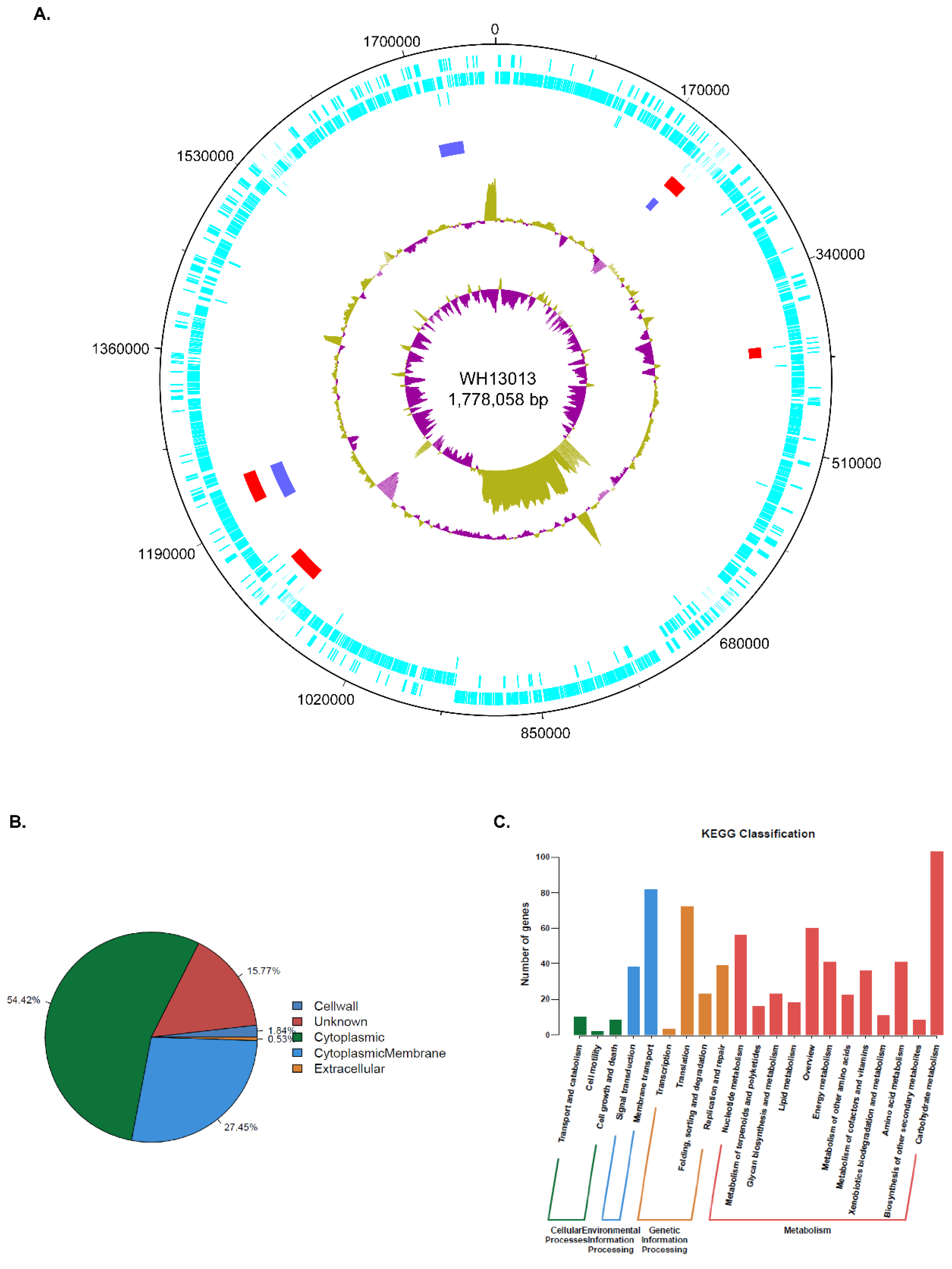
3.1. Overview of WH13013 Complete Genome Sequence
3.2. Phylogenetic Analysis of WH1303
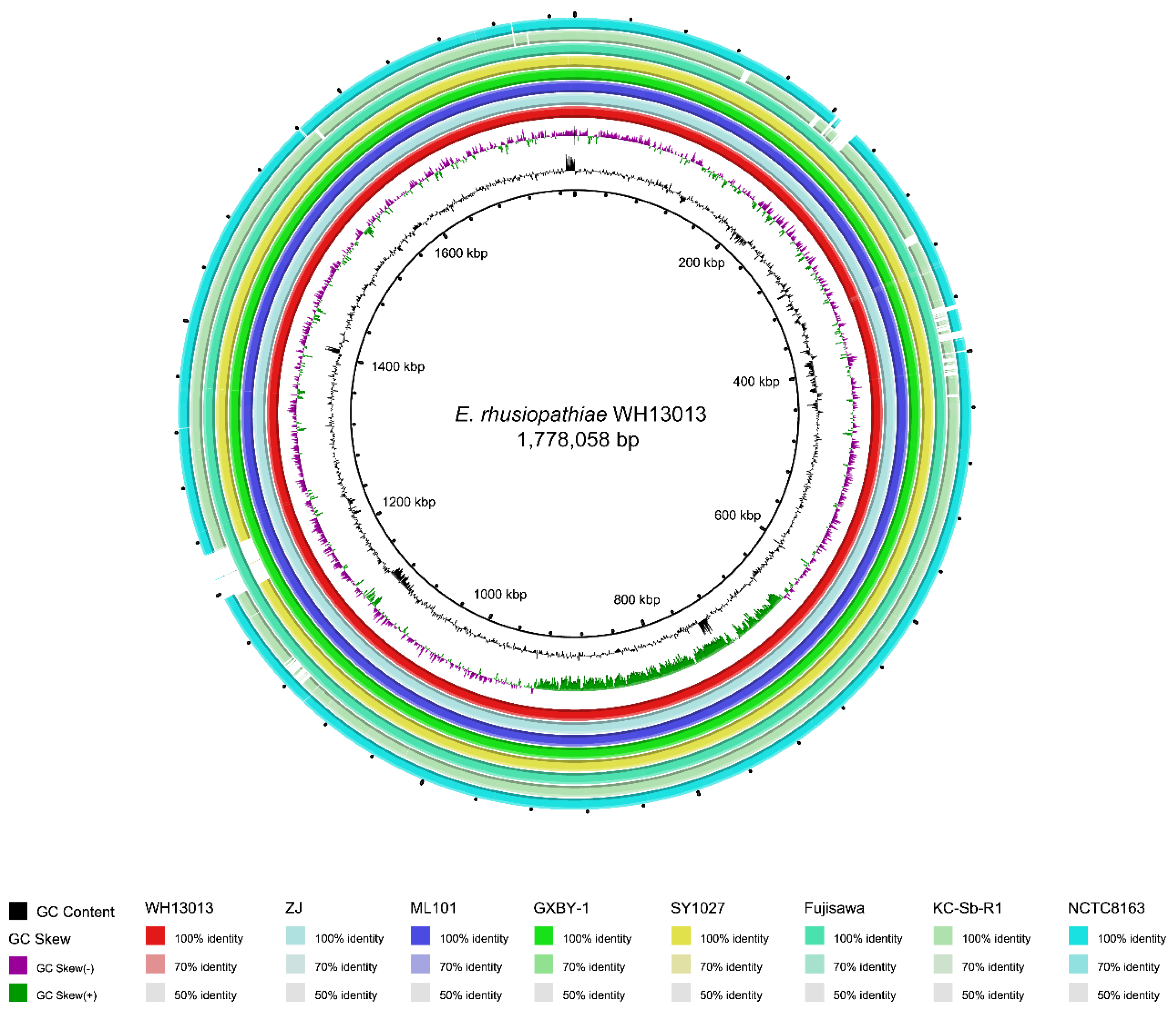
3.3. Comparative Genome Sequence Analysis of the WH13013 Genome
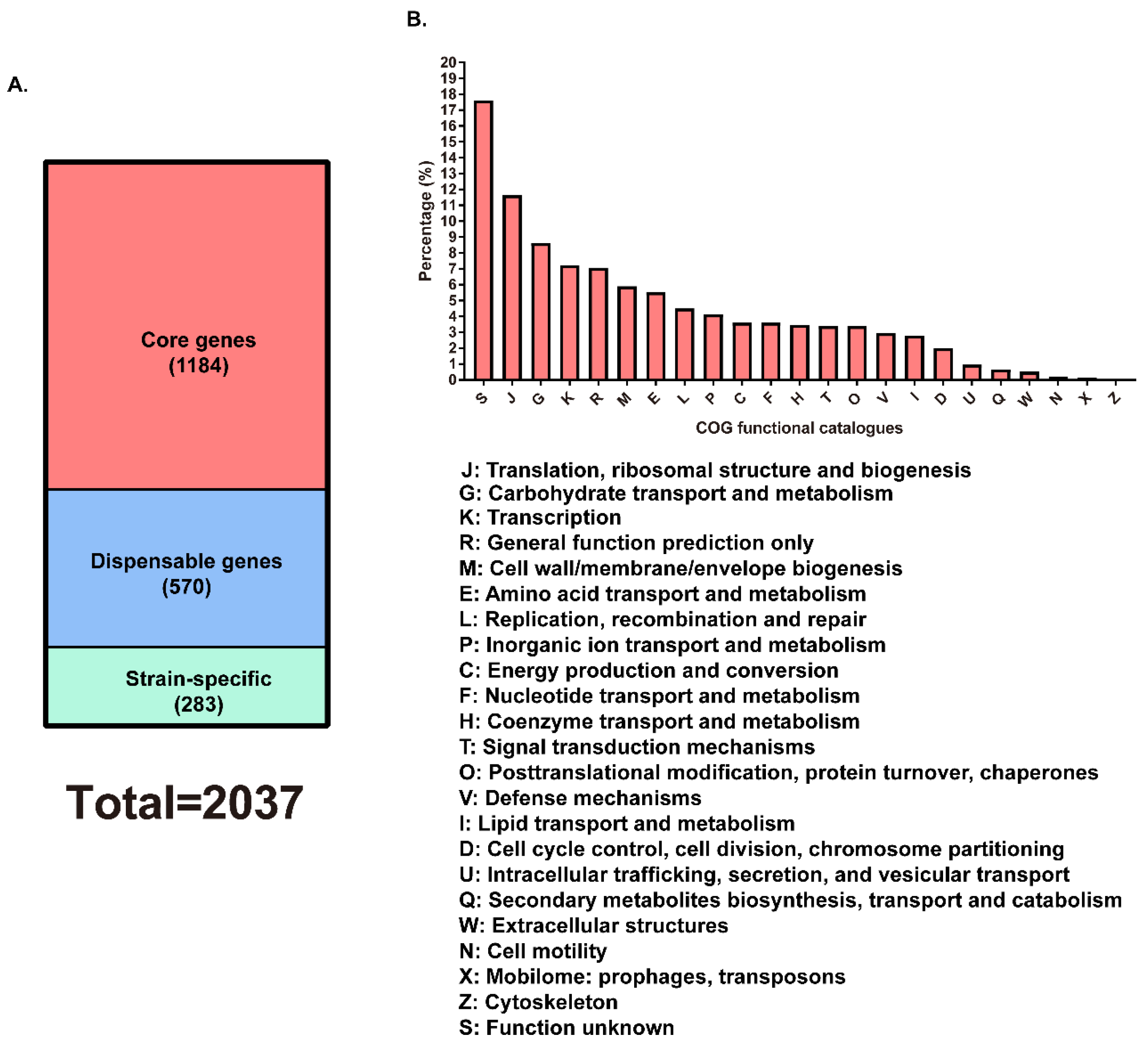
3.4. Determination of Core and Dispensable Genes
3.5. Determination of Antibiotic Resistance Genes
3.6. Determination of Virulence Factors Coding Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
References
- Wang, Q.; Chang, B.J.; Riley, T.V. Erysipelothrix rhusiopathiae. Vet. Microbiol. 2010, 140, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, D.; Zhang, J.; Yang, L.; Wang, X.; Chen, H.; Tan, C. Virulence determinants, antimicrobial susceptibility, and molecular profiles of Erysipelothrix rhusiopathiae strains isolated from China. Emerg. Microbes Infect. 2015, 4, e69. [Google Scholar] [CrossRef] [PubMed]
- Kwok, A.H.; Li, Y.; Jiang, J.; Jiang, P.; Leung, F.C. Complete genome assembly and characterization of an outbreak strain of the causative agent of swine erysipelas--Erysipelothrix rhusiopathiae SY1027. BMC Microbiol. 2014, 14, 176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Zhang, A.; Zhu, W.; Zhang, Q.; Xu, Z.; Yan, S.; Sun, X.; Chen, H.; Jin, M. iTRAQ-based quantitative proteomic analysis reveals potential virulence factors of Erysipelothrix rhusiopathiae. J. Proteom. 2017, 160, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Shimoji, Y. Pathogenicity of Erysipelothrix rhusiopathiae: Virulence factors and protective immunity. Microbes Infect. 2000, 2, 965–972. [Google Scholar] [CrossRef]
- Krasemann, C.; Muller, H.E. [The virulence of erysipelothrix rhusiopathiae strains and their neuraminidase production (author’s transl)]. Zentralbl. Bakteriol. Orig. A 1975, 231, 206–213. [Google Scholar]
- Muller, H.E.; Krasemann, C. [Immunity against Erysipelothrix rhusiopathiae infection by means of active immunization using homologous neuraminidase (author’s transl)]. Z Immun. Exp. Klin. Immunol. 1976, 151, 237–241. [Google Scholar]
- Makino, S.; Yamamoto, K.; Murakami, S.; Shirahata, T.; Uemura, K.; Sawada, T.; Wakamoto, H.; Morita, H. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb. Pathog. 1998, 25, 101–109. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Janssen, T.; Voss, M.; Kuhl, M.; Semmler, T.; Philipp, H.C.; Ewers, C. A combinational approach of multilocus sequence typing and other molecular typing methods in unravelling the epidemiology of Erysipelothrix rhusiopathiae strains from poultry and mammals. Vet. Res. 2015, 46, 84. [Google Scholar] [CrossRef] [PubMed]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liang, W.; Wang, Y.; Liu, W.; Zhang, H.; Yu, T.; Zhang, A.; Chen, H.; Wu, B. Experimental pathogenicity and complete genome characterization of a pig origin Pasteurella multocida serogroup F isolate HN07. Vet. Microbiol. 2017, 198, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Bossers, A.; Harders, F.; Lu, C.; Smith, H. Comparative genomic analysis of twelve Streptococcus suis (pro)phages. Genomics 2013, 101, 336–344. [Google Scholar] [CrossRef]
- Peng, Z.; Liang, W.; Liu, W.; Wu, B.; Tang, B.; Tan, C.; Zhou, R.; Chen, H. Genomic characterization of Pasteurella multocida HB01, a serotype A bovine isolate from China. Gene 2016, 581, 85–93. [Google Scholar] [CrossRef]
- Tang, H.B.; Xie, J.; Wang, L.; Liu, F.; Wu, J. Complete genome sequence of Erysipelothrix rhusiopathiae strain GXBY-1 isolated from acute swine erysipelas outbreaks in south China. Genom. Data 2016, 8, 70–71. [Google Scholar] [CrossRef]
- Ogawa, Y.; Ooka, T.; Shi, F.; Ogura, Y.; Nakayama, K.; Hayashi, T.; Shimoji, Y. The genome of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, reveals new insights into the evolution of firmicutes and the organism’s intracellular adaptations. J. Bacteriol. 2011, 193, 2959–2971. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, Y.; Wang, G.; Bai, J.; Wang, X.; Li, Y.; Jiang, P. Genomic and proteomic characterization of SE-I, a temperate bacteriophage infecting Erysipelothrix rhusiopathiae. Arch. Virol. 2016, 161, 3137–3150. [Google Scholar] [CrossRef]
- Bandarian, V. Radical SAM enzymes involved in the biosynthesis of purine-based natural products. Biochim Biophys. Acta 2012, 1824, 1245–1253. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Bridwell-Rabb, J. Aerobic Enzymes and Their Radical SAM Enzyme Counterparts in Tetrapyrrole Pathways. Biochemistry 2019, 58, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Manav, M.C.; Sofos, N.; Hove-Jensen, B.; Brodersen, D.E. The Abc of Phosphonate Breakdown: A Mechanism for Bacterial Survival. Bioessays 2018, 40, e1800091. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Eder, S.; Hulett, F.M. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P. J. Bacteriol. 1998, 180, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Mauel, C.; Young, M.; Margot, P.; Karamata, D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol. Gen. Genet. 1989, 215, 388–394. [Google Scholar] [CrossRef]
- Shimoji, Y.; Yokomizo, Y.; Sekizaki, T.; Mori, Y.; Kubo, M. Presence of a capsule in Erysipelothrix rhusiopathiae and its relationship to virulence for mice. Infect. Immun. 1994, 62, 2806–2810. [Google Scholar] [CrossRef]
- Timoney, J. The inactivation of Erysipelothrix rhuopathiae in macrophages from normal and immune mice. Res. Vet. Sci. 1969, 10, 301–302. [Google Scholar] [CrossRef]
- Timoney, J. The inactivation of Erysipelothrix rhusiopathiae in pig buffy-coat leucocytes. Res. Vet. Sci. 1970, 11, 189–190. [Google Scholar] [CrossRef]
- Jacques, M. Surface polysaccharides and iron-uptake systems of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 2004, 68, 81–85. [Google Scholar]



| Strain | GenBank Accession | Place of Isolation | Genome Size (bp) | G + C Content (%) | Proteins | tRNAs | rRNAs | No.s of Genomic Island | Prophage |
|---|---|---|---|---|---|---|---|---|---|
| WH13013 | CP017116 | China/Hubei | 1,778,058 | 36.5 | 1633 | 55 | 15 | 4 | 3 |
| ZJ | NZ_CP041995 | China/Sichuan | 1,945,689 | 36.5 | 1785 | 55 | 18 | 13 | 2 |
| ML101 | NZ_CP029804 | China/Hunan | 1,854,248 | 36.4 | 1699 | 55 | 15 | 10 | 1 |
| GXBY-1 | NZ_CP014861 | China/Guangxi | 1,876,490 | 36.5 | 1712 | 57 | 27 | 8 | 1 |
| SY1027 | CP005079 | China/Jiangsu | 1,752,910 | 36.4 | 1388 | 53 | 10 | 4 | 1 |
| Fujisawa | AP012027 | Japan | 1,787,941 | 36.6 | 1633 | 55 | 21 | 5 | 3 |
| KC-Sb-R1 | NZ_CP033601 | Korea/South sea | 1,771,674 | 36.6 | 1599 | 55 | 21 | 3 | 2 |
| NCTC8163 | LR134439 | United Kingdom/London | 1,770,411 | 36.6 | 1609 | 55 | 27 | 5 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Zhu, Y.; Peng, Z.; Ding, Y.; Jie, K.; Wang, Z.; Peng, Y.; Tang, X.; Wang, X.; Chen, H.; et al. Comparative Genome Analysis of a Pathogenic Erysipelothrix rhusiopathiae Isolate WH13013 from Pig Reveals Potential Genes Involve in Bacterial Adaptions and Pathogenesis. Vet. Sci. 2020, 7, 74. https://doi.org/10.3390/vetsci7020074
Yang L, Zhu Y, Peng Z, Ding Y, Jie K, Wang Z, Peng Y, Tang X, Wang X, Chen H, et al. Comparative Genome Analysis of a Pathogenic Erysipelothrix rhusiopathiae Isolate WH13013 from Pig Reveals Potential Genes Involve in Bacterial Adaptions and Pathogenesis. Veterinary Sciences. 2020; 7(2):74. https://doi.org/10.3390/vetsci7020074
Chicago/Turabian StyleYang, Longsheng, Yongwei Zhu, Zhong Peng, Yi Ding, Kai Jie, Zijian Wang, Ying Peng, Xibiao Tang, Xiangru Wang, Huanchun Chen, and et al. 2020. "Comparative Genome Analysis of a Pathogenic Erysipelothrix rhusiopathiae Isolate WH13013 from Pig Reveals Potential Genes Involve in Bacterial Adaptions and Pathogenesis" Veterinary Sciences 7, no. 2: 74. https://doi.org/10.3390/vetsci7020074
APA StyleYang, L., Zhu, Y., Peng, Z., Ding, Y., Jie, K., Wang, Z., Peng, Y., Tang, X., Wang, X., Chen, H., & Tan, C. (2020). Comparative Genome Analysis of a Pathogenic Erysipelothrix rhusiopathiae Isolate WH13013 from Pig Reveals Potential Genes Involve in Bacterial Adaptions and Pathogenesis. Veterinary Sciences, 7(2), 74. https://doi.org/10.3390/vetsci7020074






